Abstract
Colorectal cancer is the second leading cause of cancer mortality in adult Americans and is caused by both genetic and environmental risk factors. We have replicated our originally reported linkage signal at 9q22-31 by fine mapping an independent collection of colon cancer families. Then, using a custom array of single nucleotide polymorphisms (SNPs) densely spaced across the candidate region, we performed both single-SNP and moving-window association analyses to identify a colon neoplasia risk haplotype. We isolated the association effect to a five SNP haplotype centered around 98.15 megabases (Mb) on chromosome 9q. This haplotype is in strong linkage disequilibrium with the haplotype block containing HABP4 and may be a surrogate for the effect of this CD30 Ki-1 antigen. It is also in close proximity to the GALNT12, which has been recently shown to be altered in colon tumors. Finally, we used a predictive modeling algorithm to demonstrate the contribution of this risk haplotype and surrounding candidate genes in distinguishing between colon cancer cases and healthy controls. The ability to replicate this finding, the strength of the haplotype association (OR=3.68) and the accuracy of our prediction model (~60%) all strongly support the presence of a locus for familial colon cancer on chromosome 9q.
Keywords: colon cancer, linkage analysis, association analysis, risk, family cancer syndrome
Introduction
Colorectal cancer is the second leading cause of cancer mortality in adult Americans with 135,000 new cases and 57,000 deaths annually (1). Each American has, on average, a 6% lifetime risk of developing colorectal cancer and, although early stage cancers are highly curable by surgery and adjuvant chemotherapy, late stage colon cancers remain incurable (2). Both somatic and germline mutations have been associated with the development of colon cancers and their common precursor adenomatous colon polyps. However, familial colon cancers with known cause, Familial Adenomatous Polyposis (FAP) and Lynch Syndrome, also called Hereditary Nonpolyposis Colorectal Cancer (HNPCC), respectively account for <1% (3, 4) and ~5% (5, 6) of all colon cancer cases annually. This leaves a large proportion of the estimated up to 35% heritability of colorectal adenocarcinoma unexplained (7, 8).
Results of linkage and association studies further support the involvement of additional genetic variants in predisposing to colon cancer. Specifically, case-control association studies have identified loci for colon cancer on 8q24 (9, 10, 11), 9p24, 18q21, 11q23, 10p14, 8q23.3 and 15q13.3, 19p13, 20p12, 16q22, and 14q22 (10, 12, 13, 14). Family linkage studies have additionally reported linkage to 3q21-24, 7q31, 11q23 and 9q22-31 (15, 16, 17, 18, 19, 20). Not clear, however, are the penetrance and effect size of each of these identified variants, nor why they are found in some studies and not others.
It was the purpose of this study first to replicate, in an independent sample, the linkage finding at 9q22-31 that we initially identified in a genome scan of 53 kindreds with multiple colon cancer and/or advanced colon adenoma cases (20), and then confirmed by two other studies (17, 18). Indeed, we have narrowed the linkage on 9q22-31 from 13.5 to 7.7 cM and show here that the addition of 69 independent colon neoplasia kindreds increases evidence of linkage to this region. Second, we report further isolation of this effect via a family-based association analysis of over 3,000 SNPs very densely spaced across the region of interest. Third, we identify a five SNP haplotype associated with risk of early onset familial colon neoplasia and its estimated effect size in our study sample. Finally, we offer some reconciliation of the inconsistent results of the many linkage and association studies of colon cancer to date.
Methods
Replication and Localization of Linkage Signal
Sample
To further validate our initial finding of linkage to D9S1786 for familial colon neoplasia, we repeated our linkage analysis using an independent cohort of 69 colon neoplasia kindreds (herein referred to as the confirmation collection), each with at least 2 affected siblings enrolled and DNA available for genotyping. Sixty-two of these kindreds were recruited by the Colon Cancer Family Registry (Colon CFR, CCFR), an NCI-supported consortium established in 1997 to create a multinational comprehensive collaborative infrastructure for interdisciplinary studies in the genetic epidemiology of colorectal cancer. Detailed information about the Colon CFR can be found at http://epi.grants.cancer.gov/CFR/, as well as being described in detail by Newcomb et al (21). The CFR registries that were used to identify eligible cases are located at The Fred Hutchinson Cancer Research Center, University of Hawaii Cancer Research Center, Cancer Care Ontario, University of Melbourne Australasia Colorectal Cancer Family Registry, University of Southern California Consortium, and Mayo Clinic as well as a CFR data center at the University of California Irvine. This study included data from CFR participants recruited both from population-based sources and clinic-based sources.
Additionally the original collection of 53 kindreds from the Colon Neoplasia Sibling Study (CNSS) reported by Wiesner et al (20) was modified in view of updated affection status in family members; these changes resulted in a dataset (herein referred to as the revised original collection) comprising 94 discordant sibling pairs, 70 concordantly affected pairs and 21 concordantly unaffected pairs, all of whom are informative for linkage. All kindreds who self-reported to be of European descent, but collected from multiple sites, were included. Details of the enrollment procedures were published previously but, in brief, we classified individuals as affected if and only if they had a confirmed diagnosis of colorectal cancer or adenomatous polyp ≥1cm by age 65 and no inflammatory bowel disease, FAP or Lynch Syndrome/HNPCC. Persons for whom tumors were available were tested for microsatellite instability and if positive, removed from the sample. Unique to the CNSS study, individuals were classified as unaffected only if they had undergone an endoscopy and no cancer or adenomatous polyps was found, and classified as unknown if they had not been screened.
The expanded cohort (herein referred to as the combined collection) consisted of 272 sibling pairs and included the revised original collection and the new independent cohort. However, because linkage analysis and association analyses are susceptible to bias (either toward or away from the null) due to population stratification, 10 families were excluded because of non-Caucasian or incomplete ancestry information or double enrollment by two different collection sites. The final family sample therefore comprised 254 sibling pairs from 120 families (51 from the original collection, 7 additional CNSS and 62 additional from the CCFR).
Linkage Analysis
To confirm our initial linkage finding, we performed linkage analysis on these 252 siblings pairs. Rather than simply genotyping the markers identified in our original sample, we opted to genotype an additional 17 fine-mapping markers across the originally defined 13.5Mb linkage interval (average spacing of approximately 1.6 cM) in both the revised original and combined collection (Supplementary Table S1). This offered us the potential to further localize, as well as replicate, our result.
Prior to linkage analysis, the genotype data were examined for Mendelian inconsistencies using MARKERINFO in the Statistical Analysis for Genetic Epidemiology program (S.A.G.E. 5.4) and for Hardy Weinberg proportion disequilibrium in family data using FREQ (S.A.G.E. 5.4) (22). Those genotypes believed to be erroneous were removed from subsequent analysis. We then calculated multipoint identity-by-descent (IBD) sharing estimates using GENIBD (S.A.G.E. 5.4) and used those estimates to perform two tests for linkage with SIBPAL (S.A.G.E. 5.4): the weighted Haseman-Elston regression method and the mean tests for concordant affected, discordant and concordant unaffected sibs. The weighted Haseman-Elston regression method regresses on the multipoint IBD sharing estimates, using as the dependent variable a weighted average of the squared sib-pair trait difference and the squared mean-corrected sum expressed as:
where ȳ is best linear unbiased predictor including an adjustment for the population prevalence (23) and the chosen weights are optimal if the sample size is large enough. Note that the Haseman-Elston regression method relies on the presence of, at minimum, two types of sibling pairs from among concordant affected, discordant or concordant unaffected pairs. For this reason we could not perform this particular test in our confirmation sample alone as it comprised only affected sibling pairs. The mean tests however assess the statistical significance of the departure of the observed IBD sharing estimates in each of the respective pair types from what would be expected in the absence of linkage. This test was therefore conducted in the revised original, confirmation and combined collections.
Finally, using the results of these tests for linkage and the IBD sharing estimates, we ranked the families on their likelihood of being linked to this region via the Quantitative Linkage Score (QLS) proposed by Wang and Elston (24) and expressed as:
This QLS allowed us to prioritize families within which to pursue the addition of both marker genotypes and family members, where i and j are two sibs in a family and can be fixed at any value. In our case, we conservatively classified families as linked if they had positive mean and sib-specific QLS scores for all of = 0.25, 0.5, and 0.75.
Association Analysis and Risk Haplotype Identification
Sample and Genotyping
To further localize and assess risk attributable to the variant at 9q22-31, we conducted a joint family-based and case-control association study from 106 of our families comprising 222 affected and 48 unaffected sibpairs and 200 additional, independent controls. Controls were selected from individuals who presented for a colonoscopy at University Hospitals Case Medical Center in Cleveland Ohio, were independent of the families studied, had clean colonoscopies, were at least 60 years old, had no personal history of cancer, and had no more than one first-degree relative with reported cancer of any type.
We genotyped 2699 SNPs across the 13.5 cM region of interest, with an average spacing of 4,000 basepairs (4kb). The SNPs were chosen initially based on tagging, using the Human HapMap CEU (Caucasian European from Utah) samples, imposing a linkage disequilibrium (LD) threshold of r2=.8; we then added additional SNPs to achieve an average spacing of 4kb and ensure adequate coverage given strong Caucasian LD in this region. Based on the HapMap CEPH samples, tagging alone would have resulted in 1 SNP every 5.4 kb with several LD blocks spanning more than 10 kb. All SNPs had a minor allele frequency of at least 5% and were either golden-gate or double-hit validated. All SNP genotypes were collected using the Illumina Bead Station platform via the Case Comprehensive Cancer Center genotyping core.
Single SNP and Moving Window Association Analysis
We performed both single-SNP and moving-window analysis of our SNP genotype data using a regression model, based on Elston et al (25), of the form:
where for any individual i, with liability yi and j covariate values cji. In this formulation, h is the logit link function in the context of a generalized linear mixed model, under the assumption that the random effects are normally distributed, pi is a random polygenic effect, fi and f'i are random common nuclear-family effects, mi is a random marital effect, si is a random common sibship effect, and εi is a random residual individual effect, and where zi is a genotype indicator for the allele A at a diallelic locus with alleles A and B, such that, when considering only a single locus under an additive model,
In addition to testing one SNP at a time, we used a multi-locus approach to produce more robust results and potentially improve power under conditions in which multiple SNP markers are associated with the disease, or in LD with a causal locus. To simultaneously analyze multiple nearby correlated genetic variants, we incorporated moving-window approach into our family-based association method. For an appropriate window width (e.g., 5 SNPs), we moved the window from the first marker to last marker in the candidate genome region. For each window, we fitted the regression model using all markers within the window and calculated the corresponding p-values based on asymptotic theory. Assume k markers within the window. For any individual i, with trait yi and j-th covariate values cji, the regression model shown above now takes the form:
where zi is a genotype indicator as indicated above and f(g) is the smoothing function, which here we chose to be the mean of the additive genetic variants.
To identify the optimal window size while avoiding the penalty of performing additional tests, we blinded ourselves to the SNP names, and then plotted, from the results of moving windows of various sizes, the p-values for the likelihood ratio test (LRT) against the p-values for the one-degree-of-freedom Wald Chi-square test for each group of SNPs. Agreement in these two test statistics serves as an indication of stability and therefore represents the most appropriate window size. With a linear correlation coefficient of 0.98 between the Wald and LRT p–values, the window size of 5 was deemed most appropriate (compared to 0.85, 0.89, 0.93 and 0.87 for window sizes of 1, 3, 7 and 9, respectively).
Haplotype Identification
After identifying significant regions based on both our single-SNP and moving-window analyses, we constructed a risk haplotype based on the estimated odds ratios from the regression analysis outlined above. For example, if the “A” allele was coded as the reference allele and the odds ratio was greater than one, then “A” was deemed as the risk allele and if the odds ratio was less than one, then “B” was deemed the risk allele and so forth for each SNP marker in the haplotype. We were then able to verify these haplotypes in a select sample of 12 individuals from the CNSS family collection for whom we had molecular haplotypes obtained by genotyping of uniparental monochromosomal somatic cell hybrids (converted clones) as described by Yan et al. (26). We then calculated, using the entire sample, the numbers of individuals who were unambiguous carriers of two or zero copies of the risk haplotype, respectively, but included in the counts only a single count per family, even if multiple members were informative. We tested the difference in counts of the risk haplotype between cases and controls using either a Pearson's Chi-square or a Fisher's exact test, as appropriate.
Prediction Modeling
The ability of a group of SNPs to predict cancer versus no-cancer in our dataset was assessed through use of recursive feature elimination in a support vector machine framework (SVM-RFE). Weka data mining software (27) was used to perform the SVM-RFE experiments with a 10-fold cross validation. Cross validation estimates how well a given model predicts the same outcome in an unseen dataset drawn from the same statistical distribution (28). A 10-fold cross validation randomly divided the dataset into 10 equal subsets. The first 9 subsets were used as training data to construct a predictive model and the tenth used as test data. Then, the second through tenth subsets were used as training data and the first subset was used as test data. We stopped this procedure after each subset had been used as test data. The performance measures of our predictive model can be expressed by accuracy, sensitivity, and specificity on the test data. Accuracy is the probability that a subject will be correctly classified as either a case or control by a predictive model. Sensitivity refers to the probability of a positive prediction among patients with disease. Specificity measures the probability of a negative prediction among subjects without disease. A good predictive model should simultaneously optimize accuracy, sensitivity, and specificity. We determined this threshold by selecting the point on the probability output of our predictive model that gave the highest score among these performance measures. It is important to note that we performed our predictive modeling on both the full dataset (ignoring the correlation between subjects) and on a randomly selected group of independent individuals (102 cases : 201 controls).
Results
Replication and Localization of Linkage Signal
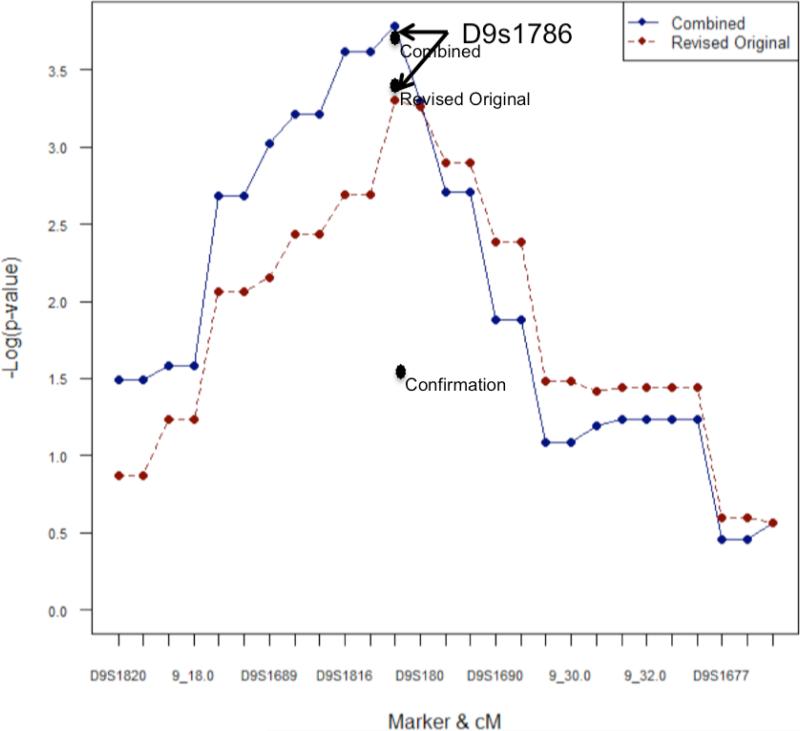
The strongest signal in the Haseman-Elston linkage analysis of the revised original and combined collection of families was with marker D9S1786 (-log p-values =3.35, and 3.38, respectively), exactly at the marker location (Figure 1). The mean test yielded p-values of 0.0005, 0.04 and 0.0001 for the revised original, confirmation and combined collections, respectively. The equivalent of a 1.1-LOD drop from the linkage maximum defined a 7.5cM (8.8Mb) linkage interval bounded by D9S1815-D9S1857 (29). As shown, this expanded colon neoplasia cohort demonstrated increased statistical significance for linkage of disease to the pre-specified marker D9S1786, with the p-value for linkage in this expanded cohort =0.00016, a 3-fold increase in significance from the value of 0.00045 seen in our initial study (20). This increase, as well as the fact that the mean-tests analysis of the confirmation sample met the P<0.05 criterion touted as necessary for significance, demonstrates confirmation of the same linkage among the newly added kinships. It additionally narrows the linkage region to an 8.8Mb interval. Also of note, in this combined collection, concordantly affected sibling pairs demonstrate an excess over 0.5 IBD sharing of 0.60, which corresponds to 40% of colon neoplasia kindreds in the expanded analysis being linked to a potentially autosomal dominant disease gene at 9q22.2-31.2 (with 95% confidence limits for this estimate now being 21%-59%). Thus, this combined collection not only provides replication of our initial finding of linkage of familial colon neoplasia to 9q22.2-31.2, but also strengthens our conclusion that this locus accounts for the development of disease in a major subgroup of colon neoplasia kindreds.
Figure 1.
Haseman-Elston and sibling-pair mean test linkage analysis and fine mapping in the revised original, confirmation and a combined collection of colon neoplasia kindreds. Points on the lines represent the –log p-values of the Haseman-Elston regression test at markers (i.e D9S1820) and intermarker distances (i.e. 9_18) across the region; Black dots represent the –log p-values of the mean allele sharing test at D9S1786. Note that there is no line for the confirmation sample as it contained only affected sibling pairs and was therefore not informative for the Haseman-Elston regression test.
Further, in our follow-up linkage analysis, 25 of our best linked CNSS families had been extended by an additional 3 affected and 42 unaffected family members not available for our originally published linkage study (20). Model-based linkage analysis of these extended families led to an increased LOD score for both the recessive and dominant model (from 3.953 to 4.277 and 2.75 to 3.01, respectively). Despite the fact that these families were identified as being linked and targeted for the addition of family members, and therefore these results are subject to the effects of ascertainment, the increase in LOD score supports the presence of a disease variant in this region.
Association Analysis and Risk Haplotype Identification
Analysis of each SNP individually did not produce any SNPs meeting a very conservative p-value of 2.8 × 10-5 after Bonferroni correction (based on the effective number of SNPs analyzed, (2699) × average proportion of LD (.67) = 1,808 independent tests). However, each of three regions, centered at approximately 92 megabases (Mb), 98Mb and 102Mb, were suggestive of association with SNPs with p-values less than 2.5 × 10-3 (Figure 2).
Figure 2.
Single SNP Association Analysis across the entire 13.5 cM region.
The moving-window analysis produced, as expected, results that were both more robust (that is, less sensitive to the requirements of asymptotic assumptions (as explained in the methods above)), more precise and, in this case, statistically significant. Although the increased statistical significance could be due to increased type I error, in view of the uniformity of the increases, it is more likely to represent a gain in power due to multiple SNPs in the same region being associated with a disease locus or in LD with a causal locus. The three regions that were statistically significant in the single SNP analysis were also significant in the moving-window analyses, with significance increasing to p < 1.0 × 10-5, p < 2.5 × 10-5, and p < 5.0 × 10-5, respectively. As can been seen in Figure 3, the moving-window approach also resulted in a smoothing of the peaks, with more associations clustering around 98Mb We further characterized these regions by examining the LD structure within and among the three regions, as well as calculating the odds ratio and 95% confidence interval for the most significant regional SNPs. Notice that, for the region at 98Mb, there was a break in statistical significance (and therefore the risk haplotype) between 98,157,208 bp and 98,296,272 bp. We therefore characterized separately the two regions: rs7865848 to rs3780442 (centered at 98,146,452 bp) and rs10818948 to rs1953087 (centered at 98,298,339 bp). There is strong linkage disequilibrium across all regions (Figure 3), and specifically between rs10820943 and rs380247 (r2>.9, D’=1, LOD>2) and between rs998952 and rs10818948 (r2>.9, D’=1, LOD>2) (Figure 3). Further, when confining the analysis to only those families most linked to this region, the association becomes stronger and spans even more SNPs, ultimately pointing to the region centered at 98.15Mb as the most likely to house a causal variant.
Figure 3.
Moving Window Association Analysis, window size of five SNPs (upper figure) compared to LD in the region (lower figure). Brackets indicate that the level of linkage disequilibrium between the three highest peaks is greater than 0.9.
This result was further supported by haplotype analysis. In fact, we found 6 cases compared to 4 controls with two copies of the five SNP risk haplotype at 98.15Mb, as well as almost twice as many controls (148 compared to 83 cases) with no copies (OR=3.68 comparing cases to controls with 2 or 0 copies). Additionally, 9 of the 12 affected persons from our most-linked families and for whom we have molecular haplotypes had at least one copy of the risk haplotype at 98.15Mb whereas only three had none. In contrast the risk haplotype in the region centered at 98.29Mb was observed in duplicate (2 copies) in almost equal numbers in cases and controls (OR=0.86). No individuals were homozygous for all risk alleles across the region and therefore could not be unambiguously classified as carrying the risk haplotype in the region centering around 92Mb; and similarly for the region around 102Mb.
To verify that the statistically predicted haplotype indeed exists, we additionally genotyped the uniparental monochromosomal somatic cell hybrids and verified that the haplotype of interest centering around 98.15Mb is indeed present. In fact, 9 of the 12 affected persons from our most-linked families for whom we have converted clones had at least one copy of the risk haplotype whereas only three had none.
Prediction modeling
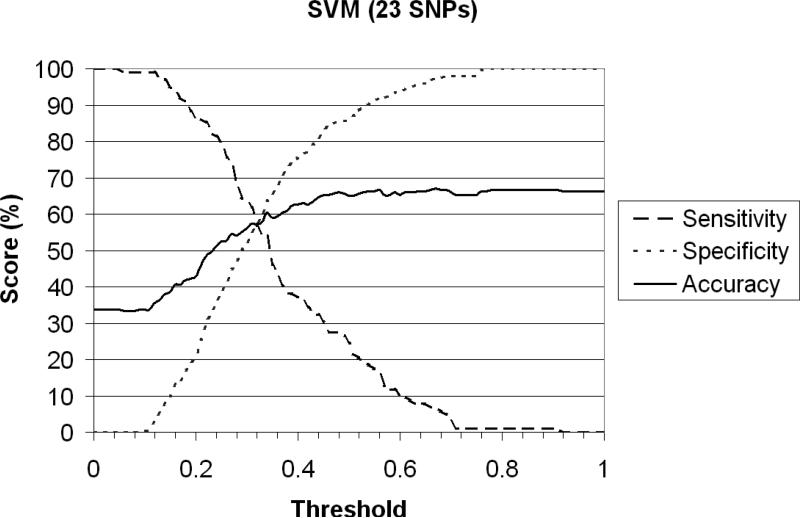
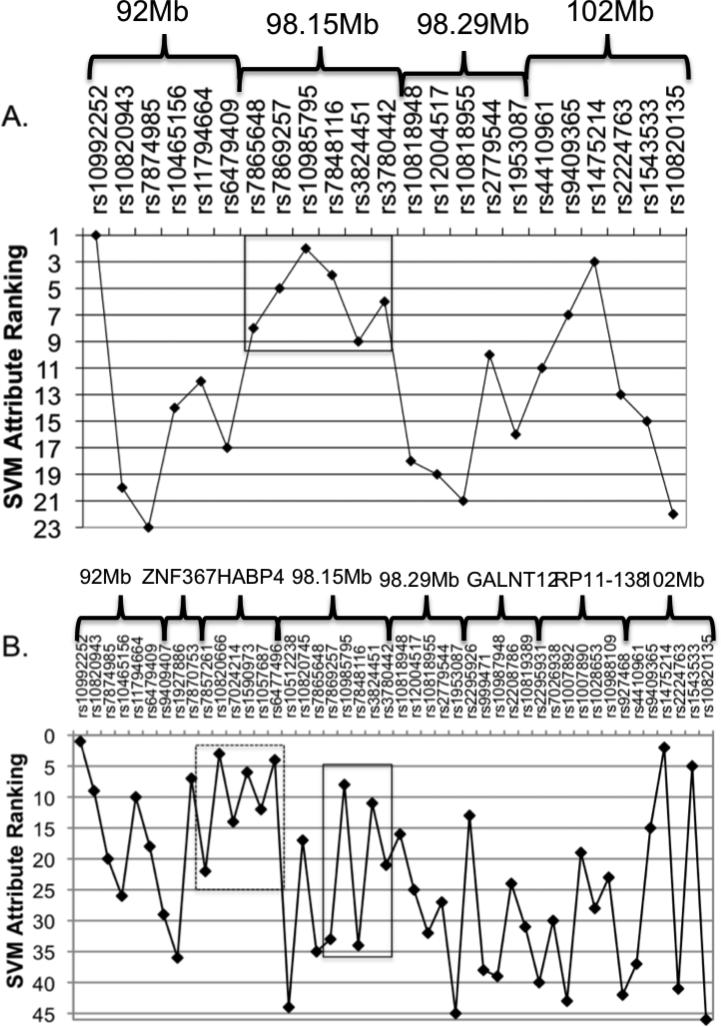
Using SVM in a recursive framework as described above, we were able to predict colon cancer using a specified subset of SNPs with sensitivity, specificity and accuracy “score” each just under 60% the point of intersection of each of the three attributes of interest (called a “threshold”) of 0.365 (Figure 4). When including all SNPs contained within the four associated regions outlined above, the subset of SNPs that best predicted colon cancer in these data span the region 98.145-98.296 (Figure 5A). This result affirmed our previous conclusion, as SVM does not depend on strength of association but rather on predictability. Additional SVM analysis incorporating SNPs within candidate genes in the region (Figure 5B) further improved these predictive models and is discussed below.
Figure 4.
Sensitivity, Specificity and Accuracy of SVM colon cancer prediction model including only SNPs in regions of significance centered at 92Mb, 98.15Mb, 98.29Mb and 102Mb. The threshold of 0.365 is the point at which the three attributes of interest intersect.
Figure 5.
Rankings by predictability A) for SNPs in the four candidate regions centered at 92 megabases (Mb), 98.15Mb, 98.29Mb and 102Mb and B) for SNPs in both the candidate regions and the surrounding candidate genes (ZNF367, HABP4, GABBR2 and GALNT12). The solid box in both A. and B. represents the risk haplotype block at 98.29Mb. The dashed box in B includes the SNPs within HABP4.
Discussion
The results presented here confirm the validity of our initially published finding of statistically significant linkage of familial colon neoplasia to a chromosome 9q22.2-31.2 disease locus. Despite the fact that some of the recent genome-wide association scans do not report association to SNPs in the 9q22.2-31.2 region, we have demonstrated a gain in statistical significance both by adding independent families and by expanding existing families where possible. These linkage results also show, as is not possible with case-control association analyses, that this signal is not due to population stratification or some other form of ascertainment bias and is therefore unlikely to be a false-positive result.
We have further isolated this signal via combined family and population-based association analysis, to a 151,602 bp region centering at 98.15 Mb. This localization was verified via linkage disequilibrium characterization and haplotype analysis. Specifically, as can be seen in Figure 3, the linkage disequilibrium structure of this region shows the strength of linkage disequilibrium between the other two regions of significance, helping to demonstrate that the region at 98.15 most likely represents the causal variant. The haplotype analysis confirms this with an OR of 2.76 for cases compared to controls when scored for 2 or 0 copies of the risk allele. Furthermore, an analysis of all individuals in the family dataset (not just the randomly selected independent cases and controls) increased the OR to 3.68. While a correlation between related individuals could inflate the OR, it may, be more conservative estimate, because of the stronger difference between related cases and controls. This result also verifies that the findings are not an artifact of our choice of controls. Finally, the predictive modeling via SVM, which has been successfully used to model other cancers, including breast (28) and esophageal cancer (30), supports this association. It is important, however, to validate any genetic association and the collection of an independent replication sample is currently underway.
Although well-supported statistically, we recognize that without biological evidence, the causal variant may only be represented by, but not actually contained within, this haplotype block. It is to our good fortune that the candidate region in which we are interested has been fairly well-characterized and houses multiple genes (ZNF367, HABP4, GABBR2 and GALNT12). Two of these plausibly have a functional effect on cancer, specifically the Hyaluronan binding protein 4 gene (HABP4), also called Ki-1/57, and the GalNAc Transferase 12 gene (GALNT12). HABP4 is a CD30 Ki-1 antigen first discovered as a marker for Reed-Sternberg cells in Hodgkin lymphoma, but was later found to be expressed in a variety of cell lines, including normal lymphocytes and monocyte-derived macrophages. The Ki-57 molecule, with which Ki-1 interacts, occurs intracellularly only in the cytoplasm, nuclear pores and the nucleus (31, 32). This antigen has also been shown to interact with the chromohelicase-DNA-binding domain protein 3 (CHD3), a nuclear protein involved in the regulation of transcription and chromatin remodeling, and the receptor of activated kinase 1 (RACK1), and further co-precipitates protein kinase C (PKC). PKC is a tumor promoter and has been extensively studied and linked to breast, bladder, skin and other forms of cancer. GALNT12 has been suggested to play a role in the initial step of mucin-type oligosaccharide biosynthesis in digestive organs (33) and has been shown to be highly expressed in digestive organs such as small intestine, stomach, pancreas and colon, and moderately expressed in testis, thyroid gland and spleen. Recent studies from our group report both rare germline GALNT12 mutations that are present in some individuals who develop colon cancer, and rare somatic GALNT12 mutations in certain colon cancer tumors (34).
It is therefore reasonable to ask if our association signal points to either of the above-mentioned genes as the most likely candidate. Five of the SNPs typed as a part of our association study lie within HABP4 (four intronic and one 3’ UTR) and 11 SNPs lie within GALNT12 (11 intronic and one synonymous coding SNP). Although none of these SNPs met our threshold for statistical significance in the association analysis, there are examples in the literature of causal variants identified within genes outside of the region of greatest statistical significance in an association study (35). Further, a comparison of the linkage-disequilibrium structure in the cases and the controls in our dataset suggests that our strongly associated haplotype may actually be a surrogate for the haplotype block spanning 98,140,446 to 98,320,232 bp, which was typed but contains less informative markers. As is shown, there is appreciable linkage disequilibrium between each of the regions identified in our association study and the haplotype block containing HABP4 in the case sample but noticeably missing from the control sample (Figure 6). Nonetheless, this haplotype block also contains the Zinc Finger Protein 367 (ZNF367) gene, a transcriptional activator of erythroid genes that has no known association to cancer. Finally, we repeated the SVM analysis including not only the SNPs within the regions of statistical significance (Figure 5A) but also the SNPs within each of the genes mentioned above (Figure 5B). None of the SNPs in ZNF367 contributed to the accuracy of the model. However, one of the 11 SNPs within GALNT12 and all five SNPs in HABP4 did increase the accuracy of the model (from 57.21 to 57.71% when adding the three SNPs in GALNT12 and from 57.21 to 60.70% when including the five SNPs in HABP4). We have not yet further explored HABP4 but have, as mentioned above, more closely examined GALNT12 and it is possible that the linkage and association signal discussed herein arises from non-coding mutations that affect expression of the GALNT12 locus. Our current studies have not however detected such an effect in lymphoblastoid cell lines from affected individuals in our linked families, and we have yet to obtain normal or malignant colon tissues from these individuals to further explore this model.
Figure 6.
LD plot of regions with significant moving-window association results and in linkage disequilibrium with candidate genes for A) cases (N=225) and B) controls (N=248). Solid ovals indicate significant regions, the solid box indicates the risk haplotype at 98.15Mb, brackets indicate genes and dashed ovals highlight regions of LD between the haplotype block containing HABP4 and the four regions of significance.
In conclusion, we note that the underlying genomic complexity of the 9q region and the differences in study design could explain the contradictory results between our analysis and other published studies. Most importantly, other linkage and association studies have used markedly different phenotype definitions, ascertainment strategies and genotyping approaches from ours. Specifically, we required that all cases have colorectal cancer, high grade dysplasia or an advanced adenoma as well as an early age of onset (<66 years) and an available affected sibling. These are more stringent criteria than just including all persons with an affected first-degree relative, because, while on average they share 50%, the amount of genetic information shared between first-degree relatives of all types is quite variable (i.e. sibs can share 0 to 100% IBD). Further, by supplementing a set of tag SNPs with additional, more uniformly spaced SNPs, we were able to capture a much larger proportion of the variability in this region. We point toward the studies by the CORGI consortium as support for these differences explaining the various results; they did not find the signal at 9q in 69 families with a mixture of colon cancer, adenomas and polyps (15), but they did replicate our findings in 57 families with >=3 affected persons, using strict age-of-onset criteria for the three phenotypes mentioned above (<75, <45, <35, respectively) (18). All of this suggests that the disease locus housed on 9q is specific to a familial syndrome with a phenotype of younger age of onset and/or severity of colon neoplasia. Finally, while the prevalence of the syndrome we describe is unknown, we suggest that the underlying variants are likely uncommon. This and further characterization of the effect at 9q are the subject of ongoing research.
Supplementary Material
Table S1: Listed are the microsatellite markers used for finemapping analysis across the 9q22-31 linkage interval. The primer sequences and genomic positions of the microsatellite markers were obtained from the UniSTS database (NCBI, build 36.3). The KG161729 marker was custom designed using the Genemachine software tool (http://genemachine.nhgri.nih.gov). One primer of each primer pair was labeled with either FAM or HEX fluorophore at the 5' end. The PCR conditions for all primer pairs included 35 cycles of 95°C for 1min, 60°C for 45 s and 72°C for 1 min. The PCR reactions were performed in a 50μl reaction volume using AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). The PCR products were purified and run on a 3130 Genetic Analyzer and genotyped using the GeneMapper software (Applied Biosystems, Foster City, CA).
Acknowledgements
We gratefully acknowledge the individuals and families who participated in this study. The results of this paper were obtained by using the program package S.A.G.E., which is supported by a U.S. Public Health Service (PHS) Resource Grant (RR03655) from the National Center for Research Resources. This work was also supported by the Prevent Cancer Foundation, the National Institutes of Health National Cancer Institute, and National Institute of General Medical Sciences PHS awards R01 CA130901, R01 CA104667, P30 CA043703, R01GM28356, and through cooperative agreements with members of the Colon Cancer Family Registry (CFR). Each CFR center that provided data for the analysis were supported as follows: Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799). Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), Seattle Colorectal Cancer Family Registry (U01 CA074794), University of Hawaii Colorectal Cancer Family Registry (U01 CA074806), University of California, Irvine Informatics Center (U01 CA078296). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CFR.
References
- 1.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics. CA Cancer J Clin. 2001;2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 2.Skibber J, Minsky B, Hoff P, DeVita V, Hellman S, Rosenberg S. Cancer: Principles and practice of oncology. Lippincott Williams and Wilkins; Philadelphia: 2001. Cancer of the colon. pp. 1216–71. [Google Scholar]
- 3.Kinzler K, Vogelstein B. The Genetic Basis of Human Cancer. McGraw-Hill; New York: 2002. Colorectal tumors. pp. 583–612. [Google Scholar]
- 4.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–79. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 5.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J NatlCancer Inst. 1995;87:1114–25. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. NEnglJ Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 8.Cannon-Albright LA, Skolnick MH, Bishop DT, Lee RG, Burt RW. Common inheritance of susceptibility to colonic adenomatous polyps and associated colorectal cancers. NEnglJ Med. 1988;319:533–7. doi: 10.1056/NEJM198809013190902. [DOI] [PubMed] [Google Scholar]
- 9.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 10.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haiman CA, Le Marchand L, Yamamato J, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poynter JN, Figueiredo JC, Conti DV, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–32. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 13.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 14.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp Z, Carvajal-Carmona L, Spain S, et al. Evidence for a colorectal cancer susceptibility locus on chromosome 3q21-q24 from a high-density SNP genome-wide linkage scan. Hum Mol Genet. 2006;15:2903–10. doi: 10.1093/hmg/ddl231. [DOI] [PubMed] [Google Scholar]
- 16.Neklason DW, Kerber RA, Nilson DB, et al. Common familial colorectal cancer linked to chromosome 7q31: a genome-wide analysis. Cancer Res. 2008;68:8993–7. doi: 10.1158/0008-5472.CAN-08-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skoglund J, Djureinovic T, Zhou XL, et al. Linkage analysis in a large Swedish family supports the presence of a susceptibility locus for adenoma and colorectal cancer on chromosome 9q22.32-31.1. J Med Genet. 2006;43:e7. doi: 10.1136/jmg.2005.033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp ZE, Carvajal-Carmona LG, Barclay E, et al. Evidence of linkage to chromosome 9q22.33 in colorectal cancer kindreds from the United Kingdom. Cancer Res. 2006;66:5003–6. doi: 10.1158/0008-5472.CAN-05-4074. [DOI] [PubMed] [Google Scholar]
- 19.Djureinovic T, Skoglund J, Vandrovcova J, et al. Swedish families with hereditary non-familial adenomatous polyposis/non-hereditary non-polyposis colorectal cancer. Gut. 2006;55:362–6. doi: 10.1136/gut.2005.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiesner GL, Daley D, Lewis S, et al. A subset of familial colorectal neoplasia kindreds linked to chromosome 9q22.2-31.2. ProcNatlAcadSciUSA. 2003;100:12961–5. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–43. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 22.S.A.G.E. Statistical Analysis for Genetic Epidemiology. 2009 Release 6.0.1: http://darwin.cwru.edu/
- 23.Sinha R, Gray-McGuire C. Haseman-Elston regression in ascertained samples: importance of dependent variable and mean correction factor selection. Hum Hered. 2008;65:66–76. doi: 10.1159/000108938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Elston RC. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet. 2007 Feb;80:353–60. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elston RC, George VT, Severtson F. The Elston-Stewart algorithm for continuous genotypes and environmental factors. Hum Hered. 1992;42:16–27. doi: 10.1159/000154043. [DOI] [PubMed] [Google Scholar]
- 26.Yan H, Papadopoulos N, Marra G, et al. Conversion of diploidy to haploidy. Nature. 2000;403:723–4. doi: 10.1038/35001659. [DOI] [PubMed] [Google Scholar]
- 27.Witten IH, Frank E. Data mining : practical machine learning tools and techniques. 2nd ed. Morgan Kaufman; Amsterdam ; Boston, MA: 2005. [Google Scholar]
- 28.Listgarten J, Damaraju S, Poulin B, et al. Predictive models for breast cancer susceptibility from multiple single nucleotide polymorphisms. Clin Cancer Res. 2004;10:2725–37. doi: 10.1158/1078-0432.ccr-1115-03. [DOI] [PubMed] [Google Scholar]
- 29.Bennewitz J, Reinsch N, Kalm E. Improved confidence intervals in quantitative trait loci mapping by permutation bootstrapping. Genetics. 2002;160:1673–86. doi: 10.1093/genetics/160.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Statnikov A, Li C, Aliferis CF. Effects of environment, genetics and data analysis pitfalls in an esophageal cancer genome-wide association study. PLoS ONE. 2007;2:e958. doi: 10.1371/journal.pone.0000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen H, Lemke H, Bredfeldt G, Konnecke I, Havsteen B. The Hodgkin-associated Ki-1 antigen exists in an intracellular and a membrane-bound form. Biol Chem Hoppe Seyler. 1989;370:409–16. doi: 10.1515/bchm3.1989.370.1.409. [DOI] [PubMed] [Google Scholar]
- 32.Froese P, Lemke H, Gerdes J, et al. Biochemical characterization and biosynthesis of the Ki-1 antigen in Hodgkin-derived and virus-transformed human B and T lymphoid cell lines. J Immunol. 1987;139:2081–7. [PubMed] [Google Scholar]
- 33.Guo JM, Zhang Y, Cheng L, et al. Molecular cloning and characterization of a novel member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett. 2002;524:211–8. doi: 10.1016/s0014-5793(02)03007-7. [DOI] [PubMed] [Google Scholar]
- 34.Guda K, Moinova H, He J, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase-12 in human colon cancers. Proc Natl Acad Sci U S A. 2009;106:12921–5. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shifman S, Johannesson M, Bronstein M, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Listed are the microsatellite markers used for finemapping analysis across the 9q22-31 linkage interval. The primer sequences and genomic positions of the microsatellite markers were obtained from the UniSTS database (NCBI, build 36.3). The KG161729 marker was custom designed using the Genemachine software tool (http://genemachine.nhgri.nih.gov). One primer of each primer pair was labeled with either FAM or HEX fluorophore at the 5' end. The PCR conditions for all primer pairs included 35 cycles of 95°C for 1min, 60°C for 45 s and 72°C for 1 min. The PCR reactions were performed in a 50μl reaction volume using AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). The PCR products were purified and run on a 3130 Genetic Analyzer and genotyped using the GeneMapper software (Applied Biosystems, Foster City, CA).