Abstract
Background
Barrett’s Esophagus (BE) is the precursor and the biggest risk factor for esophageal adenocarcinoma (EAC), the solid cancer with the fastest rising incidence in the US and western world. Current strategies to decrease morbidity and mortality from EAC have focused on identifying and surveying patients with BE using upper endoscopy. An accurate estimate of the number of patients with BE in the population is important to inform public health policy and to prioritize resources for potential screening and management programs. However, the true prevalence of BE is difficult to ascertain because the condition frequently is symptomatically silent, and the numerous clinical studies that have analyzed BE prevalence have produced a wide range of estimates. The aim of this study was to use a computer simulation disease model of EAC to determine the estimates for BE prevalence that best align with US SEER cancer registry data.
Methods
A previously developed mathematical model of EAC was modified to perform this analysis. The model consists of six health states: Normal, GERD, BE, Undetected Cancer, Detected Cancer and Death. Published literature regarding the transition rates between these states were used to provide boundaries. During the one million computer simulations that were performed, these transition rates were systematically varied, producing differing prevalences for the numerous health states. Two filters were sequentially applied to select out superior simulations that were most consistent with clinical data. First, among these million simulations, the 1,000 that best reproduced SEER cancer incidence data were selected. Next, of those 1000 best simulations, the 100 with an overall calculated BE to Detected Cancer rates closest to published estimates were selected. Finally, the prevalence of BE in the final set of best 100 simulations was analyzed.
Results
We present histogram data depicting BE prevalences for all one million simulations, the 1000 simulations that best approximate SEER data, and the final set of 100 simulations. Using the best 100 simulations, we estimate the prevalence of BE to be 5.6% [5.49–5.70%].
Conclusions
Using our model, an estimated prevalence for BE in the general population of 5.6% [5.49–5.70%] accurately predicts incidence rates for EAC reported to the US SEER cancer registry. Future clinical studies are needed to confirm our estimate.
MESH Keywords: Barrett’s esophagus, Esophageal Cancer, Adenocarcinoma, Computer Simulation, Computer Models, SEER Program
Introduction and Background
Esophageal adenocarcinoma (EAC) has the fastest rising incidence of a solid cancer in the western world [1]. Although the absolute number of EAC cases per year remains too low to screen the general population [2], targeted screening of high risk individuals may be appropriate. Heartburn, the primary symptom of gastroesophageal reflux disease (GERD), affects 60 million Americans [3] and can lead to Barrett’s esophagus (BE). BE is a pre-malignant condition associated with the greatest risk (30–125 times) of developing EAC [4]. Because of the significant number of individuals affected by GERD and BE, the management of these patients has become a significant public health issue.
The incidence of BE is debated, partially because it is symptomatically silent and the potential impact of publication bias [5]. An accurate estimate of the number of patients in the population with BE is important to aid in the prioritization of medical resources and to inform public health policy. Also, a better estimate of BE prevalence would help to assess the feasibility, effectiveness, and cost-effectiveness of a screening and management program for BE; the ultimate goal of such a screening program would be to prevent EAC morbidity and mortality.
Numerous studies have analyzed the prevalence of BE but with a wide range of results [6–15]. The two largest (approximately 1000 patients) and more methodologically rigorous studies have also produced widely divergent results. A Swedish study attempted to recruit patients from a community sample and found 1.6% of those who had endoscopic screening had histologically confirmed BE [6]. A second study had a similar number of subjects, but a potentially biased group, as patients were recruited to undergo upper endoscopic screening for BE among individuals who were undergoing screening colonoscopies. This analysis was performed in the US and found a BE prevalence of 6.8% [7], which is more consistent with other recent US studies. The discrepancy between these two studies’ findings has been discussed with possible suggested etiologies including differences in the populations studied and endoscopy and biopsy techniques [16]; however, the true prevalence continues to be debated.
Simulation models can be useful in various scenarios, including this particular circumstance, where clinical studies that provide more conclusive data are not feasible and/or the existing data is conflicting. A model can synthesize and integrate the data that is available and can be used to perform various analyses. This includes using the model to make projections by extrapolation, or increasing the understanding or “filling in” areas of the natural history of a disease that are lacking data by interpolation.
The aim of this study was to use a mathematical simulation model of the natural history of esophageal adenocarcinoma (Esophageal Adenocarcinoma Model, EACMo) to estimate the BE prevalence that best aligns with published clinical data, particularly, National Cancer Institute (NCI) Surveillance Epidemiology and End Results (SEER).
Materials and Methods
Model Structure
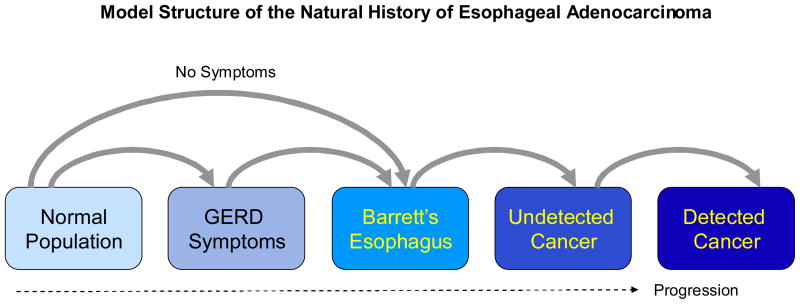
In order to gain a better understanding of EAC, and specifically BE prevalence, a mathematical model was developed. The Markov model used in our analysis is a scaled down version of a larger population model of EAC that was constructed to explore and analyze the rising incidence of EAC [17]. The model consists of 6 health states: Normal, GERD Symptoms, BE, Undetected Cancer, Detected Cancer, and Death (see Figure 1). The model was programmed in C# using the Microsoft.NET Version 2.0 Framework (Redmond, WA).
Figure 1.
The flow diagram above depicts the different health states and transition probabilities used in the simulation model. All of these states may also transition to death from natural causes.
Fundamentally, the model is comprised of two primary components: the model structure or the various health states; and the transition rates or probabilities between them. The rectangles in Figure 1 represent the various health states and the arrowed lines connecting them represent transition probabilities, or the chance or probability that a simulated patient in a specific health state would leave one state to enter the other state.
Approximately a million simulations were performed, each using a different set of transition probabilities. The simulation would begin with the hypothetical cohort of patients at age 0 and track them as they aged from birth to age 80 or death, whichever came first. After each cycle, or time increment, the simulation model would keep track of the fraction of patients in each health state. Because each simulation had a different and unique set of transition probabilities, the number (or percentage) of individuals in each health state by cycle would reflect these differences and also be unique. Using this method, our million simulation produced a wide range of BE prevalences.
Model Inputs and Assumptions
Several assumptions were made in the construction of our model. Details of many of the assumptions follow below and also refer to a summary table (Table 1) which presents model input estimates and references to publications upon which they are based.
Table 1.
Model Inputs: Selected Parameters Estimates
This table summarizes input parameter assumptions used in the simulation model.
LSBE vs SSBE
Although not depicted in the figure, the health state representing BE was further sub-categorized as Long Segment (LSBE) and Short Segment Barrett’s esophagus (SSBE). The prevalence of SSBE compared to SSBE was fixed at a 3:1 ratio [6,7,14]. The progression rates from LSBE versus SSBE to cancer were assumed to be 2:1 based on a surface area rationale [18].
BE Prevalence
Additionally, the BE prevalence was assumed to increase with age [17]. This was achieved by having the transition probabilities that governed entry into the BE health state (Normal to BE and GERD to BE) increase with age.
Undetected Cancer
As with any cancer, many cases of EAC may be dormant or asymptomatic for many years prior to detection. Accordingly, an Undetected Cancer state was included in the model. Because an undetected state is by nature unobservable, transitions surrounding it are intrinsically difficult to quantify. We estimated that the sojourn time, which we defined as the time for an undetected cancer to become detected, was in the range of 3–6 years [19,20].
Select Population
Our model simulated only white males in the US as this is the group with the greatest risk for both BE and EAC, and as a consequence, also the group most studied with the most published data.
Parameter Search for Transition Probabilities
There is limited published data regarding the transition probabilities between the health states that precede EAC. However, even if “optimal” clinical data were available (e.g. a large, representative cohort of patients with BE followed over a long period of time with intensive or relatively frequent endoscopic surveillance), the exact time of transition from one health state to another within the surveillance interval would be difficult to determine; the inaccuracies associated with health state determination by endoscopic biopsies (false negative and false positive diagnoses) add additional uncertainty. These factors make estimating the five transition probabilities between health states within the natural history model challenging.
In an attempt to address the significant uncertainty in transition probability estimation, a parameter search process was performed where the five transition probabilities within the model are varied systematically while constrained to a plausible range guided by the literature (parameter space grid search; [21,22]) to produce a large number of distinct parameters or transition probability sets. Each distinct transition probability set was used to perform a single run of the model or a simulation, producing unique model outputs or distributions of health states. For our model, the outputs were GERD Symptom, BE, and Undetected CA prevalences and Detected CA incidences. Restated from a “big picture” perspective, each simulation represents a distinct potential depiction for the natural history of EAC (including the prevalence of BE), and by systematically varying the transition probabilities, a comprehensively exhaustive range of possibilities are covered.
Additional details of parameter search and other technical details of model constructions can be found in our esophageal adenocarcinoma policy model manuscript which focuses on model development, calibration, and validation [17].
Criteria Used for Superior Simulation Selection
After running approximately a million simulations using the numerous distinct transition probability sets, a systematic process to select the superior simulations was necessary. Two sequential filters or criteria were applied to determine the superior simulations among the large pool of samples. The purpose of these filters was to identify those simulations that were more consistent with specific published data and reality. The first and primary filter used to assess simulation quality was how well simulation results approximated or reproduced NCI SEER reported EAC incidences by age in 2005 [2]. The second filter was based on a published meta-analysis that estimated the progression rate from BE to EAC at approximately 0.5% per year, after attempting to adjust for publication bias [5].
As described, when running the simulations, the transition probabilities were systematically varied (grid search pattern) producing a comprehensive range of model results. To quantitatively assess which simulations (based on the particular transition probability sets) best approximated SEER data, a Chi-Squared Goodness of Fit (GOF) test was performed and a score calculated for each simulation. The simulations with the best GOF scores (top 0.1% or 1000 simulations) were chosen for further analysis.
Of these 1000 selected simulations, the second criterion or filter was applied. The BE to cancer progression rates were calculated for each of the remaining simulations and those that were closest to the published rates of 0.5% (top 10% or 100 simulations) were selected to produce the final group that were deemed to be the best by our described selection process.
The final group of superior or the 100 best simulations were then analyzed to determine the BE prevalences that were estimated by each simulation; averages and ranges were calculated.
3. Results
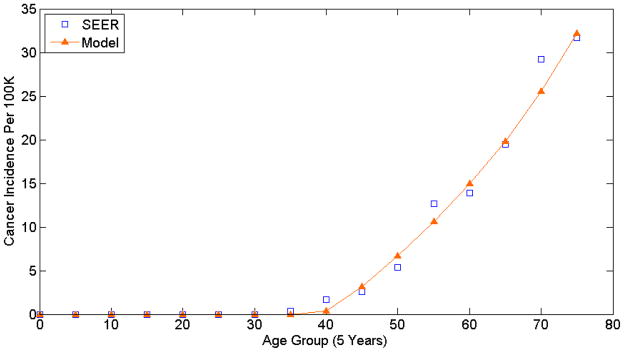
As previously described in the Methods section, the first selection criterion was to assess each simulation’s fit to SEER cancer incidence and choose the superior 1000 among the pool of a million simulations. Figure 2 is a plot of the model’s projected cancer incidence versus SEER cancer incidence presented by age group; the model data corresponds to the simulation with the best fit to SEER data (best GOF score). On inspection, the model output or projected EAC incidence approximates SEER cancer incidence well.
Figure 2.
This plot compares the Model versus SEER cancer incidences for the simulation with the top GOF score.
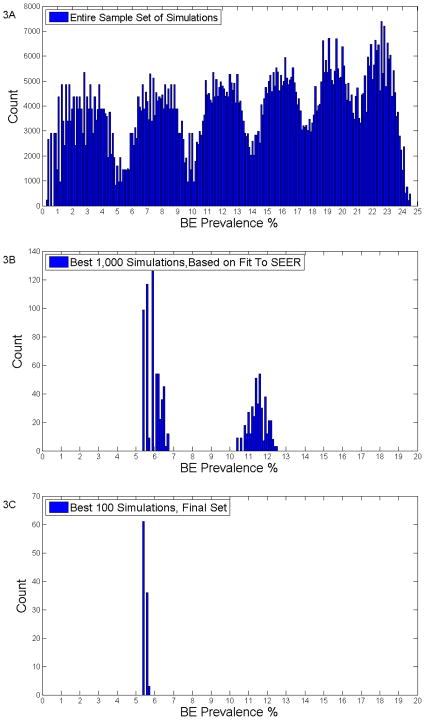
Figure 3 depicts the distributions of BE prevalence for all and selected simulations. Figure 3A is a histogram representation of the distribution of BE prevalences for all one million simulations prior to any filtering. The range is quite broad, from 0.4% to 24.5%, but is consistent with the wide range found in clinical studies [6–15].
Figure 3.
The top graph (3A) depicts the BE prevalences of all the simulations prior to any filtering. The middle graph (3B) show the BE prevalences of the 1000 simulations that best fit SEER data. The bottom graph (3C) shows the BE prevalences for the final, best 100 simulations.
Figure 3B graphs the superior 1000 simulation after the first filter of best fit to SEER data is applied. The results appear to have a bimodal distribution with one peak at a BE prevalence of 5.9% and a second peak at 11.6% with an overall range between 5–12%. The bimodal distribution or peaks are most likely an artifact of the granularity of the grid search (i.e. if the grids in the search had been smaller or finer in granularity then the distribution would have appeared flatter). This explanation also applies to the numerous peaks seen in Figure 3A.
The distribution of BE prevalences for the best 100 simulations are graphically displayed in Figure 3C and demonstrates a narrow single peak. The mean average BE prevalence was 5.57% (SD=0.10 while the median value was 5.49%). 95% of the distribution was within the range of 5.488–5.700%; see Table 2.
Table 2.
BE Prevalence Distributions
The above table depicts the BE prevalence value’s mean, median, and ranges after both of the filters were applied to the simulations.
| Mean (SD) | 5.571 (0.10) |
| Median | 5.49 |
| Ranges | Min Prev | Max Prev |
|---|---|---|
| 50% | 5.489 | 5.698 |
| 75% | 5.488 | 5.699 |
| 90% | 5.488 | 5.700 |
| 95% | 5.488 | 5.700 |
4. Discussion
The incidence of EAC has been rising dramatically and much of the efforts to date have focused on identifying and surveying patients with BE, which is the greatest risk factor for developing EAC [4]. The number of individuals in the U.S. with BE is difficult to estimate because a substantial proportion of patients do not have symptoms [23] and a consensus regarding screening guidelines does not exist [24]. Clinical studies that have analyzed the prevalence of BE have produced a wide range of estimates [6–15]. Among these numerous studies of varying design and populations, the two largest and methodologically rigorous analyses have produced widely divergent estimates. A Swedish study which attempted to approximate a community sample by recruiting volunteers for endoscopic screening estimated BE population prevalence at 1.6% [6]. A similar sized study performed in the U.S. which was performed in patients who were undergoing screening colonoscopies found a substantially higher prevalence of 6.8% [7]. Our millions simulations were comprised of BE prevalences ranging from 0.4–25%, but our final set of the best 100 simulations produced a precise range between 5.49–5.70% with a mean value of 5.57%. Additionally, our estimate for BE prevalences were calculated across the entire population from birth to age 80 in our simulations, where BE prevalence was assumed to increase with age; if our simulations had used a population that excluded younger patients (published studies were performed in older patients), our estimates for BE prevalence would presumably have been higher.
Because of the acknowledged uncertainties in the model (see limitations below), we do not believe that the final estimate produced by our exploratory analysis of ~5.6% should be considered the definitive prevalence. A more reasonable interpretation of our findings might interpret them in the context of prior clinical data, specifically the divergent estimates of 1.6% and 6.8%. Our analysis, which was performed independently of both studies, is quite close to the 6.8%. Furthermore, our model’s estimate would be higher than 5.6% if it were restricted to an older patient group similar to the one in which this clinical study was performed. The true prevalence of BE may lie closer to 6.8% rather than 1.6%.
Limitations
A limitation to our analysis is the uncertainty in both the model structure, which is a simplification of the natural history of EAC, and the limitation of available data that we based model inputs upon. Our analysis attempted to determine which estimates of BE prevalence were most consistent with the better clinical data that was available (e.g. SEER). An inherent limitation to this methodology is that our estimates were biased or constrained to those estimates that have been published. We acknowledge the exploratory nature of our analyses.
We attempted to synthesize and integrate the available data to produce a model that was clinically realistic. However, as all disease models, ours was a simplification of reality. For example, our model did not include dysplasia states within BE (e.g. low grade and high grade dysplasia) and did not include regression between health states. We did not believe that these clinical realities were directly relevant to the goals of our analysis, which was to determine an estimate for BE prevalence, and so they were not included. The primary benefit of maintaining simplicity was model transparency. This allows the model and the analysis performed to be more comprehensible to both the investigators and readers. However, future analysis may include dysplastic states, particularly if they are directly relevant to the aims or hypothesis of the study.
Our analysis used the most recent SEER data from 2005 for white males. SEER data is based on a sample that represents 26% of the US population, while not perfect, is a significant strength of our analysis. We did not try to incorporate the secular trends and potential cohort effects that factor into the significant rise witnessed in EAC incidence over the past three decades. This simplification was necessary to perform our relatively simple and exploratory analysis.
Additionally, we chose to focus on white males as the majority of published data focused on this group and analyses on non-white and females would have introduced even more uncertainty into the analyses.
In conclusions, we performed an exploratory analysis where a simulation model of esophageal adenocarcinoma was used to estimate the prevalence of BE. Our result of 5.6% is more consistent with the 6.8% estimate from the analysis by Rex et al. [7] than the 1.6% estimate by Ronkainen et al. [6]. The prevalence of BE prevalence is central to determining the effectiveness and cost-effectiveness of current and future screening and management programs for BE and EAC. Consequently, our results could be useful to inform clinical and policy decisions. Future clinical studies are necessary to confirm the prevalence of BE in the population, but our results provide additional support to the 6.8% estimate.
Acknowledgments
Grant support: NIH/NCI Grant CA107060 (C.H.)
Abbreviations
- GERD
gastroesophageal reflux disease
- BE
Barrett’s esophagus
- EAC
Esophageal Adenocarcinoma
- SEER
Surveillance, Epidemiology and End Results
- NCI
National Cancer Institute
Footnotes
No potential financial conflicts to report.
References
- 1.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white americans by sex, stage, and age. Journal of the National Cancer Institute. 2008 doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, Lewis DR, Clegg L, Eisner MP, Reichman M, Edwards BKe. Seer cancer statistics review, 1975–2005. National Cancer Institute; Bethesda, MD: 2008. http://seer.cancer.gov/csr/1975-2005/ based on November 2007 SEER data submission, posted to the SEER web site. [Google Scholar]
- 3.Locke GR, 3rd, Talley NJ, Fett SL, Zinsmeister AR, Melton LJ., 3rd Prevalence and clinical spectrum of gastroesophageal reflux: A population-based study in olmsted county, minnesota. Gastroenterology. 1997;112:1448–1456. doi: 10.1016/s0016-5085(97)70025-8. [DOI] [PubMed] [Google Scholar]
- 4.Williamson WA, Ellis FH, Jr, Gibb SP, Shahian DM, Aretz HT, Heatley GJ, Watkins E., Jr Barrett’s esophagus. Prevalence and incidence of adenocarcinoma. Arch Intern Med. 1991;151:2212–2216. doi: 10.1001/archinte.151.11.2212. [DOI] [PubMed] [Google Scholar]
- 5.Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in barrett’s esophagus? Gastroenterology. 2000;119:333–338. doi: 10.1053/gast.2000.9302. [DOI] [PubMed] [Google Scholar]
- 6.Ronkainen J, Pertti A, Storskrubb T, Johansson S-E, Linde T, Bolling-Sternevald E, Vieth M, Stolte M, Talley NJ, Agreus L. Prevalence of barrett’s esophagus in the general population: An endoscopic study. Gastroenterology. 2005;129:1825–1831. doi: 10.1053/j.gastro.2005.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Rex DK, Cummings OW, Shaw M, Cumings MD, Wong RK, Vasudeva RS, Dunne D, Rahmani EY, Helper DJ. Screening for barrett’s esophagus in colonoscopy patients with and without heartburn. Gastroenterology. 2003;125:1670–1677. doi: 10.1053/j.gastro.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Cameron A. Barrett’s esophagus: Age, prevalence, and extent of columnar epithelium. Gastroenterology. 1992;103:1241–1245. doi: 10.1016/0016-5085(92)91510-b. [DOI] [PubMed] [Google Scholar]
- 9.Clark GWB, Ireland AP, Peters JH, Chandrasoma P, DeMeester TR, Bremner CG. Short-segment barrett’s esophagus: A prevalent complication of gastroesophageal reflux disease with malignant potential. Journal of Gastrointestinal Surgery. 1997;1:113–122. doi: 10.1016/s1091-255x(97)80098-4. [DOI] [PubMed] [Google Scholar]
- 10.O’Connor JB, Falk GW, Richter JE. The incidence of adenocarcinoma and dysplasia in barrett’s esophagus: Report on the cleveland clinic barrett’s esophagus registry. Am J Gastroenterol. 1999;94:2037–2042. doi: 10.1111/j.1572-0241.1999.01275.x. [DOI] [PubMed] [Google Scholar]
- 11.Corley DA, Levin TR, Habel LA, Weiss NS, Buffler PA. Surveillance and survival in barrett’s adenocarcinomas: A population-based study. Gastroenterology. 2002;122:633–640. doi: 10.1053/gast.2002.31879. [DOI] [PubMed] [Google Scholar]
- 12.Corey KE, Schmitz SM, Shaheen NJ. Does a surgical antireflux procedure decrease the incidence of esophageal adenocarcinoma in barrett’s esophagus? A meta-analysis. American Journal of Gastroenterology. 2003;98:2390–2394. doi: 10.1111/j.1572-0241.2003.08702.x. [DOI] [PubMed] [Google Scholar]
- 13.Westhoff B, Brotze S, Weston A, McElhinney C, Cherian R, Mayo MS, Smith HJ, Sharma P. The frequency of barrett’s esophagus in high-risk patients with chronic gerd. Gastrointestinal Endoscopy. 2005;61:226–231. doi: 10.1016/s0016-5107(04)02589-1. [DOI] [PubMed] [Google Scholar]
- 14.Gerson LB, Shetler K, Triadafilopoulos G. Prevalence of barrett’s esophagus in asymptomatic individuals. Gastroenterology. 2002;123:461–467. doi: 10.1053/gast.2002.34748. [DOI] [PubMed] [Google Scholar]
- 15.Pera M. Trends in incidence and prevalence of specialized intestinal metaplasia, barrett’s esophagus, and adenocarcinoma of the gastroesophageal junction. World Journal of Surgery. 2003;27:999–1008. doi: 10.1007/s00268-003-7052-2. [DOI] [PubMed] [Google Scholar]
- 16.Rex DK, Shaw M, Wong R. Prevalence of barrett’s esophagus. Gastroenterology. 2006;130:1373–1374. doi: 10.1053/j.gastro.2006.02.046. author reply 1374–1375. [DOI] [PubMed] [Google Scholar]
- 17.Hur C, Hayeck TJ, Yeh JM, Richards E, Spechler SJ, Gazelle GS, Kong CY. Development, calibration, and validation of a us population-based simulation model of esophageal adenocarcinoma. 2009 doi: 10.1371/journal.pone.0009483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menke-Pluymers MB, Hop WC, Dees J, van Blankenstein M, Tilanus HW. Risk factors for the development of an adenocarcinoma in columnar-lined (barrett) esophagus. The rotterdam esophageal tumor study group. Cancer. 1993;72:1155–1158. doi: 10.1002/1097-0142(19930815)72:4<1155::aid-cncr2820720404>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Guanrei Y, Songliang Q, He H, Guizen F. Natural history of early esophageal squamous carcinoma and early adenocarcinoma of the gastric cardia in the people’s republic of china. Endoscopy. 1988;20:95–98. doi: 10.1055/s-2007-1018145. [DOI] [PubMed] [Google Scholar]
- 20.Provenzale D, Kemp JA, Arora S, Wong JB. A guide for surveillance of patients with barrett’s esophagus. Am J Gastroenterol. 1994;89:670–680. [PubMed] [Google Scholar]
- 21.Yeh JM, Kuntz KM, Ezzati M, Hur C, Kong CY, Goldie SJ. Development of an empirically calibrated model of gastric cancer in two high-risk countries. Cancer Epidemiol Biomarkers Prev. 2008;17:1179–1187. doi: 10.1158/1055-9965.EPI-07-2539. [DOI] [PubMed] [Google Scholar]
- 22.Lee JM, Kopans DB, McMahon PM, Halpern EF, Ryan PD, Weinstein MC, Gazelle GS. Breast cancer screening in brca1 mutation carriers: Effectiveness of mr imaging--markov monte carlo decision analysis. Radiology. 2008;246:763–771. doi: 10.1148/radiol.2463070224. [DOI] [PubMed] [Google Scholar]
- 23.Chak A, Faulx A, Eng C, Grady W, Kinnard M, Ochs-Balcom H, Falk G. Gastroesophageal reflux symptoms in patients with adenocarcinoma of the esophagus or cardia. Cancer. 2006;107:2160–2166. doi: 10.1002/cncr.22245. [DOI] [PubMed] [Google Scholar]
- 24.Shaheen N, Ransohoff DF. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: Scientific review. Jama. 2002;287:1972–1981. doi: 10.1001/jama.287.15.1972. [DOI] [PubMed] [Google Scholar]
- 25.El-Serag HB. Time trends of gastroesophageal reflux disease: A systematic review. Clinical Gastroenterology and Hepatology. 2007;5:17–26. doi: 10.1016/j.cgh.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Terry P, Lagergren J, Wolk A, Nyren O. Reflux-inducing dietary factors and risk of adenocarcinoma of the esophagus and gastric cardia. Nutrition and Cancer. 2000;38:186–191. doi: 10.1207/S15327914NC382_7. [DOI] [PubMed] [Google Scholar]
- 27.Collen MJ, Abdulian JD, Chen YK. Gastroesophageal reflux disease in the elderly: More severe disease that requires aggressive therapy. American Journal of Gastroenterology. 1995;90:1053–1057. [PubMed] [Google Scholar]
- 28.Mohammed I, Cherkas LF, Riley SA, Spector TD, Trudgill NJ. Genetic influences in gastro-oesophageal reflux disease: A twin study. Gut. 2003;52:1085–1089. doi: 10.1136/gut.52.8.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiocca JC, Olmos JA, Salis GB, Soifer LO, Higa R, Marcolongo M. Prevalence, clinical spectrum and atypical symptoms of gastro-oesophageal reflux in argentina: A nationwide population-based study. Alimentary Pharmacology and Therapeutics. 2005;22:331–342. doi: 10.1111/j.1365-2036.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 30.El-Serag HB, Petersen NJ, Carter J, Graham DY, Richardson P, Genta RM, Rabeneck L. Gastroesophageal reflux among different racial groups in the united states. Gastroenterology. 2004;126:1692–1699. doi: 10.1053/j.gastro.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 31.Isolauri J, Laippala P. Prevalence of symptoms suggestive of gastroesophageal reflux disease in an adult population. Annals of internal medicine. 1995;27:67–70. doi: 10.3109/07853899509031939. [DOI] [PubMed] [Google Scholar]
- 32.Talley NJ, Zinsmeister AR, Schleck CD, Melton LJ., 3rd Dyspepsia and dyspepsia subgroups: A population-based study. Gastroenterology. 1992;102:1259–1268. [PubMed] [Google Scholar]
- 33.Diaz-Rubio M, Moreno-Elola-Olaso C, Rey E, Locke GR, 3rd, Rodriguez-Artalejo F. Symptoms of gastro-oesophageal reflux: Prevalence, severity, duration and associated factors in a spanish population. Alimentary Pharmacology and Therapeutics. 2004;19:95–105. doi: 10.1046/j.1365-2036.2003.01769.x. [DOI] [PubMed] [Google Scholar]