Abstract
Type 2 diabetes (T2D) is strongly linked to obesity and an adipose tissue unresponsive to insulin. The insulin resistance is due to defective insulin signaling, but details remain largely unknown. We examined insulin signaling in adipocytes from T2D patients, and contrary to findings in animal studies, we observed attenuation of insulin activation of mammalian target of rapamycin (mTOR) in complex with raptor (mTORC1). As a consequence, mTORC1 downstream effects were also affected in T2D: feedback signaling by insulin to signal-mediator insulin receptor substrate-1 (IRS1) was attenuated, mitochondria were impaired and autophagy was strongly upregulated. There was concomitant autophagic destruction of mitochondria and lipofuscin particles, and a dependence on autophagy for ATP production. Conversely, mitochondrial dysfunction attenuated insulin activation of mTORC1, enhanced autophagy and attenuated feedback to IRS1. The overactive autophagy was associated with large numbers of cytosolic lipid droplets, a subset with colocalization of perlipin and the autophagy protein LC3/atg8, which can contribute to excessive fatty acid release. Patients with diagnoses of T2D and overweight were consecutively recruited from elective surgery, whereas controls did not have T2D. Results were validated in a cohort of patients without diabetes who exhibited a wide range of insulin sensitivities. Because mitochondrial dysfunction, inflammation, endoplasmic-reticulum stress and hypoxia all inactivate mTORC1, our results may suggest a unifying mechanism for the pathogenesis of insulin resistance in T2D, although the underlying causes might differ.
INTRODUCTION
Type 2 diabetes (T2D), which is most often coupled with obesity, is characterized by an early and marked insulin resistance in the adipose tissue and as a consequence is associated with disturbed cellular and whole-body metabolism. The insulin resistance is due to dysfunctions in intracellular signal transduction, but the molecular details remain unclear. A recurring theme is an impaired phosphorylation of the insulin receptor substrate-1 (IRS1) protein by the activated insulin receptor (1–5). Largely on the basis of results from animal studies, this impairment has, in turn, been ascribed to increased phosphorylation of IRS1 at serine residues and to low levels of IRS1 protein (reviewed in [6]). In contrast, we previously found that in adipocytes from patients with T2D (5), in healthy lean volunteers after overeating (7), and in adipocytes made insulin resistant with the adipokine retinol-binding protein-4 (RBP4) (8), a feedback loop to phosphorylation of IRS1 at serine 307 (corresponding to serine 302 in the murine sequence) is strongly attenuated. Because phosphorylation of IRS1 at serine 307 appears to enhance the phosphorylation of IRS1 at tyrosine by the insulin receptor (5,9–11), attenuation of this positive feedback loop could explain the poor activation of IRS1 in T2D. The feedback signal to phosphorylation at serine 307 is inhibited by rapamycin (5,9), a very specific inhibitor of the protein kinase target of rapamycin (TOR), which indicates a direct involvement of the mammalian (m) ortholog mTOR in the feedback signal. Serine 307 is located in a consensus sequence for phosphorylation by protein kinase B or p70 ribosomal protein S6 kinase-1 (S6K1), and the mTOR-downstream S6K1 has been reported to phosphorylate serine 307 in vitro (12). The fact that S6K1 is an important insulin-controlled serine-307 protein kinase was further indicated by small-interfering RNA–mediated knockdown of S6K1 in TSC2−/− mouse embryo fibroblasts, which inhibited the phosphorylation at serine 307 in response to insulin (12). It appears that phosphorylation of IRS1 at serine 307 may indirectly promote the phosphorylation of IRS1 at tyrosine, through inhibition of the dephosphorylation by phosphotyrosine protein phosphatases (13).
TOR coordinates control of cell growth and metabolism in accordance with nutrient availability in unicellular organisms. During evolution of multicellular organisms this control was seized by insulin and other growth factors. However, the ancient ability of TOR to sense nutrient levels in cells independently of insulin is retained in multicellular organisms, including man, giving TOR a key role in cellular control of metabolism and cell growth, as well as tolerance to starvation through control of autophagy. In mammalian cells mTOR, in complex with the protein raptor (mTORC1), is activated by insulin and the insulin receptor substrate-1 (IRS1) via either or both of the two signaling branches of insulin that lead to activation of protein kinase B/Akt or the Map-kinase ERK1/2, respectively. By responding to amino acid and energy levels in the cell, mTORC1 thus integrates insulin signaling with nutrient availability to control cellular processes such as cell growth, protein synthesis, mitochondrial function and autophagy (Figure 1A), reviewed in (14). In several studies, in particular on animals after high-fat feeding regimens, insulin resistance has been coupled with hyperactive mTORC1 (reviewed in [6]). However, because mTORC1 mediates the positive feedback signal to phosphorylation of IRS1 at serine 307, we wanted to further investigate the function of mTORC1 and its role in insulin resistance and T2D in human fat cells.
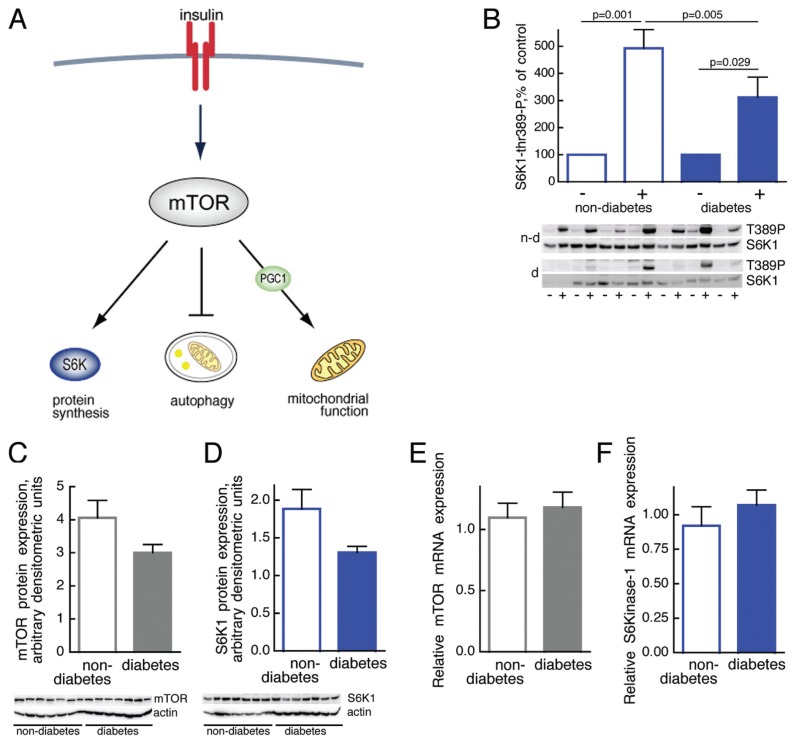
Figure 1.
Impaired mTOR activation in T2D. (A) Outline of major mTOR functions. (B) Phosphorylation of S6K1 in response to insulin stimulation of adipocytes from seven diabetic (d, filled bars; age 36–76, average 62.6 years; BMI 29.0–48.0, average 38.0 kg/m2) and seven nondiabetic subjects (n-d, open bars; age 46–61, average 52.7 years; BMI 20.8–32, average 25.4 kg/m2). Adipocytes were incubated with (+) or without (–) 10 nmol/L insulin for 10 min, when cells were subjected to SDS-PAGE and immunoblotting with antibodies against phospho-threonine(389)S6K1. Phosphorylation was normalized to the amount of S6K1 protein. Results are given as % of control without insulin, for each subject, mean ± SE. Immunoblots show the state of phosphorylation of S6K1 (T389P) and amount of S6K1 protein. (C,D) Abundance of mTOR (C) or S6K1 (D) protein in adipocytes from seven diabetic (filled bars; age 32–80, average 51.9 years; BMI 27.5–47.9, average 38.8 kg/m2) and seven nondiabetic subjects (open bars; age 27–71, average 51.4 years; BMI 18.7–32.9, average 32.6 kg/m2). Adipocytes were subjected to SDS-PAGE and immunoblotting with antibodies against mTOR or S6K1. The amount of mTOR or S6K1 protein was normalized to the amount of actin in each sample. Results are given as arbitrary densitometric units, mean ± SE. (E,F) Abundance of mTOR (E) or S6K1 (F) mRNA. Total RNA was extracted from isolated adipocytes from eight nondiabetic subjects (open bars; age 22–73, average 48.9 years; BMI 20–32.9, average 25.3 kg/m2) and eight patients with T2D (filled bars; age 32–63, average 48.1 years; BMI 29–45.4, average 40.0 kg/m2). The amount of mTOR or S6K1 mRNA was determined by RT-PCR. Results are given as amount of indicated mRNA in relation to GAPDH mRNA, mean ± SE.
MATERIALS AND METHODS
Subjects
Informed consent was obtained from all study participants, and all procedures were approved by the ethics committee, Linköping University, and were performed in accordance with Declaration of Helsinki. Subcutaneous fat was obtained from patients undergoing elective abdominal surgery during general anesthesia. A slice of subcutaneous tissue from skin to muscle fascia was excised. Subjects undergoing elective abdominal surgery at the University Hospital in Linköping-Norrköping were recruited consecutively. To ensure inclusion only of patients with T2D related to obesity, study patients were selected when diagnosed with T2D and obesity/overweight (body-mass index [BMI] > 27). In the comparison group, the only selection criterion for nondiabetic subjects was that they had not received a diagnosis of diabetes. We did not match subjects for obesity or BMI, the diabetic patients were thus on average more obese than the nondiabetic subjects. Nondiabetic subjects and patients with diabetes were selected only for absence or presence of the disease, respectively. Thus, there were some obese insulin-resistant subjects in the nondiabetic comparison group, but this method of selection also allows for a wide significance of the results. Whole-body insulin sensitivity (Quantitative Insulin Sensitivity Check Index [QUICKI] [15]) was determined from concentrations of insulin and glucose in fasting plasma samples from nondiabetic subjects characterized in Supplemental Table S1.
Materials
Rabbit anti-mTOR (#2972), rabbit anti–phospho-p70S6K-thr389 (#9205), rabbit anti-70S6K (#9202), and rabbit anti–phosphoS6-ser235/236 (#2211) and anti–phospho-IRS1-ser302 (#2384, murine sequence) were from Cell Signaling Technology (Danvers, MA, USA). Rabbit anti-LC3 (sc-28266) and goat anti-actin (sc-1616) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit anti-LC3 (PM036) was from Medical & Biological Laboratories (Naka-ku, Japan). Monoclonal anti-UQCRC2 (A11143) was from Molecular Probes (Eugene, OR, USA). Antiperilipin antibodies were a kind gift of C. Londos (NIH, Bethesda, MD, USA).
Isolation and Incubation of Adipocytes
Adipocytes were isolated from subcutaneous adipose tissue by collagenase (Type 1, Worthington Biochemical, Lakewood, NJ, USA) digestion as described (16). Cells were treated and incubated in supplemented Krebs-Ringer solution as described (17).
Fluorescence Microscopy
To quantify LC3 we used immunofluorescence microscopy. Cells were fixed with 3% paraformaldehyde for 20 min and attached to poly-L-lysine–coated coverslips. Cells were then permeabilized and blocked in 0.1% saponin; 5% normal goat serum in: 137 mmol/L NaCl, 2.7 mmol/L KCl, 10 mmol/L Na2HPO4 and 1.8 mmol/L KH2PO4, at 37°C for 1 h; and then incubated with anti-LC3 antibody at 4°C for 20 h. Cells were then washed and incubated with fluorescent secondary antibody (Alexa fluor488; Molecular Probes, Eugene, OR, USA). Confocal scanning microscopy was performed with a Nikon D-Eclipse CI (Nikon, Tokyo, Japan). To determine the amount of LC3-puncta, a fixed threshold was set in Adobe Photoshop Elements 4.0 and the mean fluorescence intensity was quantified with Volocity Quantitation (ver. 4.2.0, Improvision, Coventry, UK).
To quantify the amount of mitochondria, cells were incubated with 200 nmol/L Mitotracker Green (Molecular Probes) for 30 min, and analyzed with a Nikon D-Eclipse C1. Mean fluorescence intensity was quantified with Volocity Quantitation.
Lipofuscin particles were identified by confocal microscopy as Cu2+-quenched autofluorescent particles exhibiting emission throughout the commonly used excitation spectrum ranging from 480 to 630 nm. The number of lipofuscin particles clustered around the nucleus was counted in 20 cells per patient.
Electron Microscopy
Pieces of adipose tissue were fixed in 3.5% (w/v) paraformaldehyde with 0.1% (w/v) picric acid (Zambonis fixative [18]) for 1 h, cut into approximate 1-mm3 pieces and fixed for 2.5 h in the same solution. Tissue pieces were then fixed with 2% (v/v) glutaraldehyde in cacodylate buffer at 4°C overnight, followed by 1% (w/v) OsO4 in cacodylate buffer at 4°C overnight. To enhance contrast, fixed tissue pieces were incubated in 1% (w/v) uranyl acetate in H2O for 2 h at 4°C. Samples were then dehydrated in increasing concentrations of ethanol and then propylene oxide. Epon 812-like resin Agar 100 (Agar Scientific, Essex, UK) was gradually infiltrated and allowed to harden for 48 h at 64°C. Ultrathin sections (65 nm) were collected on copper grids, briefly exposed to 0.4% (w/v) lead citrate solution to further enhance contrast, and examined by transmission electron microscopy (JEOL JEM-1230, Tokyo Japan). Cells were imaged at 80,000× magnification by using a SC1000 Orius CCD camera (Gatan, Pleasanton, CA, USA).
To quantify the number of autophagosomes, mitochondria with cristae, or cytosolic lipid droplets, thin sections covering a whole adipocyte were examined and organelles counted. To quantify the volume fraction of mitochondria with developed cristae, the area of the thin section covered by such mitochondria was determined in the section of each cell with Volocity Quantitation.
SDS-PAGE and Immunoblotting
Cell incubations were terminated by separating cells from medium by using centrifugation through dinonylphtalate. To minimize postincubation signaling and protein modifications, which can occur during immunoprecipitation, cells were immediately dissolved in SDS and β-mercaptoethanol with protease and protein phosphatase inhibitors, frozen within 10 s and thawed in boiling water for further processing (16) for SDS-PAGE and immunoblotting (17). The amount of mTOR, S6K1 and UQCRC2 protein were normalized to actin, and phosphorylation of S6K1, S6 and IRS1 was normalized to the amount of S6K1, S6, and IRS1, respectively, in each sample. Data were normalized to percent of maximal effect.
mRNA Isolation and Analysis by Real-Time PCR
RNA was prepared from isolated adipocytes by Trizol extraction (Qiagen, Hilden, Germany) and solid-face extraction by using RNeasy MinElute Cleanup columns (Qiagen). Reagents for real-time PCR analysis of PGC1α, mTOR, S6K1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Assays-on-Demand and TaqMan Universal PCR Master Mix) were from Applied Biosystems (Foster City, CA, USA). GAPDH was used as an internal reference to normalize expression levels between samples. First strand cDNA was synthesized from 2 μg of adipocyte total RNA by using SuperScript III First Strand Synthesis SuperMix for quantitative RT-PCR according to manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). All real-time PCR measurements were performed in duplicate (Applied Biosystems 7900 Real-Time PCR System) with default cycle parameters.
Glucose Uptake
After transfer of cells to medium without glucose, cells were incubated with 10 nmol/L insulin for 15 min, when glucose transport was determined as uptake of 50 μmol/L (10 μCi/mL) 2-deoxy-d-[1-3H]glucose (19), and then incubated 30 min. Uptake was linear for at least 30 min.
Determination of ATP
For determination of ATP, samples of 200 μL cells at 10% (v/v) were incubated in triplicate with or without 5 μmol/L chloroquine, as indicated, for 18 h. Cells were transferred to 600 μL lysis buffert (E194A; Promega, Madison, WI, USA) and passed three times through a 25-gauge needle. Trichloroacetic acid, 400 μL 5% (w/v), was added immediately, and samples were put on ice. ATP concentration was measured with a luciferase-based assay (ENLITEN ATP assay system; Promega).
Statistical Significance
The statistical significance of differences between groups or controls and treatments were analyzed with the Student t test or linear regression analysis, as appropriate, with P < 0.05 considered significant. Nonsignificance (P > 0.05) is not indicated in figures.
All supplementary materials are available online at www.molmed.org.
RESULTS
Insulin Control of mTORC1 in T2D
We examined mTORC1 in isolated adipocytes obtained from obese patients with T2D and compared these with cells from nondiabetic subjects. In diabetic cells the activation of mTORC1 by insulin assessed as insulin-stimulated phosphorylation of S6K1 (Figure 1B) was attenuated. However, neither the amount of mTOR protein (Figure 1C) nor mTOR mRNA (Figure 1E) was significantly different. Likewise, the S6K1 protein (Figure 1D) and mRNA levels (Figure 1F) were not significantly different in the diabetic cells, although there was a clear tendency towards reduced levels of both mTOR and S6K1 protein.
Autophagy in T2D
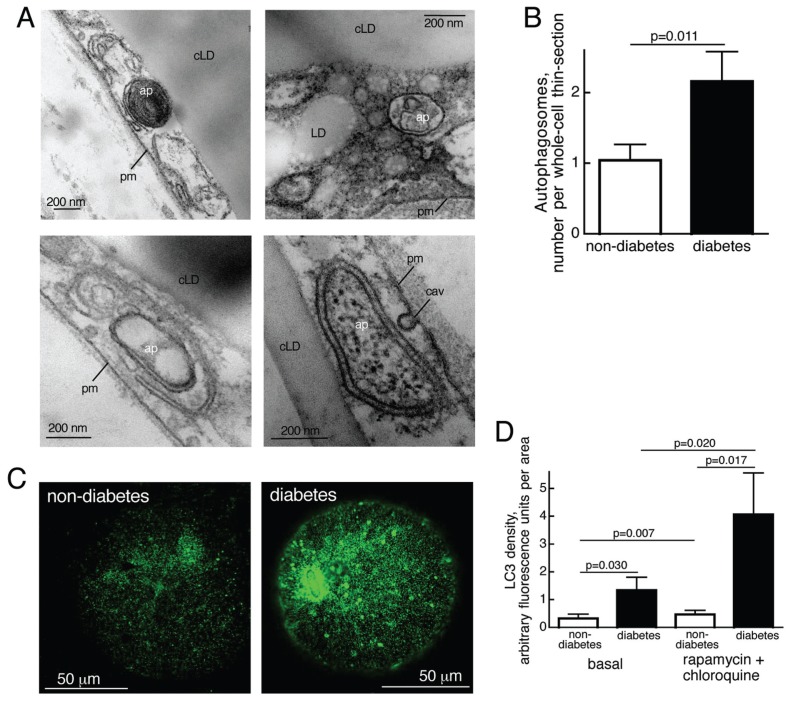
As insulin inhibits autophagy through activation of mTORC1 (20) (Figure 1A), the attenuated mTORC1 in diabetic cells should in consequence be paralleled by enhanced autophagy. During autophagy cellular material is enclosed in a double-membrane autophagosome that eventually fuses with lysosomes to degrade enclosed content within a single-membrane autolysosome. By transmission electron microscopy (TEM) of thin sections of adipocytes we identified (Figure 2A) and quantified characteristic autophagosome-related structures (Figure 2B). The number of autophagosomes was significantly higher in the diabetic adipocytes, indicating that autophagy is increased in T2D, in accordance with the attenuated mTORC1 activity. As a general cellular response to nutrient deprivation, autophagy secures an uninterrupted supply of micronutrients, thus safeguarding cell survival. Indeed, the diabetic adipocytes exhibited a significant, albeit moderate, dependence on autophagy to maintain the cellular concentration of ATP (see for example 21), as evidenced by a decrease of the ATP concentration after inhibition of autophagic digestion (Supplemental Figure S1).
Figure 2.
Enhanced autophagy in T2D. (A) Autophagosome-related structures visualized in thin sections of adipocytes by transmission electron microscopy; ap, autophagosome; cLD, central lipid droplet; LD, cytosolic lipid droplet; pm, plasma membrane; cav, caveolae. (B) Number of autophagosome-related structures in adipocytes from five nondiabetic (open bar; age 49–76, average 63.6 years; BMI 26.7–34.9, average 29.1 kg/m2) and five diabetic (filled bar; age 32–63, average 46.6 years; BMI 37.2–45.4, average 40.9 kg/m2) subjects determined by transmission electron microscopy of thin sections of five different whole cells from each subject. Results are given as number of autophagosome-related structures per whole-cell thin-section, mean ± SE. (C) Immunofluorescence confocal microscopy of nondiabetic (left) and diabetic (right) adipocytes that were incubated with 50 nmol/L rapamycin and 5 μmol/L chloroquine for 18 h, when LC3 was detected. (D) Autophagic activity, amount and turnover of particulate LC3. Adipocytes were isolated from seven nondiabetic subjects (open bars; age 28–85, average 53.3 years; BMI 19–35, average 26.1 kg/m2) and seven patients with T2D (filled bars; age 32–69, average 48.4 years; BMI 29–45.4, average 39.5 kg/m2) and incubated, with or without rapamycin and chloroquine, as in (C), when LC3 was determined by immunofluorescence microscopy. We analyzed 20 randomly selected cells per subject and condition. Results are given as fluorescence per imaged area of the cells, mean ± SE.
During autophagosome biogenesis, the protein LC3/atg8 is incorporated in the membrane of the forming autophagosome, and enhanced autophagic activity in the diabetic adipocytes was evidenced by an increased punctate distribution of LC3/atg8 in these cells (Figure 2C, D). Furthermore, after inhibition of mTORC1 with the specific inhibitor rapamycin, in the presence of chloroquine, LC3 puncta increased dramatically in the diabetic cells compared with nondiabetic cells (Figure 2D). To ascertain the rapamycin-and chloroquine-induced increase in endogenous LC3 puncta, we also used antibodies against a different LC3 epitope and obtained similar results (not shown). Chloroquine inhibits the continuous lysosomal breakdown of LC3, which indicates increased flow through the autophagosomal system in diabetic cells (22). The effect of rapamycin also indicates a strongly upregulated potential for increased autophagy in the diabetic cells.
Autophagy and Accumulation of Lipofuscin in T2D
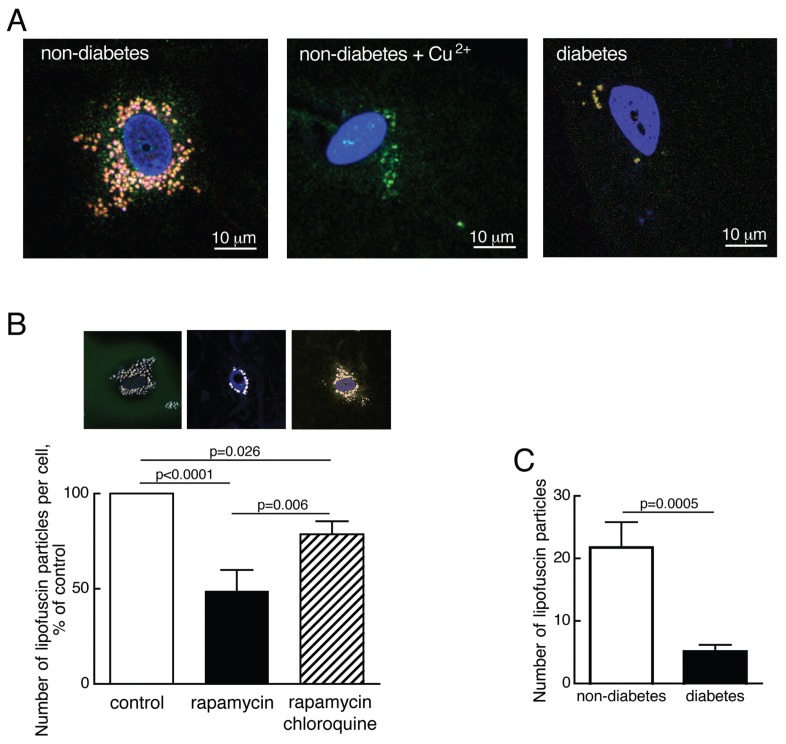
Human adipocytes displayed an accumulation of lipofuscin-like particles, identified by confocal fluorescence microscopy as Cu2+-quenched broad-spectrum autofluorescent particles (Figure 3A). Lipofuscin accumulation is indicative of inhibited autophagy in human fibroblasts (23), and we found that activation of autophagy by inhibition of mTORC1 with rapamycin reduced the number of lipofuscin-like particles in the human adipocytes (Figure 3B). This reduction was inhibited by the lysosomal inhibitor chloroquine (Figure 3B), demonstrating that the particles were degraded by an autophagic/lysosomal process. In accordance with attenuated mTOR activity and enhanced autophagy in the diabetic cells, the number of lipofuscin particles was considerably reduced in adipocytes from patients with T2D (Figure 3A, C).
Figure 3.
Reduced number of lipofuscin particles in T2D. (A) Visualization of autofluorescent lipofuscin particles in adipocytes from a nondiabetic subject (left), after treatment with 5 mmol/L CuSO4, 50 min (middle), and from a diabetic subject (right), using confocal fluorescence microscopy. Cell nuclei were stained with TO-PRO3 (blue). (B) Quantification of lipofuscin particles in adipocytes from 10 nondiabetic subjects (age 34–80, average 60.7 years; BMI 21–35, average 24.9 kg/m2). Cells were incubated without additions (control, open bar), with 50 nmol/L rapamycin (filled bar) or with 50 nmol/L rapamycin and 5 μmol/L chloroquine (hatched bar) for 18 h and then the number of lipofuscin particles was determined by fluorescence microscopy. Microscopic image of one cell after the respective treatment is shown above the corresponding graph bar. In each condition 20 cells were analyzed per subject. Results are given as number of lipofuscin particles per cell as a percentage of that in controls, for each subject, mean ± SE. (C) Quantification of lipofuscin particles in adipocytes from 17 nondiabetic subjects (open bar; age 34–80, average 56.7 years; BMI 21–38.3, average 26.9 kg/m2) and 10 diabetic patients (filled bar; age 46–76, average 60.9 years; BMI 27–66.7, average 39.7 kg/m2); 20 cells were analyzed per patient. Results are given as number of lipofuscin particles per cell, mean ± SE.
Mitochondria and Mitochondrial Function in T2D
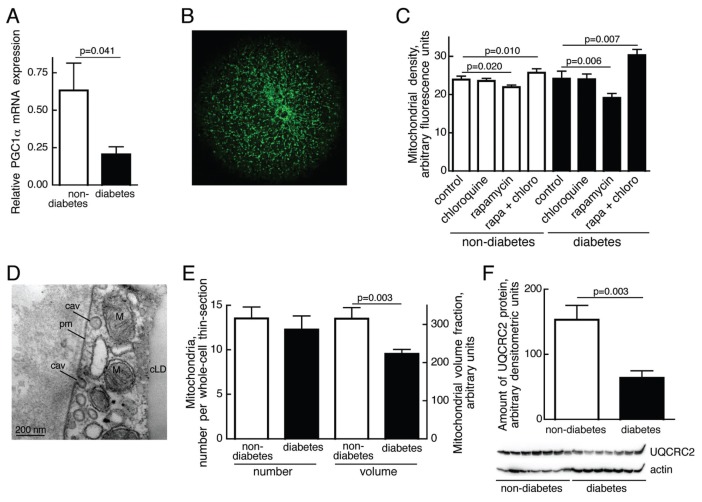
In addition to promoting protein synthesis via S6K1 and inhibiting autophagy, mTORC1 stimulates mitochondrial biogenesis (Figure 1A). In accordance with attenuated activation of mTORC1, we found that peroxisome proliferator-activated receptor-γ coactivator-1 (PGC1α) mRNA was down-regulated in adipocytes from patients with T2D compared with nondiabetic subjects (Figure 4A). We also quantified the amount of mitochondria (Figure 4B, C), determined as accumulation of Mitotracker Green (which accumulates in mitochondria independent of membrane potential and thus measures the content of mitochondria independent of function). Using this method of measurement we observed little difference in the amount of mitochondria between cells from patients with diabetes and the non-diabetic comparison group (Figure 4C). However, after stimulating autophagy by rapamycin-inhibition of mTORC1, the amount of mitochondria was reduced in nondiabetic and, more so, in diabetic adipocytes (Figure 4C). Inhibition of lysosomal digestion with chloroquine in rapamycin-treated cells restored mitochondrial volume to supranormal levels in the diabetic cells (Figure 4C), indicating increased autophagic turnover and increased potential for autophagy of mitochondria in diabetic adipocytes.
Figure 4.
Mitochondria in T2D. (A) Relative abundance of PGC1α mRNA. Total RNA was extracted from isolated adipocytes from eight nondiabetic subjects (open bars; age 22–73, average 48.9 years; BMI 20–32.9, average 25.3 kg/m2) and eight patients with T2D (filled bars; age 32–63, average 48.1 years; BMI 29–45.4, average 40.0 kg/m2). The amount of PGC1α mRNA was determined by RT-PCR. Results are given as amount of PGC1α mRNA in relation to GAPDH mRNA, mean ± SE. (B) Mitochondria constitute a tubular network in human adipocytes. Fluorescence microscopic image of an adipocyte incubated with Mitotracker Green. (C) Density of mitochondria determined as Mitotracker Green fluorescence. Adipocytes from eight nondiabetic (open bars; age 27–71, average 47 years; BMI 18.7–32.9, average 25.7 kg/m2) and eight diabetic subjects (filled bars; age 32–63, average 46.5 years; BMI 33.2–47.9, average 41.3 kg/m2) were incubated without additions (control), with 5 μmol/L chloroquine, with 50 nmol/L rapamycin, or with 50 nmol/L rapamycin and 5 μmol/L chloroquine for 18 h, and then total mitochondrial volume was determined by fluorescence microscopy. Twenty cells per subject and condition were analyzed. Results are given as fluorescence per imaged area of the cells, mean ± SE. (D) Mitochondria (M) in the cytosolic space between plasma membrane (pm) and central lipid droplet (cLD) visualized by transmission electron microscopy; cav, caveolae. (E) Morphometric analyses of number or volume fraction of mitochondria with developed cristae in adipocytes from five non-diabetic (open bars; age 49–76, average 63.6 years; BMI 26.7–34.9, average 29.1 kg/m2) and five diabetic (filled bars; age 32–63, average 46.6 years; BMI 37.2–45.4, average 40.9 kg/m2), determined by transmission electron microscopy of thin sections of five different whole cells from each subject. Results are given as number of mitochondria per whole-cell thin section, mean + SE; or as mitochondrial volume fraction (that is, area per whole-cell thin section) in arbitrary units, mean ± SE. (F) Amount of oxidative phosphorylation protein in adipocytes from seven nondiabetic (open bar; age 27–71, average 51.4 years; BMI 18.7–32.9, average 32.6 kg/m2) and seven diabetic (filled bar; age 32–80, average 51.9 years; BMI 27.5–47.9, average 38.8 kg/m2) subjects. Cells were subjected to SDS-PAGE and immunoblotting with antibodies against the electron transport chain protein ubiquinol-cytochrome c oxidoreductase (complex III, core protein 2, UQCRC2). The amount of UQCRC2 protein was normalized to the amount of actin in each sample. Results are given as arbitrary densitometric units, mean ± SE.
By using electron microscopy of adipocyte thin-sections (Figure 4D), we observed that the number of mitochondria with cristae was not different (Figure 4E). However, the volume fraction, determined from TEM, of mitochondria with cristae was substantially reduced in T2D cells (Figure 4E). Impaired mitochondrial function in the diabetic adipocytes was also demonstrated by a reduced amount of the nuclear-encoded mitochondrial inner membrane protein UQCRC2 (Figure 4F). The UQCRC2 gene contains a consensus cis-regulatory element (ATGGCG) for control by YY1-PGC1α (24) in the first intron. Taken together our findings indicate that the total amount of tubular mitochondria is not affected, whereas functional mitochondrial tubules with cristae are considerably reduced in the diabetic state, in accordance with little PGC1α (25) and attenuated mTORC1 activity.
Cytosolic Lipid Droplets in T2D
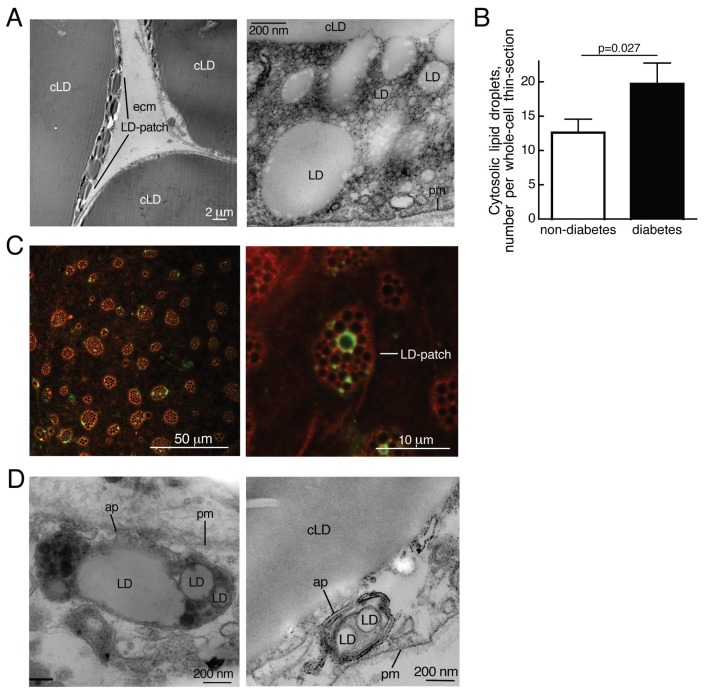
By using TEM we noted an increased number of cytosolic lipid droplets in diabetic compared with nondiabetic adipocytes (Figure 5A, B). A subpopulation of cytosolic lipid droplets was also associated with the autophagy marker protein LC3, which then colocalized with the lipid-droplet marker protein perilipin (Figure 5C). Most interestingly, the lipid droplets were found in patches by TEM (Figure 5A) and, more globally, by confocal fluorescence microscopy (Figure 5C). These patches may be areas where lipolysis takes place, because a high rate of lipolysis by triacylglycerol lipase will require the extended surface expansion provided by fragmentation of the large central lipid droplet, and autophagy may take part in the process. This notion was supported by the frequent occurrence of lipid-droplet–containing autophagosomes (Figure 5D).
Figure 5.
Increased number of lipid droplets in T2D. (A) Lipid droplets (LD) visualized by transmission electron microscopy. Left, parts of three cells are seen; in the leftmost cell there are small lipid droplets in the cytosol separate from the central lipid droplet (cLD). LD-patch, patch of cytosolic lipid droplets; pm, plasma membrane; ecm, extracellular matrix. (B) Amount of cytosolic lipid droplets (>100 nm) in adipocytes from five nondiabetic subjects (open bar; age 49–76, average 63.6 years; BMI 26.7–34.9, average 29.1 kg/m2) and five diabetic patients (filled bar; age 32–63, average 46.6 years; BMI 37.2–45.4, average 40.9 kg/m2) determined by transmission electron microscopy of thin sections of five different whole cells from each subject, as described in Experimental Procedures. Results are given as number of cytosolic droplets per whole-cell thin section, mean ± SE. (C) Autophagy of lipid droplets visualized by immunofluorescence confocal microscopy. Adipocytes were incubated with 50 nmol/L rapamycin and 5 μmol/L chloroquine for 18 h, when cells were incubated with antibodies against perilipin (red) and LC3 (green). Confocal section close to the plasma membrane shown. LD-patch, patch of cytosolic lipid droplets. (D) Autophagy of lipid droplets visualized by transmission electron microscopy; ap, autophagosome with lipid droplets; LD, lipid droplets; cLD, central lipid droplet; pm, plasma membrane.
Insulin Control of mTORC1 in a Cohort of Nondiabetic Subjects
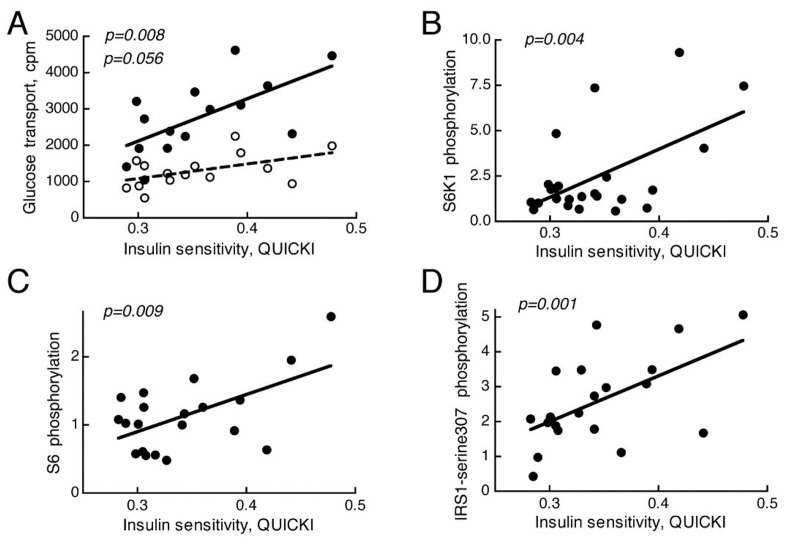
To validate our findings we examined mTORC1 activation by insulin in a larger cohort of 26 nondiabetic subjects (Supplemental Table S1). We found that, similar to the effects of insulin on glucose uptake (Figure 6A), the effects of insulin on phosphorylation of the mTORC1 substrate S6K1 (Figure 6B) correlated with whole-body insulin sensitivity over a very wide range of insulin sensitivity/resistance. This relationship was further mirrored by the phosphorylation of the S6K1 substrate ribosomal protein S6 (Figure 6C). The findings indicate that activation of mTORC1 in the adipose tissue by insulin is dependent on a subject’s insulin resistance rather than a T2D diagnosis. We have previously demonstrated that the positive feedback in insulin signaling, to phosphorylation of IRS1 at serine 307 (human sequence, corresponding to murine serine 302), is attenuated in adipocytes from patients with T2D (5,8). Inhibition of the feedback signal by rapamycin indicates the involvement of mTORC1 in the feedback loop (5,9), and it has been suggested that phosphorylation at serine 307 serves to integrate nutrient availability with insulin signaling (9). Similar to the activity of mTORC1, the ability of insulin to stimulate the feedback phosphorylation at serine 307 of IRS1 in adipocytes was directly related to the insulin sensitivity of the subjects donating the cells (Figure 6D).
Figure 6.
Correlation of insulin-stimulated mTORC1 activity or glucose transport to whole body insulin sensitivity (QUICKI). Adipocytes from 26 nondiabetic subjects (Supplemental Table S1) were examined. (A) Glucose transport into isolated adipocytes was determined, in triplicate, as uptake of 2-deoxy-D-glucose with (filled circles) or without (open circles) preincubation with 10 nmol/L insulin for 15 min. (B–D) The effect of insulin on the phosphorylation of S6K1 (B), S6 (C), and IRS1 at serine-307 (D) were determined in isolated adipocytes after incubation with or without 10 nmol/L insulin for 10 min, when cells were subjected to SDS-PAGE and immunoblotting with antibodies against phospho-threonine(389)S6K1 (B), phosphoserine(235/236)S6 (C), or phosphoserine(307, human sequence)IRS1 (D). Results are given as fold over of controls without insulin, mean of triplicates.
Effects of Mitochondrial Inhibition on mTORC1
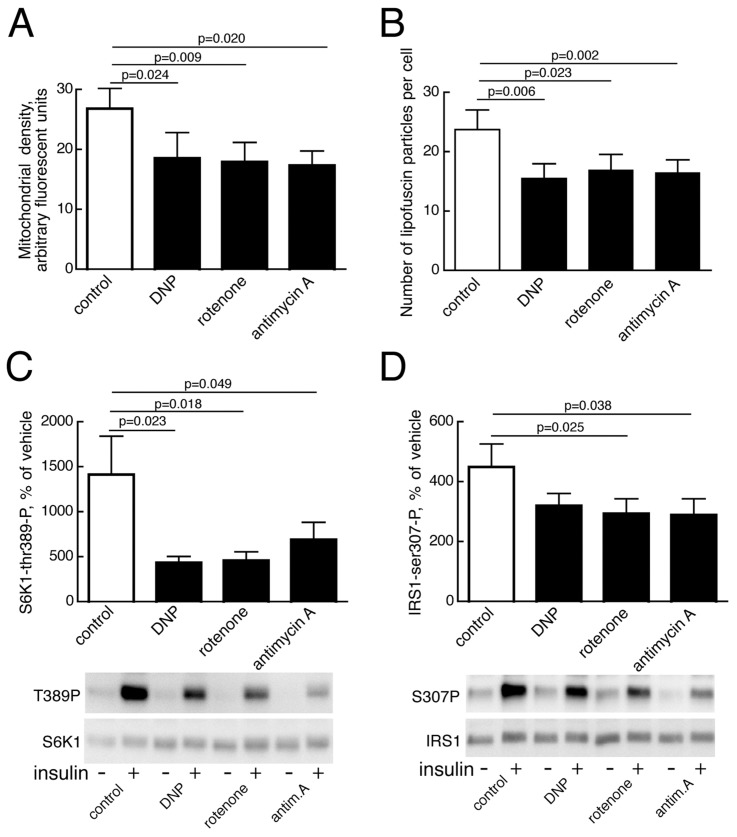
The etiology of the attenuated mTORC1 activity remains elusive, but defects in mitochondria and reduced ATP production are known to induce autophagy, perhaps via AMP-kinase inhibition of mTORC1 (26,27). We found that inhibition of mitochondrial function, by low concentrations of inhibitors specific for different stages of mitochondrial oxidative phosphorylation, significantly reduced the total volume of mitochondria in adipocytes from nondiabetic subjects (Figure 7A). The number of lipofuscin particles was also significantly reduced after incubation with these mitochondrial inhibitors (Figure 7B), indicating that mitochondrial dysfunction activates general autophagy, as well as mitophagy. The active autophagy shows that the inhibitors were not toxic to the cells. Mitochondria inhibitors also significantly inhibited mTORC1 activation by insulin (Figure 7C). This finding was further reflected in a reduced insulin feedback signal from mTORC1 to phosphorylation of IRS1 at serine 307 (human sequence) (Figure 7D).
Figure 7.
Effects of inhibition of mitochondrial function on mitophagy, autophagy and mTORC1 activity. (A) Density of mitochondria determined as Mitotracker fluorescence after inhibition of mitochondrial function. Adipocytes from six nondiabetic subjects (age 46–71, average 60.8 years; BMI 21.5–31.6, average 26.7 kg/m2) were incubated with low concentration of dinitrophenol (50 μmol/L, DNP), rotenone (1 μmol/L), antimycin A (10 nmol/L), or vehicle (0.1 % ethanol, control) for 18 h, then mitochondrial density was determined by fluorescence microscopy. 20 cells per subject were analyzed. Results are given as mitochondria density in arbitrary fluorescence units, mean ± SE. (B) Number of lipofuscin particles per cell was determined after inhibition of mitochondrial function, as in (A), in adipocytes from five nondiabetic subjects (age 42–71, average 54.2 years; BMI 22.2–27.2, average 24.0 kg/m2). 20 cells per subject were analyzed. Results are given as number of lipofuscin particles per cell, mean ± SE. (C) Insulin-stimulated activation of mTORC1, determined as phosphorylation of S6K1, was assayed after incubating adipocytes from 11 nondiabetic subjects (age 35–82, average 56.0 years; BMI 22.2–31.6, average 26.2 kg/m2) with mitochondrial inhibitors, as in (A). Cells were then incubated without (−) or with (+) 10 nmol/L insulin for 10 min, when cells were subjected to SDS-PAGE and immunoblotting with antibodies against phospho-threonine(389)S6K1. Results are given as percent of without insulin, mean ± SE. The immunoblots of one experiment representing one subject are shown. (D) Insulin-stimulated phosphorylation of IRS1 at serine 307 (human sequence) was analyzed after incubating adipocytes from 12 nondiabetic subjects (age 35–82, average 53.9 years; BMI 22.2–31.6, average 26.5 kg/m2) with mitochondrial inhibitors, as in (A). Cells were then incubated without (−) or with (+) 10 nmol/L insulin for 10 min, when cells were subjected to SDS-PAGE and immunoblotting with antibodies against phosphoserine(307, human sequence)IRS1. Results are given as percent of without insulin, mean ± SE. The immunoblots of one experiment representing one subject are shown.
DISCUSSION
mTORC1
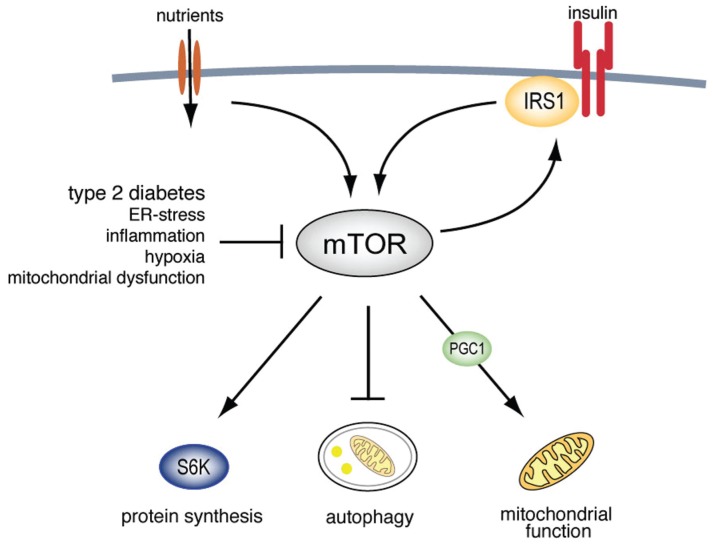
Our conclusion that mTORC1 activation by insulin is attenuated in adipocytes from obese patients with T2D is based on direct measurements of the major functions of mTORC1 signaling activity (Figure 8): (a) Attenuated effects of insulin on the state of phosphorylation of the major mTORC1 substrate S6K1, as well as (b) on two downstream substrates of S6K1: the ribosomal protein S6 and IRS1 at serine 307 (human sequence); (c) increased autophagic activity (see below); and (d) impaired mitochondria (see below). We, moreover, validated the attenuation of the mTORC1 pathway in a cohort of nondiabetic subjects exhibiting a wide range of insulin sensitivities, which indicates that attenuated mTORC1 is a property of the insulin-resistant state and does not require full-blown T2D with impaired function of the insulin-producing β cells. We further describe the mechanisms causing attenuation of mTORC1 below.
Figure 8.
Schematic overview of mTOR involvement in insulin signaling and dysregulation in T2D.
Others have reported that excess nutrients and other manipulations that cause overactivation of mTORC1-S6K1 induce insulin resistance (6,28–30). These studies have usually related hyperactive mTORC1-S6K1 to increased phosphorylation of IRS1 at serine, in particular at serine 312 (human sequence, corresponding to murine serine 307), which has been attributed to an inhibition of IRS1 signaling. The inhibitory effect of phosphorylation of IRS1 at serine 312 has, however, recently been refuted. Knock-in of IRS1 serine312alanine mutant in mice demonstrated a negative, rather than a positive, effect on IRS1 signaling, which aggravates the insulin resistance of high-fat feeding (31). Hence it appears that insulin feedback signaling to phosphorylation of IRS1 at serines 307 (5,9,11), 312 (31), and 323 (11) (human sequence), all requiring mTORC1 activity, sustain positive-feedback signals. As a consequence, hypoactive mTORC1 should be associated with impaired insulin signaling and insulin resistance, in accordance with our findings here. A dependence on mTORC1 for insulin signaling in vivo in human subjects has been demonstrated by the treatment with rapamycin, which is linked to inhibition of IRS1 and induction of insulin resistance (32,33).
mTORC1 and Autophagy
A direct consequence of attenuation of the mTORC1 pathway in insulin resistance, and an important aspect of insulin resistance in adipocytes, is enhanced autophagy. We demonstrated enhanced autophagy in the adipocytes from obese patients with T2D along several different lines of evidence: (a) Electron microscopic identification of an increased number of autophagosomes; (b) increased turnover and particle association of the autophagy protein LC3, which indicates an increased flow through the autophagosomal system; (c) a partial dependence on autophagy for production of ATP; (d) reduced density of lipofuscin particles, apparently by autophagosomal degradation; (e) reduced mitochondrial volume and increased potential for autophagy of mitochondria. It is interesting that endoplasmic-reticulum stress, induced by thapsigargin, decreased the amount of insulin receptor in 3T3-L1 adipocytes, similar to the situation in T2D (7), and that this was reversed by inhibition of autophagy with 3-methyladenine (34). These authors also reported that the LC3-II/LC3-I ratio was increased in the adipose tissue of db/db mice, indicating increased autophagic activity. Increased autophagy has recently also been demonstrated in the insulin-producing β cells of db/db mice as well as after high-fat feeding of mice (35).
A compromised differentiation of adipocytes in autophagy-deficient mice (36) as well as in adipose-specific autophagy-deficient mice (37) has precluded assessment of the role of autophagy in fatty-acid release in those animals. In rat hepatocytes, however, components of the autophagic pathway were recently shown to associate with lipid droplets and to control lipolysis (38). It is possible that the perceived starvation in adipocytes from patients with T2D increases autophagy to enhance hydrolysis of stored triacylglycerol, thus also contributing to the elevation of fatty acids in blood that is characteristic of T2D. Indeed, adipocytes from T2D patients exhibited (a) an increased number of cytoplasmic lipid droplets, consistent with fragmentation of large lipid droplets during lipolysis in 3T3-L1 adipocytes (39,40); (b) lipid-droplet–containing autophagosomes; and (c) in a subpopulation of cytoplasmic lipid droplets, LC3 colocalized with perilipin. It will be interesting to examine to what extent autophagy contributes to the systemic release of fatty acids from the adipose tissue, in healthy subjects and in patients with diabetes.
An interesting corollary to enhanced autophagy was our finding that cellular lipofuscin particles can be degraded after stimulating autophagy. Lipofuscin has been identified as nondegradable remnants of lysosomal digestion, which accumulate over the lifespan of an organism to eventually cause cell death and set a limit to the survival of the organism (reviewed in [41,42]). Our finding that stimulation of autophagy can dispose of the lipofuscin could have direct implications for our understanding of how lysosomal storage diseases associated with cellular deposits of waste products or misfolded proteins can be treated. Furthermore, by removing misfolded proteins (43), enhanced autophagy may, in the insulin-resistant diabetic state, have a protective function to alleviate endoplasmic reticulum stress. In contrast to the reduced levels of lipofuscin in the insulin-resistant adipocytes, it has been reported that destruction of the insulin-producing β cells in rats by streptozotocin increases the amount of lipofuscin in erythrocytes (44) and neurons (45).
mTORC1 and Mitochondria
There is much evidence for reduced mitochondrial function and PGC1α in obesity and T2D (46–54). In accordance with this evidence we found mTOR activity, PGC1α, the electron transport protein UQCRC2, and the volume of mitochondria with cristae to be coordinately downregulated in diabetic adipocytes.
Inhibition of mTORC1, and thus stimulation of autophagy, directly impacted the amount of mitochondria in human adipocytes, especially in the diabetic cells. Inversely, inhibition of mitochondrial function stimulated autophagy, apparently through inhibition of mTORC1. Reciprocal control between mTORC1 and mitochondria would serve a critical function to coordinate mitochondrial activity and protein synthesis and growth. On one hand, mTOR controls mitochondrial oxidative function through the transcription factor PGC1α (24), thereby determining the balance between mitochondrial and nonmitochondrial production of ATP (55). On the other hand, mTOR has been found localized to mitochondria (55,56), and was sensitive to perturbation of mitochondrial function (see for example [55,57]). In this context it should be noted that adipocytes from subjects with T2D release more lactate (58), consistent with the reduced activity of mTORC1 and its role in aerobic ATP production. The central role of PGC1α as a mediator between mTOR and the mitochondria has been directly demonstrated by insulin resistance causing mitochondrial dysfunction in C2C12 cells, whereas overexpression of PGC1α rescues insulin signaling and mitochondrial function (59). Knockout of raptor in mice has, however, provided conflicting results. With the expected decrease in oxidative capacity and mitochondrial gene expression in muscle-specific raptor knockout mice (60), while adipocyte-specific raptor knockout mice exhibited induction of UCP1 expression and characteristics of brown adipocytes (61). mTORC1 is required for proper differentiation of adipocytes, and transdifferentiation of white adipocytes to brown adipocytes makes it difficult to draw any conclusions from these knockout mice regarding the normal function of mTORC1 and autophagy in fully differentiated adipocytes. Although knockout of either raptor (60) or rictor (62) in skeletal muscle generates mice with impaired glucose tolerance, knockout of mTOR does not affect glucose tolerance (63), further attesting to the difficulty of predicting and interpreting findings from these animals. However, although differentiation of adipocytes was impaired in adipose-specific autophagy-deficient animals, UCP1 was not induced (37). The animals were nevertheless resistant to high-fat-diet–induced obesity and were also more insulin sensitive.
mTORC1 and the Insulin-Resistant State
To conclude, adipocytes from patients with T2D exhibit hypoactive mTORC1 signaling, increased autophagic activity and impaired mitochondrial function, which is consistent with an attenuated positive-feedback loop to IRS1 (Figure 8). Our findings enforce the notion of mTORC1 as a central integrator of insulin signaling and cellular energy metabolism in human adipocytes. On one hand, exogenous substrate availability through insulin signals an anabolic state and activates mTORC1 to stimulate protein synthesis and aerobic ATP production. On the other hand, shortage of cellular substrate or dysfunctional oxidative phosphorylation inhibits mTORC1, which inhibits insulin signaling through reduced positive feedback to phosphorylation of IRS1 at serine 307 (human sequence).
The role of TOR in coordinating nutrient and insulin/growth-factor signaling is demonstrated by the finding that either insulin or nutrient availability determines the size of the Drosophila fly. Low dTOR in the fat body of Drosophila induces a state of pseudo-starvation and cellular dependence on autophagy, in spite of the availability of plenty of extra-cellular nutrients, with attenuated growth-factor signaling and systemic release of growth inhibitory signals (64). In human adipose tissue, by analogy, the impaired insulin signaling and mTORC1 activity in T2D set the adipocyte in a state of starvation and growth inhibition, which may induce the release of systemic insulin-resistance–inducing adipokines (65). Interestingly, investigators recently found that the Drosophila fat body releases NLaz to systemically inhibit insulin signaling (66). The human ortholog to NLaz is RBP4, which is released from human adipose tissue and can induce insulin resistance (8,67).
Cause or Consequence?
Although our findings add novel and crucial information to enhance understanding of the defect mechanisms behind insulin resistance in T2D, the origin of the defect remains to be identified, if one exists. It is indeed possible that causes vary from individual to individual, but owing to the interrelatedness of the cellular functions affected, consequences will be identical. Mitochondrial dysfunction, reactive oxygen species production/oxidative stress, endoplasmic-reticulum stress, genotoxic stress, hypoxia, cellular energy shortage, and inflammation can all impinge on mTOR, IRS1 phosphorylation, insulin signaling, mitochondria and autophagy (Figure 8).
Supplemental Data
ACKNOWLEDGMENTS
We thank Östergötland County Council, Novo Nordisk Foundation, Swedish Diabetes Association, and Swedish Research Council for financial support. The authors have no competing financial interests. We are grateful to BA Fredriksson for help with thin-sectioning of cells.
Footnotes
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Online address: http://www.molmed.org
REFERENCES
- 1.Kim Y-B, Nikoulina SE, Ciaraldi TP, Henry RR, Kahn BB. Normal insulin-dependent activation of Akt/protein kinase B, with diminished activation of phosphoinositide 3-kinase, in muscle in type 2 diabetes. J. Clin. Inv. 1999;104:733–41. doi: 10.1172/JCI6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodyear L, et al. Insulin receptor phosphorylation, insulin receptor substrate-1 phosphorylation and phosphatidylinositol 3-kinase activity are decreased in intact skeletal muscle strips from obese subjects. J. Clin. Inv. 1995;95:2195–204. doi: 10.1172/JCI117909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cusi K, et al. Insulin resistance differentially affects the PI 3-kinase and MAP kinase-mediated signaling in human muscle. J. Clin. Inv. 2000;105:311–20. doi: 10.1172/JCI7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björnholm M, Kawano Y, Lehtihet M, Zierath J. Insulin receptor substrate-1 phosphorylation and phosphatidylinositol 3-kinase acitivity in skeletal muscle from NIDDM subjects after in vivo insulin stimulation. Diabetes. 1997;46:524–7. doi: 10.2337/diab.46.3.524. [DOI] [PubMed] [Google Scholar]
- 5.Danielsson A, Öst A, Nystrom FH, Strålfors P. Attenuation of insulin-stimulated insulin receptor substrate-1 serine 307 phosphorylation in insulin resistance of type 2 diabetes. J. Biol. Chem. 2005;280:34389–92. doi: 10.1074/jbc.C500230200. [DOI] [PubMed] [Google Scholar]
- 6.Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Danielsson A, et al. Short-term over-eating induces insulin resistance in fat cells in lean human subjects. Mol. Med. 2009;15:228–34. doi: 10.2119/molmed.2009.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Öst A, et al. Retinol-binding protein-4 attenuates insulin-induced phosphorylation of IRS1 and ERK1/2 in primary human adipocytes. FASEB J. 2007;21:3696–704. doi: 10.1096/fj.07-8173com. [DOI] [PubMed] [Google Scholar]
- 9.Giraud J, Leshan R, Lee Y-H, White MF. Nutrient-dependent and insulin-stimulated phosphorylation of insulin receptor substrate-1 on serine 302 correlates with increased insulin signaling. J. Biol. Chem. 2004;279:3447–54. doi: 10.1074/jbc.M308631200. [DOI] [PubMed] [Google Scholar]
- 10.Danielsson A, Nystrom FH, Strålfors P. Phosphorylation of IRS1 at serine 307 and serine 312 in response to insulin in human adipocytes. Biochem. Biophys. Res. Commun. 2006;342:1183–7. doi: 10.1016/j.bbrc.2006.02.075. [DOI] [PubMed] [Google Scholar]
- 11.Weigert C, et al. Interplay and effects of temporal changes in the phosphorylation state of serine-302, -307, and 318 of insulin receptor substrate-1 on insulin action in skeletal muscle cells. Mol. Endocrinol. 2008;22:2729–40. doi: 10.1210/me.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrington LS, et al. The TSC1–2 tumor suppressor controls insulin–PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–23. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paz K, et al. Phosphorylation of insulin receptor substrate-1 (IRS-1) by protein kinase B positively regulates IRS-1 function. J. Biol. Chem. 1999;274:28816–22. doi: 10.1074/jbc.274.40.28816. [DOI] [PubMed] [Google Scholar]
- 14.Wullschleger S, Loewith R, Hail MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Katz A, et al. Quantitative Insulin Sensitivity Check Index: a simple, accurate method for assessing insulin sensitivity in humans. J. Clin. Endocrinol. Metab. 2000;85:2402–10. doi: 10.1210/jcem.85.7.6661. [DOI] [PubMed] [Google Scholar]
- 16.Strålfors P, Honnor RC. Insulin-induced dephosphorylation of hormone-sensitive lipase: correlation with lipolysis and cAMP-dependent protein kinase activity. Eur. J. Biochem. 1989;182:379–85. doi: 10.1111/j.1432-1033.1989.tb14842.x. [DOI] [PubMed] [Google Scholar]
- 17.Danielsson A, et al. Insulin resistance in human adipocytes downstream of IRS1 after surgical cell isolation, but at the level of phosphorylation of IRS1 in type 2 diabetes. FEBS J. 2005;272:141–51. doi: 10.1111/j.1432-1033.2004.04396.x. [DOI] [PubMed] [Google Scholar]
- 18.Stefanini M, deMartino C, Zamboni L. Fixation of ejaculated spermatozoa for electron microscopy. Nature. 1967;216:173–174. doi: 10.1038/216173a0. [DOI] [PubMed] [Google Scholar]
- 19.Frost SC, Kohanski RA, Lane MD. Effect of phenylarsine oxide on insulin-dependent protein phosphorylation and glucose transport in 3T3-L1 adipocytes. J. Biol. Chem. 1987;262:9872–9876. [PubMed] [Google Scholar]
- 20.Kanazawa T, et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J. Biol. Chem. 2004;279:8452–8459. doi: 10.1074/jbc.M306337200. [DOI] [PubMed] [Google Scholar]
- 21.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroikin Y, Dalen H, Lööf S, Terman A. Inhibition of autophagy with 3-methyladenine results in impaired turnover of lysosomes and accumulation of lipofuscin-like material. Eur. J. Cell Biol. 2004;83:583–590. doi: 10.1078/0171-9335-00433. [DOI] [PubMed] [Google Scholar]
- 24.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC1alpha transcriptional complex. Nature. 2007;450:736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 25.Vercauteren K, Gleyzer N, Scarpulla RC. Short hairpin RNA-mediate silencing of PRC (PGC-1-related coactivator) results in a severe respiratory chain deficiency associated with the proliferation of aberrant mitochondria. J. Biol. Chem. 2009;284:2307–19. doi: 10.1074/jbc.M806434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meley D, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 2006;281:34870–9. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 28.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–5. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 29.Tremblay F, et al. Overactivation of S6 kinase 1 as a cause of human insulin resistance during increase amino acid availabiltiy. Diabetes. 2005;54:2674–84. doi: 10.2337/diabetes.54.9.2674. [DOI] [PubMed] [Google Scholar]
- 30.Tremblay F, Gagnon AM, Veilleux A, Sorisky A, Marette A. Activation of the mammalian Target of rapamycin pathway acutely inhibits insulin signaling to Akt and glucose transport in 3T3-L1 and human adipocytes. Endocrinology. 2005;146:1328–37. doi: 10.1210/en.2004-0777. [DOI] [PubMed] [Google Scholar]
- 31.Copps KD, et al. Irs1 serine 307 promotes insulin sensitivity in mice. Cell Metab. 2009;11:84–92. doi: 10.1016/j.cmet.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiPaolo S, Teutonico A, Leogrande D, Capobianco C, Schena PF. Chronic inhibition of mammalian target of rapamycin signaling downregulates insulin receptor substrates 1 and 2 and AKT activation: a crossroad between cancer and diabetes? J. Am. Soc. Nephrol. 2006;17:2236–44. doi: 10.1681/ASN.2006030196. [DOI] [PubMed] [Google Scholar]
- 33.Teutonico A, Schena PF, Paolo SD. Glucose metabolism in renal transplant recipients: effect of calcineurin inhibitor withdrawal and conversion to sirolimus. J. Am. Soc. Nephrol. 2005;16:3128–35. doi: 10.1681/ASN.2005050487. [DOI] [PubMed] [Google Scholar]
- 34.Zhou L, et al. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol. Pharmacol. 2009;76:596–603. doi: 10.1124/mol.109.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increases of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–32. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Singh R, et al. Autophagy regulates adipose tissue mass and differentiation. J. Clin. Inv. 2009;119:3329–39. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19860–5. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 2005;280:42325–35. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 40.Londos C, et al. On the control of lipolysis in adipcytes. Ann. N. Y. Acad. Sci. 1999;892:155–68. doi: 10.1111/j.1749-6632.1999.tb07794.x. [DOI] [PubMed] [Google Scholar]
- 41.Terman A, Gustafsson G, Brunk UT. Autophagy, organelles and ageing. J. Pathol. 2007;211:134–43. doi: 10.1002/path.2094. [DOI] [PubMed] [Google Scholar]
- 42.Gray DA, Woulfe J. Lipofuscin and aging: a matter of toxic waste. Sci. Aging Knowledge Environ. 2005;2005:1–5. doi: 10.1126/sageke.2005.5.re1. [DOI] [PubMed] [Google Scholar]
- 43.Gamerdinger M, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain SK, Levine SN, Duett J, Hollier B. Reduced vitamin E and increased lipofuscin products in erythrocytes of diabetic rats. Diabetes. 1991;40:1241–4. doi: 10.2337/diab.40.10.1241. [DOI] [PubMed] [Google Scholar]
- 45.Sugaya A, Sugimioto H, Mogi N, Tsujigami H, Deguchi S. Experimental diabetes accelerates accumulation of fluorescent pigments in rat trigeminal neurons. Brain Res. 2004;999:132–4. doi: 10.1016/j.brainres.2003.11.033. [DOI] [PubMed] [Google Scholar]
- 46.Morino K, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J. Clin. Inv. 2005;115:3587–93. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 2004;350:664–71. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietiläinen KH, et al. Global transcript profiles of fat monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 51.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–50. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 52.Krook A, Digby J, O’Rahilly S, Zierath JR, Wallberg-Henriksson H. Uncoupling protein 3 is reduced in skeletal muscle of NIDDM patients. Diabetes. 1998;47:1528–31. doi: 10.2337/diabetes.47.9.1528. [DOI] [PubMed] [Google Scholar]
- 53.Stump CS, Short KR, Bigelow ML, Schimke JM, Nair KS. Effect of insulin on human skeletal muscle mitochondrial ATP production, protein synthesis, and mRNA transcripts. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7996–8001. doi: 10.1073/pnas.1332551100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semple RK, et al. Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. Int. J. Obesity. 2004;28:176–9. doi: 10.1038/sj.ijo.0802482. [DOI] [PubMed] [Google Scholar]
- 55.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006;281:27643–52. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 56.Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin-associated protein associates with mitochondria and senses omsotic stress via mitochondrial dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4319–24. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D-H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 58.Digirolamo M, Newby FD, Lovejoy J. Lactate production in adipose tissue: a regulated function with extra-adipose implications. FASEB J. 1992;6:2405–12. doi: 10.1096/fasebj.6.7.1563593. [DOI] [PubMed] [Google Scholar]
- 59.Pagel-Langenickel I, et al. PGC-1alpha integrates insulin signaling, mitochondrial regulation, and bioenergetic function in skeletal muscle. J. Biol. Chem. 2008;283:22464–72. doi: 10.1074/jbc.M800842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–24. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Kumar A, et al. Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances basal glycogen synthase activity. Mol. Cell. Biol. 2008;28:61–70. doi: 10.1128/MCB.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Risson V, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 2009;187:859–74. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colombani J, et al. A nutrient sensor mechanism controls drosophila growth. Cell. 2003;114:739–49. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 65.Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J. Clin. Inv. 2006;116:1767–75. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hull-Thopson J, et al. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Gen. 2009;5:e1000460. doi: 10.1371/journal.pgen.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Q, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–62. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.