Abstract
Three-dimensional (3D) nanoscale structures of the fission yeast, Schizosaccharomyces pombe, can be obtained by full-field transmission hard x-ray microscopy with 30 nm resolution using synchrotron radiation sources. Sample preparation is relatively simple and the samples are portable across various imaging environments, allowing for high throughput sample screening. The yeast cells were fixed and double stained with Reynold’s lead citrate and uranyl acetate. We performed both absorption contrast and Zernike phase contrast imaging on these cells in order to test this method. The membranes, nucleus and subcellular organelles of the cells were clearly visualized using absorption contrast mode. The x-ray images of the cells could be used to study the spatial distributions of the organelles in the cells. These results show unique structural information, demonstrating that hard x-ray microscopy is a complementary method for imaging and analyzing biological samples.
Keywords: X-ray microscopy, tomography, imaging, Schizosaccharomyces pombe, yeast subcellular structure
Introduction
New imaging methods have greatly advanced our understanding of cell structure and function. However, no single imaging method proves to be the perfect solution. For example, light microscopy is probably the most well-known and most-used tool to study live cells, but its resolution is limited by the wavelength of the light. Electron microscopy provides high resolution information but requires elaborate sample processing such as dehydration, embedding and thin sectioning of samples [1]. Soft x-ray microscopy and tomography is an exciting technique for quantitatively imaging whole, hydrated cells in 3D [2–10]. With resolution of several tens of nanometers, up to 10 μm of sample penetration range, as well as contrast from the water window, soft x-ray microscopy is a unique tool to complement existing imaging techniques such as light and electron microscopy. Since each of these imaging methods has its pros and cons, scientists often combine several techniques to achieve a better understanding of how cells organizes and functions in a living organism. For instance, high numerical aperture cryogenic light microscopy with correlated soft x-ray tomography is a new multimodal methodology for cellular imaging [11, 12].
Full field hard X-ray microscopy is another excellent complementary imaging technique because of the high penetration power, ease of sample preparation, and high throughput for sample screening due to the ease of use and portability of the sample across various imaging techniques, including scanning x-ray fluorescence and atomic force microscopy. Thanks to the advances of x-ray optics [13–15], synchrotron radiation based transmission hard x-ray microscopes have already achieved 30–50 nm resolution [16–19]. Hard x-ray microscopes have been used for materials science [20, 21], industrial application [22, 23], environmental samples [24, 25], as well as biological specimens such as bone [19, 26]. Soft x-ray microscopy using current state-of-the-art Fresnel zone-plates (FZPs) has achieved a maximum spatial resolution of 15 nm [15]. Using zone plates with better than 25 nm resolution, the depth of field is smaller than the thickness of most eukaryotic cells [12]. Hard x-ray microcopy has the potential to examine thicker cells and tissues as well as to benefit from increasing spatial resolution. In addition, hard x-ray microscopy has been an excellent tool for investigating mineralized tissues such as bone, but there is very little absorption by soft matter. The image contrast for low-Z elements such as carbon decreases as photon energy increases.
The fission yeast Schizosaccharomyces pombe is a unicellular eukaryote which has long been an important model organism in studies of the cell cycle, mitosis, chromosome dynamics, and epigenetics. Since the whole genome sequence was completed in 2001, S. pombe has also been employed to study other important issues such as cytokinesis, DNA repair, mRNA processing and so on, due to its unique characteristics. S. pombe has been extensively studied by different techniques. In this work we have used Zernike phase contrast, and fixation and heavy metal stains to generate absorption contrast to collect images of S. pombe to demonstrate this methodology.
Materials and methods
Preparation of cells
Yeast, S. pombe, were cultured in liquid YPD (1% yeast extract, 2% bapto peptone, 2% glucose) at 25°C overnight. Then the culture was mixed with an equal volume of 5.0% glutaraldehyde in 0.2 M PIPES for 75mins at 4°C, rinsed in PIPES buffer 3 times and post-fixed with 2% osmium tetroxide (in PIPES) for 1 hour at room temperature. After rinsing 2 times with PIPES buffer, the cells were stained with Reynolds’ lead citrate [27] for 20 mins, dehydrated in a graded ethanol series of 50–100% concentration, and subsequently stained with 2 % uranyl acetate for 10 mins. The cells were mounted onto a silicon nitride membrane of 100 nm thickness and dried in the air.
X-ray microscope and tomographic reconstruction
The experiments were performed on the transmission hard x-ray microscope at 5.4 keV on beamline 6-2 at the Stanford Synchrotron Radiation Lightsource (SSRL). This system, based on an Xradia lab source [28], uses elliptical capillary condensers and zone plate optics. It is capable of absorption contrast imaging from 5–14 keV and Zernike phase contrast at 8 and 5.4 keV. The detailed schematic experimental setup of this x-ray microscope is described elsewhere [19, 26, 29]. At 5.4 keV, the field of view is 30 μm with a 2K x 2K CCD detector. The recent addition of a zone plate with 30 nm outermost width has resulted in a spatial resolution of ≤ 30 nm.
For tomography, 151 10-second images were collected at 1° intervals from −75° to +75°. The time required to collect the tomographic data was less than 30 minutes. During the acquisition, parameters such as defocus and horizontal specimen shift were controlled automatically by the Xradia software [28]. At regular intervals, the sample was moved out and 10 projection images were collected and averaged to generate a background reference. The negative logarithm of the tomographic data divided by the background reference (−ln(I/I0)) provided raw absorption data for further quantification [30]. The top of the yeast cell was used to align the data, and tomographic data were reconstructed using Xradia software [28]. The 3D volume reconstruction was performed using a standard filtered-back-projection algorithm [31].
Results and discussion
X-ray tomography using synchrotron radiation source is a unique tool to investigate the 3D information of thick specimens, with relatively simple and portable sample preparation. The yeast cells were chemically treated so in order to obtain high quality x-ray images in absorbance contrast, which produces quantifiable data for further analysis.
Absorption contrast and Zernike phase contrast imaging
We performed both absorption contrast and Zernike phase contrast imaging at 5.4 keV on yeast cells to compare these methods. In absorption contrast and Zernike phase contrast images of yeast cells during division (Fig 1a and b), a large difference is seen. Numerous organelles inside the yeast cells were visible in the absorption contrast image (Fig. 1a). In contrast we did not observe organelles when the same cells were analyzed using phase contrast imaging (Fig. 1b). In the phase contrast image, the cells appeared to have bright boundaries which made the cells appear larger than in absorption contrast (the so-called “halo effect”). After 3D reconstruction, a reconstructed slice from tomography of the yeast cells in absorption contrast mode revealed detailed internal structures of the cells (Fig. 1c). However, in a reconstructed slice of the cells in phase contrast mode (Fig. 1d) only several bright spots could be found, and these were blurred by halos.
Fig. 1.
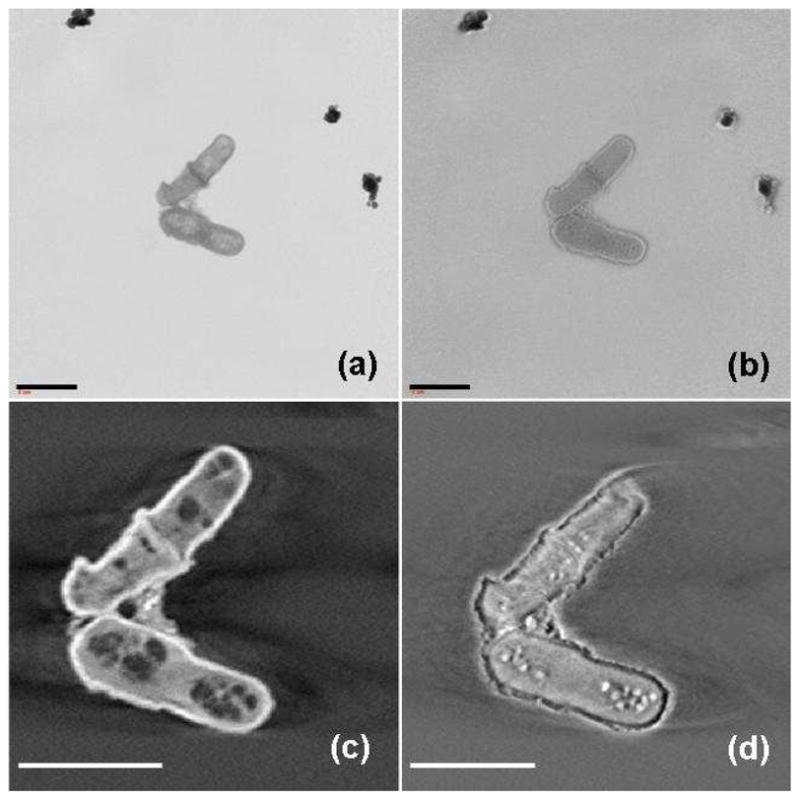
Reference-corrected x-ray images of yeast cells during division (a) in absorption contrast and (b) in Zernike phase contrast. A reconstructed slice from tomography of the yeast cells (c) in absorption contrast and (d) in Zernike phase contrast. The scale bar is 5 microns.
Zernike phase contrast microscopy was first employed to increase the image contrast of transparent specimens in optical microscopy [32]. Similarly, for x-ray microscopy, the absorption contrast of light elements decreases rapidly as the x-ray energy increases. Zernike phase contrast techniques were later applied both to soft x-ray [33] and hard x-ray microscopy [34], respectively. Although the boundary of the specimen can be enhanced by phase contrast, the observed intensity in the image contains both amplitude and phase information, which makes it hard to quantitatively analyze the data. In previous work, we found that the halo effect is obvious in areas with highly different indices of refraction [24]. In this work we have used heavy element staining to enhance the contrast of the features in the cells, and thus the quality of absorption contrast images is better than phase contrast at 5.4 keV. In the following sections we will focus on the subcellular structures seen using absorption contrast mode.
Cellular structures of S. pombe
Combining simple chemical treatment with absorption contrast imaging at 5.4 keV, the ultrastructural details of S. pombe were well delineated. The tomographic results of five yeast cells with diameter of 2.5–3.5 μm (Fig. 2) clearly show the differences inside cells progressed through different stages of the cell cycle; from a small initially formed S. pombe (Fig. 2a) to increased in size (Fig. 2b), fully-grown (Fig. 2c) and beginning replication (Fig. 2d). Fig. 2e is a dividing yeast cell, in which we can see clearly the septum at the cell center between two daughter cells. The first column in Fig. 2 is the projection image of each cell, in which the exact boundaries of the organelles were not clearly identifiable. After x-ray tomographic reconstruction, we are able to reveal the 3D subcellular structure of the yeast cell through both the reconstructed slices (2nd to 5th column in Fig. 2) and 3D renderings (6th column in Fig. 2).
Fig. 2.
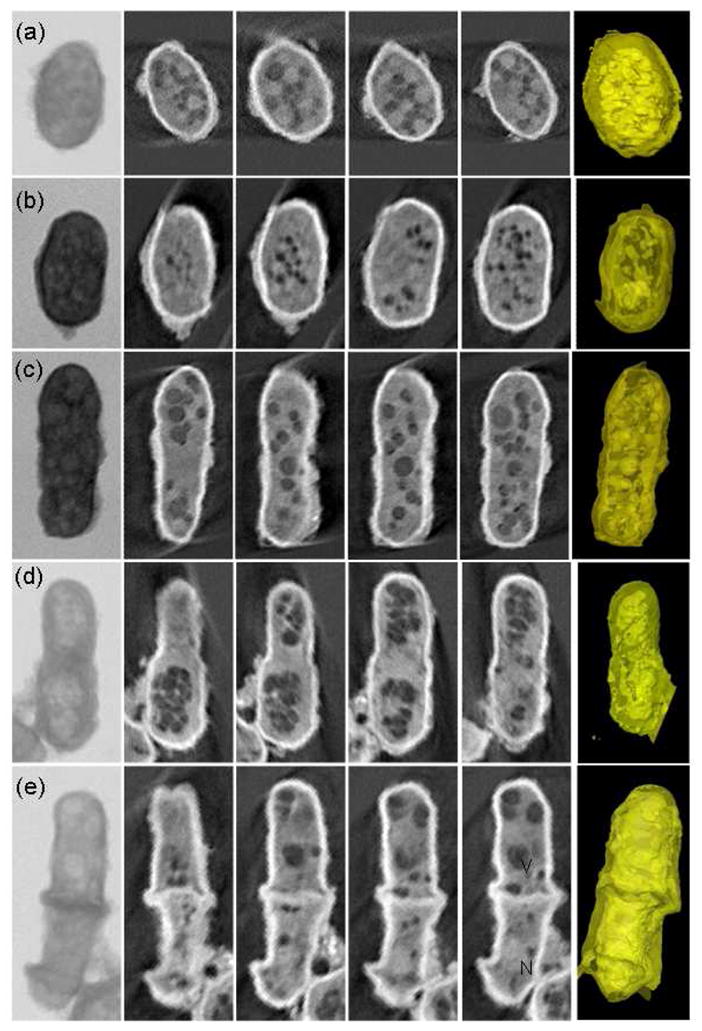
Absorption contrast tomographic results of five single cells with diameter of 2.5–3.5 μm progressed through different stages of the cell cycle. The first column is the projection x-ray image of each yeast cell. The 2nd to 5h columns are reconstructed slices through the corresponding reconstruction data showing different regions of the cells. Each slice is about 30nm thick. The 6th column contains the 3D renderings of the cells.
The details of the yeast cells can be seen clearly. Some cellular structures are easily identified, such as the cell wall, nucleus and large vacuoles. At the present time, assignment of other structures seen with x-ray tomography is complex. It will require more than just direct comparison with transmission electron microscopy (TEM). The standard TEM techniques require further sample preparation such as embedding, thin section and staining, which could cause differences both in density and structure. In order to aid in assignment, we could combine other techniques and methods such as scanning x-ray fluorescence microprobe and atomic force microscopy. The sample preparation techniques for hard x-ray microscopy are quite compatible with these other methods. Other techniques such as light microscopy, fluorescence imaging, and immunolabeling have also been used in conjunction with x-ray microscopy and employed to identify the subcellular structures. In a recent paper, using TEM Ubaidus and his colleagues immunolocalized the fibroblast growth factor (FGF) 23 and dentin matrix protein 1 in bone samples. They proved that the synthesis of FGF23 is mainly controlled by the arrangement of the osteocytic lacunar canalicular systems [35]. We could similarly extend protein localization into three dimensions and further quantify the proteins in the biological samples based on this work using x-ray microscopy.
Chemical treatment of yeast cells
An important prerequisite for cell imaging is to preserve the native state of biological samples during sample preparation. For chemical treatment, we used fixation and heavy metal staining to improve the image contrast. Progressive substitution of water with ethanol followed by air drying was employed at the end. Straight chemical fixation would lead to cytoplasmic contraction and organelle redistribution. Ethanol dehydration and air-drying would cause precipitation on membranes and the cytoskeleton, which would result in considerable collapse and shrinkage of cells. All these procedures could introduce artifacts. Therefore, the critical limitation of this method is the structural damage of the cell sample. For instance, we can see a shrinkage edge at the bottom part of the cell (Fig. 2e). Cryofixation and freeze-substitution is a much better method for preserving the cellular ultrasturcture, which has been used successfully in soft x-ray tomographic imaging of S. pombe [7]. It can be considered as a good alternative method to prepare samples in the future. In order to obtain good signal-to-noise ratio, the x-ray dose needed for collecting tomographic data would eventually cause radiation damage by breaking the structures and chemical bonds in the sample. Although chemical fixation of cells can help prevent radiation damage, we chose a suitably short exposure time to minimize the potential cell damage. In addition, mounting the specimen in a cryogenic stage is the most efficient way to resolve frozen-hydrated cells and tissue without inducing significant structural changes, and also to prevent cells from entering a dormant state [7].
Although the artifacts are unavoidable, this won’t be too significant for pombe cells, since they are so small. The results above show well defined cellular structure. However, the unpredictable accumulation of staining metal leads to problems in interpretation. It is still hard to exactly distinguish all the visible organelles. We could not simply compare the results with those from other techniques such as TEM. All the chemical treatment procedures here originated from the well-established protocol of TEM specimen preparation. Thus the TEM results still can provide some clues for assigning organelles. The organelles could be assigned based on their characteristic ultrastructures such as distribution, number and shape. Since the samples were mounted on the SiN3 substrate, they can be used on the same mount for imaging with complementary methods such as XRF, AFM, immunolabeling. The resolution of x-ray microscopy also limits the interpretation. The future improvement of resolution would reduce the difficulties in identifying organelles.
Conclusions
In summary we have successfully imaged subcellular structure in S. pombe combining chemical treatment with synchrotron-based transmission x-ray microscopy in absorption contrast. The 3D cellular structures have been revealed with excellent resolution. This method has the potential of observing thick cells and unmineralized biological samples. With the advances in fabrication of x-ray optics, it could also benefit from increasing spatial resolution in the future.
Acknowledgments
This project was supported by the 985 project of the State Ministry of Education, the National Natural Science Foundation of China (10675113, 10734070) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KJXC2-YW-N43). SSRL is supported by the Department of Energy, Office of Basic Energy Sciences. The transmission x-ray microscope is supported by NIH/NIBIB grant number 5R01EB004321.
References
- 1.Rosier DJDE, Klug A. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 2.Kirz J, Jacobsen C, Howells M. Quart Rev biophys. 1995;28:33–130. doi: 10.1017/s0033583500003139. [DOI] [PubMed] [Google Scholar]
- 3.Schneider G. Ultramicroscopy. 1998;75:85–104. doi: 10.1016/s0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 4.Weiss D, Schneider G, Niemann B, et al. Ultramicroscopy. 2000;84:185–197. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
- 5.Weiss D, Schneider G, Vogt S, et al. Nuclear Instrum Methods Phys Res A. 2001;467:1308–1311. [Google Scholar]
- 6.Meyer-Ilse W, Hamamoto D, Nair A, et al. J Microsc. 2001;201:395–403. doi: 10.1046/j.1365-2818.2001.00845.x. [DOI] [PubMed] [Google Scholar]
- 7.Larabell CA, Le Gros MA. Mol Biol Cell. 2004;15:957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Gros MA, McDermott G, Larabell CA. Current Opinion in Structural Biology. 2005;15:593–600. doi: 10.1016/j.sbi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Gu WW, Etkin LD, Le Gros MA, et al. Differentiation. 2007;75:529–535. doi: 10.1111/j.1432-0436.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson DY, McDermott G, Etkin LD, et al. J Struct Biol. 2008;162:380–386. doi: 10.1016/j.jsb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Gros MA, McDermott G, Uchida M, et al. J Microsc-Oxford. 2009;235:1–8. doi: 10.1111/j.1365-2818.2009.03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott G, Knoechel CG, Le Gros MA, et al. Trends in Cell Biology. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai B, Yun WB, Legnini D, et al. Appl Phys Lett. 1992;61:1877–1879. [Google Scholar]
- 14.Yun WB, Lai B, Cai Z, et al. Rev Sci Instrum. 1999;70:2238–2241. [Google Scholar]
- 15.Chao W, Harteneck BD, Liddle JA, et al. Nature. 2005;435:1210–1213. doi: 10.1038/nature03719. [DOI] [PubMed] [Google Scholar]
- 16.Chu YS, Yi JM, De Carlo F, et al. Appl Phys Lett. 2008;92:103119. [Google Scholar]
- 17.Yin GC, Song YF, Tang MT, et al. Appl Phys Lett. 2006;89:221122. [Google Scholar]
- 18.Tian Y, Li W, Chen J, et al. Rev Sci Instrum. 2008;79:103708. doi: 10.1063/1.3002484. [DOI] [PubMed] [Google Scholar]
- 19.Andrews JC, Brennan S, Patty C, et al. Synchrotron Radiation News. 2008;21:17–26. doi: 10.1080/08940880802123043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Wu C, Tian, et al. Appl Phys Lett. 2008;92:233104. [Google Scholar]
- 21.Li W, Wang N, Chen J, et al. Appl Phys Lett. 2009;95:053108. [Google Scholar]
- 22.Yin GC, Tang MT, Song YF, et al. Appl Phys Lett. 2006;88:241115. [Google Scholar]
- 23.Levine ZH, Kalukin AR, Frigo SP, et al. Appl Phys Lett. 1999;74:150–152. [Google Scholar]
- 24.Chen J, Li W, Liu Y, et al. Journal of Physics: Conference Series. 2009;186:012005. [Google Scholar]
- 25.Patty C, Barnett B, Mooney B, et al. Environ Sci Technol. 2009;43:7397–7402. doi: 10.1021/es901076q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews JC, Brennan S, Liu Y, et al. Journal of Physics: Conference Series. 2009;186:012081. [Google Scholar]
- 27.Reynolds ES. J Cell Biol. 1963;17:208–213. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tkachuk A, Duewer F, Cui H, et al. Z Kristallogr. 2007;222:650–655. [Google Scholar]
- 29.Andrews JC, Brennan S, Pianetta P, et al. Journal of Physics: Conference Series. 2009;186:012002. [Google Scholar]
- 30.Andrews JC, Pianetta P, Almeida E, et al. Microscopy and Microanalysis. 2010;16(3) doi: 10.1017/S1431927610000231. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kak AC, Slaney M. Principles of Computerized Tomographic Imaging. IEEE; New York: 1988. [Google Scholar]
- 32.Zernike F. Z Tech Phys. 1935;11:454–457. [Google Scholar]
- 33.Schmahl G, Rudolph D, Guttmann P, et al. Rev Sci Instrum. 1995;66:1282–1286. [Google Scholar]
- 34.Neuhäusler U, Schneider G, Ludwig W, et al. J Phys D: Appl Phys. 2003;36:A79–A82. [Google Scholar]
- 35.Ubaidus S, Li M, Sultana S, et al. J Electron Microsc. 2009;58:381–392. doi: 10.1093/jmicro/dfp032. [DOI] [PubMed] [Google Scholar]


