Abstract
Hereditary colorectal cancer develops through a series of well-defined genetic and histological changes. However, elucidation of the canonical pathway based on hereditary colorectal cancer has not provided a clear explanation of the molecular mechanisms of sporadic colorectal cancer. To identify the alterative pathways involved in sporadic colorectal tumorigenesis, we performed gene expression analysis in patients with sporadic colorectal tumors. A comparison analysis of gene expression profiles revealed a pattern of upregulation of small proline rich repeat protein 3 (SPRR3) in tumor samples. SPRR3 has previously been reported to be downregulated in esophageal cancer. However, in the present study, we observed that SPRR3 was strongly upregulated in 31 of 35 samples of sporadic colorectal tumors (88%). We also determined that overexpression of SPRR3 not only accelerates colorectal cancer cell proliferation but also is associated with lymphovascular invasion in colorectal cancer. Moreover, AKT was activated and p53 levels were decreased in cells that overexpressed SPRR3. In contrast to the pattern seen in esophageal cancer, these results suggest that increased expression of SPRR3 is involved in colorectal tumorigenesis.
INTRODUCTION
The occurrence of colorectal cancer has gradually increased over the last decade, and colorectal carcinoma was the third most common cancer in Korea in 2005 (1). Although several oncogenes and tumor-suppressor genes are known to be involved in the progression of hereditary colorectal cancer, the molecular changes associated with sporadic colorectal cancer are not yet understood (2–4). Genome-wide gene expression analysis is the method most widely used to investigate how cancer-related genes are altered during tumorigenesis (5–7). We previously identified several genes associated with molecular changes in sporadic colorectal cancers by microarray analysis of the gene expression profiles of sporadic colorectal cancer patients. When we investigated these genes, we found that SPRR3 (small proline rich repeat protein 3) was a protein associated with the molecular changes and clinicopathological features of this disease (8). SPRR proteins are known to be markers for terminal squamous cell differentiation, but they also function in nonsquamous tissues (9). For example, SPRR1A is induced by and is protective against ischemic stress in cardiomyocytes (10). It has recently been reported that SPRR3 is upregulated in celiac disease and also by prolonged exposure to cyclic strain (11,12). However, SPRR3 is frequently downregulated in esophageal squamous cell carcinoma (ESCC) and it has been shown to suppress the tumorigenicity of ESCC cells (13–15).
AKT has emerged as a key molecule mediating multiple signaling pathways. AKT was discovered as an oncogene and is involved in metabolic functions, cell survival and the cell cycle (16). Recent studies have shown that AKT activates Mdm2 to accelerate p53 ubiquitination (17–19).
In this study, we showed that SPRR3 is dramatically upregulated in colorectal cancer. The ectopic expression of SPRR3 accelerates cell proliferation through AKT activation in SW480 colorectal cancer cells. Moreover, overexpressed SPRR3 is associated with lymphovascular invasion in colorectal cancer. Our results suggest that SPRR3 is highly expressed in colorectal tumors and may potentiate tumorigenesis in colorectal cancer.
MATERIALS AND METHODS
Patients, Tissue Samples and Cell Lines
For the initial screening of SPRR3 expression, 35 colorectal cancer tissue samples were randomly chosen from the archival samples of the Asan Medical Center (Seoul, Korea). For the clinical validation, a total of 128 sporadic colorectal cancer patients, including 123 patients who underwent curative surgery (R0 resection) and 5 who underwent palliative surgery, were consecutively recruited (Table 1). Patients with hereditary non-polyposis colorectal cancers or familial adenomatous polyposis and patients who had received preoperative chemoradiation therapy were excluded. Primary colorectal adenocarcinoma tissue and the adjacent normal colonic mucosa tissue (at least 10 cm from tumor) were obtained during surgery; ESCC tissue and normal esophageal epithelium samples were obtained from the Biological Resources Center (Department of Pathology at the Asan Medical Center, Seoul, Korea). All patients provided written informed consent, and the study protocol was approved by the Institutional Review Board for Human Genetic and Genomic Research, in accordance with the Declaration of Helsinki. FHC, a colorectal cell line, was obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). CCD841 cells were kindly provided by SY Rha (Yonsei University, Seoul, Korea). The AMC5 cell line was established in our laboratory (20). All other colorectal cancer cells were obtained from ATCC, and cultured at 37°C in a 5% CO2 incubator and maintained as recommended by ATCC in RPMI1640, minimal essential medium, or Dulbecco’s modified Eagle’s medium, containing 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA).
Table 1.
Baseline demographic and biological features of 128 colorectal cancer patients.
| Features | No. of patients (missing values) |
|---|---|
| Sex, male/female | 64/64 |
| Age, years, mean ± SD | 59 ± 12 |
| Stage,a I/II/III/IV | 11/42/55/20 |
| Preoperative serum carcinoembryonic antigen, ≤6/>6 ng/mL | 89/39 |
| Tumor characteristics | |
| Site,b right colon/left colon/rectum | 25/41/62 |
| Long diameter, ≤4/>4 cm | 38/90 |
| Growth pattern, expanding/infiltrative | 109/19 |
| Differentiation, well and moderately differentiated/poorly differentiated and mucinous | 115/13 |
| Lymphovascular invasion, no/yes | 67/61 |
| Either MLH1 or MSH2 expression, no/yes | 9/84 (35) |
Cancer staging according to the American Joint Committee on Cancer (sixth edition, 2001; http://www.cancerstaging.org).
Right colon, cecum–splenic flexure of the transverse colon; left colon, splenic flexure of the transverse colon–sigmoid colon.
Cloning and Transfection
The full-length cDNA encoding SPRR3 was generously provided by M-R Wang (Peking Union Medical College, Beijing, China). SPRR3 cDNA was amplified by polymerase chain reaction and subcloned into hemagglutinin (HA)-tagged pcDNA3 (pHA-SPRR3) (Invitrogen) for the generation of stable transfectants. SW480 cells were transfected with pHA-SPRR3 using lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). After selection with G418 (1 mg/mL) for 10 d, stable expression of SPRR3 (SW480/SPRR3) was confirmed by Western blot analysis with anti-HA antibody. A control cell line was established by transfecting an empty expression vector, pcDNA3.1 (Invitrogen). The SPRR3 small interfering RNA (siRNA) (5′-CAGACAAGCCCUUGAGAA-3′) and nontargeting scrambled siRNA (5′-CCUACGCCACCAAUUUCGU-3′) were synthesized from the Bioneer (Daejeon, Korea) and transfected into cells with lipofectamine 2000 (Invitrogen). Knock-down efficiency was evaluated by Western blot analysis with anti-SPRR3 antibody.
Western Blot Analysis
All tissues and cells were extracted in 2 × Laemmli sample buffer (62.5 mmol/L Tris-HCl, 25% glycerol, 2% sodium dodecyl sulfate (SDS), 5% 2-mercaptoethanol, 0.01% Bromophenol Blue) (BioRad, Hercules, CA, USA), separated by SDS–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes. After blocking with skim milk in Tris-buffered saline Tween-20 (10 mmol/L Tris-HCl, 0.1 mol/L NaCl2, 0.1% Tween-20, pH 7.4), membranes were incubated with specific primary antibodies (anti-SPRR3 antibody was from Proteintech Group [Chicago, IL, USA]; anti-AKT, anti–phospho-AKT, anti-ERK, anti–phospho-ERK, anti–phospho-p38 and anti-p53 antibodies were from Cell Signaling Technology [Beverly, MA, USA]; anti-Actin antibody was from Chemicon International [Temecula, CA, USA]; anti-HA antibody was from Santa Cruz Biotechnology [Santa Cruz, CA, USA]; anti-mdm2 and anti–phospho-mdm2 antibodies were from BD [San Jose, CA, USA] and Calbiochem [La Jolla, CA, USA]). Then the membranes were incubated with horse radish peroxidase–conjugated secondary antibodies (Pierce, Rockford, IL, USA).
Immunohistochemistry
Paraffin-embedded tissue cores obtained from carcinomas were used to construct tissue microarrays and arrayed onto recipient paraffin blocks by using a precision instrument (Beecher Instruments, Sun Prairie, WI, USA). Tissue array blocks containing core cylinders were subjected to immune staining based on the labeled streptavidin–biotin method with a DAKO LSAB® kit (DAKO, Carpinteria, CA, USA), by using monoclonal antibodies against SPRR3 (Protein-tech Group, Chicago, IL, USA), MLH1 and MSH2 (G168-15 and G219-1129; BD PharMingen, San Diego, CA, USA). The SPRR3 cytoplasmic expression was classifed into two categories depending on the degree. Category I was formed by two grades, negative expression at ≤10% and positive expression at >10% staining cells; category II included two grades, downregulation at ≤50% and overexpression at >50% staining cells. The MLH1-and MSH2-stained cells were divided into two grades, negative at ≤10% and positive at >10% cells with nuclear staining. All hematoxylineosin and immunohistochemical staining results were confirmed by two pathologists.
MTT and Flow Cytometry Assay
SW480 and SW480/SPRR3 cells were seeded into a 96-well plate. The cell proliferation rate was measured by a plate reader (absorbance 450) after incubation with Cell-counting kit 8 (CCK8) solution reagent (10 μL) (Dojindo Laboratories, Kumamoto, Japan) for 2 h. For cell-cycle distribution analysis, SW480 and SW480/SPRR3 cells were resuspended in ice-cold phosphate-buffered saline, fixed with 70% ethanol on ice, and incubated with propidium iodide solution (50 μg/mL) (Sigma, St Louis, MO, USA) for 30 min. After washing with phosphate-buffered saline, the cells were analyzed on a flow cytometer using CellQuest software (Becton Dickinson, Franklin Lakes, NJ, USA)
Colony-Formation Assay
A colony-formation assay was performed by using a Cytoselect 96-well cell-transformation assay kit according to the manufacturer’s protocol (Cell Biolabs, San Diego, CA, USA). Briefly, SW480 and SW480/SPRR3 cells (1 × 103) were seeded into the individual wells of a 96-well plate containing 0.6% agar/RPMI1640 medium. After culture for 14 d, the cells were solubilized, lysed and detected with CyQuant GR dye by using a fluorescence plate reader with a 485/520-nm filter (Molecular Devices, Sunnyvale, CA, USA), or the colonies were visualized by crystal violet staining (Sigma).
Statistical Analysis
Presented bar data show standard error of the mean (SEM). Significant differences among groups were determined using the Student t test. SPRR3 expression was compared with demographic and biological parameters by cross-table analysis performed using Fisher exact or Pearson χ2 test, depending on statistical validity. We verified potential variables by multivariate analysis using binary logistic regression. Statistical significance was defined as a P-value < 0.05, and all calculations were performed by using SPSS software (version 14; SPSS Inc., Chicago, IL, USA).
All supplementary materials are available online at www.molmed.org.
RESULTS
SPRR3 Is Upregulated in Colorectal Cancer
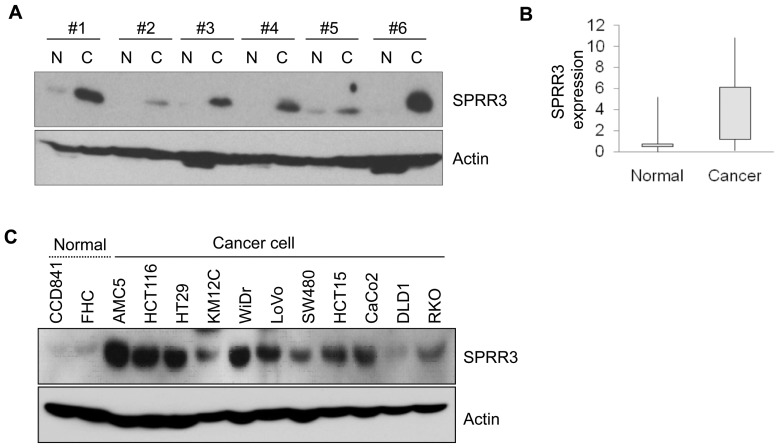
To investigate the molecular changes associated with colorectal tumorigenesis, we analyzed gene expression patterns in 84 patients with sporadic colorectal cancer by using a microarray (8). Among 1263 differentially expressed genes, SPRR3 was shown to be associated with molecular changes, and we therefore examined the expression level of SPRR3 in colorectal cancers. Samples of the tumor tissues and the surrounding normal tissues were obtained at the time of surgery. Among 35 colorectal cancer patients, SPRR3 was highly expressed in tumors compared with the adjacent normal mucosa in 31 cases (88%) (Figure 1A, B). Next, the analysis of expression levels was extended to colorectal cell lines. Consistent with our findings in tissues, SPRR3 was weakly expressed in normal cell lines including CCD841 and FHC cells, but its expression was increased in most of the colorectal cancer cell lines except DLD1 cells (Figure 1C).
Figure 1.
SPRR3 is upregulated in colorectal cancer. The expression of SPRR3 was assessed by Western blotting with SPRR3 antibody in colorectal tissues (T, tumor tissue; N, corresponding normal tissue) (A) and in several colorectal cell lines (C). Actin was used as a loading control. The expression levels of SPRR3 in normal and cancer tissues were quantified by densitometric image scanning of the Western blots. The expression ratio was normalized by Actin (n = 35) (B).
The two categories of SPRR3 expression were associated with demographic and biological features (Figure 2, Table 2). We found that SPRR3 expression was closely correlated with lymphovascular invasion (LVI) of tumor cells, as seen in the 1.5-and 2.1-fold greater positive expression and overexpression of the SPRR3 in tumors with LVI compared with tumors without LVI (P = 0.002 and 0.004, respectively). In a multivariate analysis, which included possibly potent demographic and biological features, tumors with LVI showed significant correlation with positive expression and overexpression of SPRR3 (relative risk, 6.204 and 3.161; 95% confidence interval, 2.108–18.261 and 1.233–8.105; P = 0.001 and 0.017, respectively). These results show that SPRR3 is upregulated and may play a role in colorectal cancer invasion. SPRR3 expression was also associated with female sex and with tumors showing mismatch repair (MMR) defects (P < 0.05).
Figure 2.
Representative photomicrographs of immunohistochemical staining for SPRR3 in colorectal cancer tissues. Cytoplasmic expression of SPRR3 in colorectal cancer tissues was examined by immunohistochemical analysis on a tissue microarray, showing down-regulation (left) and overexpression (right) in colorectal cancer tissues.
Table 2.
SPRR3 expression associated with demographic and biological parameters in 128 sporadic colorectal cancers
| Percentage of patients with SPRR3 expressiona |
||||
|---|---|---|---|---|
| Parameters | Positive expression | P valueb | Overexpression | P valueb |
| Sex, male/female | 59.4/75 | 0.045c | 25/39.1 | 0.065 |
| Age, years, ≤50/>50 | 73.3/65.3 | 0.279 | 30/32.7 | 0.486 |
| Stage,d I + II/III + IV | 66/68 | 0.482 | 26.4/36 | 0.171 |
| Tumor characteristics | ||||
| Site, right colon/left colone | 76/65 | 0.211 | 40/30.1 | 0.235 |
| Growth pattern, expanding/infiltrative | 67.9/63.2 | 0.436 | 33.9/21.1 | 0.201 |
| Differentiation, well and moderately differentiated/poorly differentiated and mucinous | 67/69.2 | 0.569 | 29.6/53.8 | 0.075 |
| Lymphovascular invasion, no/yes | 55.2/80.3 | 0.002c | 20.9/44.3 | 0.004c |
| Either MLH1 or MSH2 expression, no/yes | 77.8/70.2 | 0.484 | 66.7/32.1 | 0.048c |
Category I includes 2 grades, negative at ≤10% and positive at >10% staining cells, and category II includes 2 grades, downregulation at ≤50% and overexpression at >50% staining cells, respectively.
Cross-table analysis using Fisher exact test.
P < 0.05.
Cancer staging according to the American Joint Committee on Cancer (sixth edition, 2001).
Right colon, cecum–splenic flexure of the transverse colon; left colon, descending colon–the rectum.
Overexpression of SPRR3 Accelerates Cell Proliferation
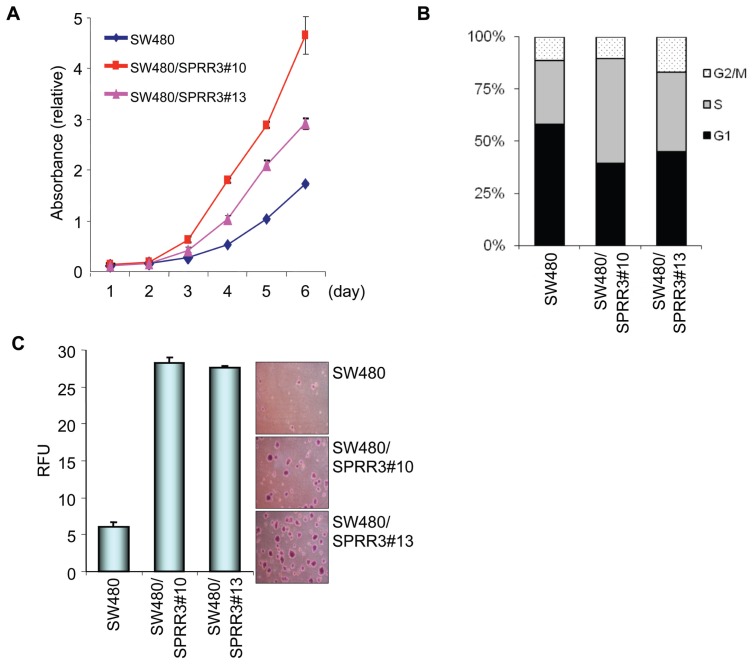
The significant overexpression of SPRR3 in colorectal cancer led us to investigate the effect of overexpressed SPRR3 on the proliferation rate of colon cancer cells. An SPRR3-overexpressing cell line was generated from SW480 human colon cancer cells (SW480/SPRR3 clone). As shown in Figure 3A, the ectopic expression of SPRR3 dramatically increased the proliferation rate of SW480 cells in a time-dependent manner. To determine whether the increased proliferation rate was due to acceleration of the cell cycle, we next examined the effect of SPRR3 on the distribution of cell-cycle phases. The flow cytometer results indicated that overexpression of SPRR3 in SW480 cells caused enhanced accumulation of cells in the G2/M or S phase compared with SW480 control cells (Figure 3B). We further explored the effect of SPRR3 on anchorage-independent growth by conducting a soft agar assay (Figure 3C). Colony formation was increased in the SW480/SPRR3 cells compared with control SW480 cells, suggesting that increased SPRR3 expression potentiates the growth of colorectal cancer cells.
Figure 3.
SPRR3 increases cell proliferation. The cell proliferation rates of control SW480 cells and SW480 cells stably expressing SPRR3 (SW480/SPRR3) were measured by a CCK8 cell proliferation assay. The results are presented in terms of the daily proliferation rate (A). The SW480 and SW480/SPRR3 cells were harvested and stained with propidium iodide and then they were subjected to a flow cytometer for cell cycle analysis. The bars represent the cell population at each stage of the cell cycle (B). Anchorage-independent cell growth was measured by a fluorescence plate reader after SW480 and SW480/SPRR3 (1 × 104) cells were seeded and cultured for 14 d. The colonies were visualized by crystal violet staining (C). RFU, relative fluoresce unit. The data are presented as the mean ± SEM.
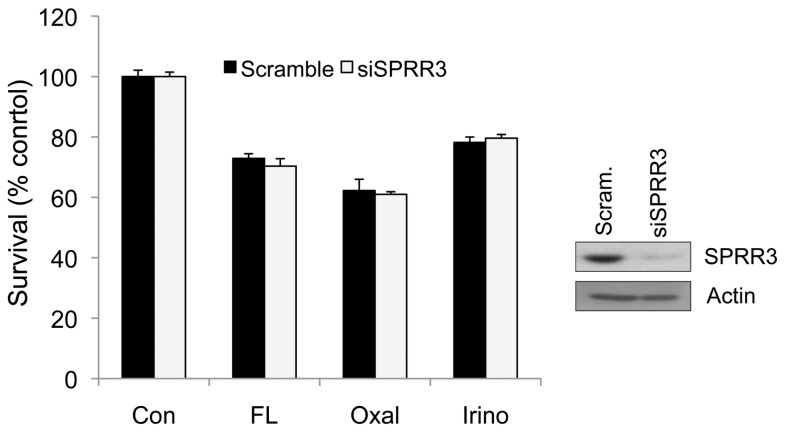
It has previously been shown that SPRR3 expression is reduced in esophageal cancer. To investigate the effect of downregulated SPRR3 in colorectal cancer cells, SPRR3 siRNA was used to specifically knock down SPRR3 expression. SW480 cells were transiently transfected with either nontargeting scrambled siRNA or SPRR3-specific siRNA, and the effects on cell death were determined. Unlike the findings in ESCC, our results showed that the reduced SPRR3 expression did not significantly affect cell death in SW480 colorectal cancer cells (Figure 4).
Figure 4.
Reduced expression of SPRR3 does not affect cell proliferation and cell death in colorectal cancer cells. The SW480 cells were transfected with control scrambled siRNA (Scram) or SPRR3 siRNA (siSPRR3). Reduced SPRR3 expression was confirmed by Western blot analysis with anti-SPRR3 antibody. Two days after siRNA transfection, cells were exposed to 50 ug/mL 5 FU with 10 ug/mL Leucovorin (FL), 30 ug/mL Oxaliplatin (Oxal) or 30 ug/mL Irinotecan (Irino) for 24 h and then cell viability was determined by MTT assay. The values are mean ± SEM (n > 3 times).
AKT Is Activated in SPRR3-Overexpressing Cells
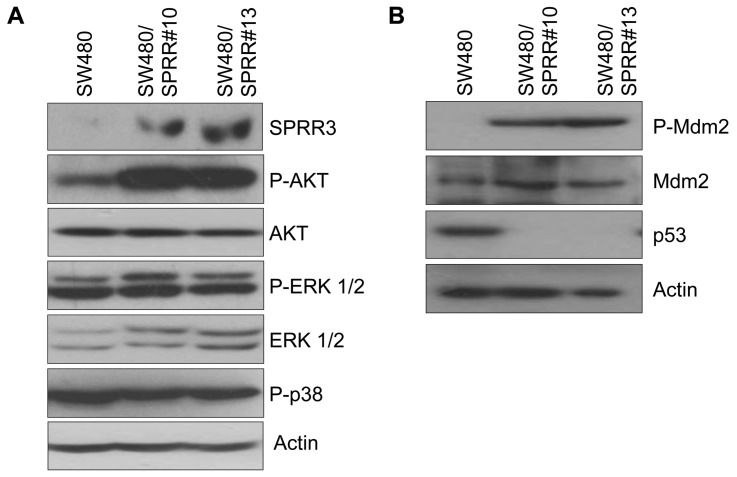
To determine the functional role of SPRR3 in colorectal cell proliferation, we investigated possible involvement of the AKT or MAP-kinase pathways in SPRR3-mediated cell proliferation. AKT and MAP kinase both play pivotal roles in fundamental cellular functions such as cell proliferation, migration and apoptosis. As shown in Figure 4, the overexpression of SPRR3 in SW480 cells led to a dramatic increase in AKT phosphorylation. There was no change, however, in ERK1/2 or p38 MAP kinase (Figure 5A). The AKT pathway is associated with tumorigenesis in many cancers, including colon cancer, and AKT has been shown to regulate the activity of various proteins associated with oncogenesis. Several research groups have recently shown that AKT phosphorylates Mdm2 to promote p53 degradation. The tumor suppressor p53 is a transcription factor that can induce either cell-cycle arrest or cell death, and p53 is expressed at very low levels or mutated in many types of cancer. We therefore examined the expression of phospho-Mdm2 and p53 in association with SPRR3 expression. Interestingly, phosphorylation of Mdm2 was highly increased, but p53 levels were decreased in SPRR3-overexpressing SW480 cells (Figure 5B). These results suggest that the AKT pathway is involved in SPRR3-mediated proliferation of colorectal cancer cells.
Figure 5.
The AKT signaling pathway is activated in SPRR3-expressing cells. The changes in the expression of several signaling molecules involved in the cell cycle or cell proliferation were evaluated by Western blot analysis with the indicated antibodies in SPRR3-expressing cells (A and B). And SPRR3 stable expression (SPRR3) was detected with anti-HA antibody. Actin was used as an internal loading control.
DISCUSSION
Loss of SPRR3 expression is frequently observed in ESCC, where it is known to inhibit tumorigenesis (Supplementary Figure 1) (13–15). However, the clinicopathological characteristics of SPRR3 have not been investigated in other types of cancers, including colorectal cancer. We have previously reported the association of several genes with molecular alterations in colorectal tumorigenesis (8). Surprisingly, in contrast to the findings in ESCC, SPRR3 was frequently and dramatically upregulated in colorectal cancers. Expression analysis with tumor samples from colorectal patients showed that the rate of SPRR3 upregulation in colorectal tumors was almost 88% (Figure 1). The analysis of the gene expression pattern in 35 colorectal cancer cases showed that SPRR3 was increased in 31 cases. Moreover, most of the colorectal cancer cell lines analyzed also exhibited an increase in the expression of SPRR3. The immunohistochemical analysis of SPRR3 expression revealed that up-regulated SPRR3 may be involved in LVI in colorectal cancer. A previous related study showed that the ratio of GATA6, which is a regulator of epithelial differentiation, to SPRR3 can significantly discriminate between normal tissue and esophageal adenocarcinoma (21). Venous and lymphatic invasion are crucial steps in the formation of micrometastases and eventual macroscopic tumor growth in a distant organ (22). LVI may thereby enhance existing staging systems for analyzing individual prognostic features (23). SPRR proteins are differentially expressed in different tissues, suggesting that they have distinct tissue-specific functions. Therefore, the differential expression of SPRR3 in colorectal and esophageal cancers might be due to different tissue-specific expression patterns. Another possible explanation of these differences is that transcriptional regulation of SPRR3 may vary according to tissue type.
Alterations in Wnt/β-catenin (CTNNB1) signaling and in MMR genes have been shown to contribute to the development of colorectal cancer. In the current study, SPRR3 overexpression was associated with deficient MLH1 and MSH2 expression, which was possibly related to the frequent AXIN2 mutations reported in MMR-deficient cells (24). AXIN2 is known to have an important role in the regulation of the stability of CTNNB1 in the Wnt-signaling pathway. However, the association of SPRR3 expression with MMR defects and female sex requires further verification in a larger cohort of colorectal cancer patients. Because the CTNNB1 complex regulates the expression of several genes that have been implicated in colorectal tumorigenesis, including Myc and Cox2 (25–27), it is possible that expression of SPRR3 might be regulated by the CTNNB1-mediated pathway in colorectal cancer. Although we have not identified the factors involved in the regulation of SPRR3 expression, the underlying mechanisms should be examined further to elucidate the function of SPRR3 in colorectal tumorigenesis.
We also investigated the effect of ectopic expression of SPRR3 in the potentiation of colorectal tumorigenisis, and we found that SW480/SPRR3 cells showed accelerated cell proliferation compared with SW480 cells. Further evidence for a function for SPRR3 in tumorigenesis was provided by the soft agar assay. SW480/SPRR3 cells showed a high anchorage-independent proliferation rate, supporting the hypothesis that increased SPRR3 plays a role in colorectal tumorigenesis. It has been reported that SPRR1A protein is upregulated in, and is protective against, hypoxia-induced stress (10). To further elucidate the mechanisms of molecular control of SPRR3 in colorectal cancer, we next examined the effect of SPRR3 on AKT signaling. AKT signaling has been implicated in angiogenesis and tumor invasion as well as in tumor cell survival (28–29). AKT regulates cell growth by modulating several key proteins such as p21, p27, GSK3β and Mdm2 (17,30–32). Because the ectopic expression of SPRR3 markedly enhances AKT phosphorylation in colorectal cancer cells, we further explored SPRR3-mediated AKT signaling. AKT activation is mainly induced by growth-factor stimulation, which is negatively regulated by PTEN. Therefore, we investigated the expression of PTEN and phospho-PI3K in SPRR3-overexpressing cells, and found that these proteins were not significantly altered by SPRR3 expression (data not shown). It was recently reported that increased α1β1-integrin enhances SPRR3 expression (33), and integrin could induce AKT activation through the integrin-associated protein ILK (integrin-linked kinase) (34,35). The molecular mechanisms underlying the activation of AKT by SPRR3 are not clear, and further investigation is therefore needed to elucidate the specific mechanisms. We also screened the AKT downstream molecules and found that Mdm2 is a key molecule in SPRR3-mediated stimulation of cell proliferation. AKT promotes the degradation of p53 through the phosphorylation of Mdm2 (17–19), and p53 controls cell proliferation by inducing either growth arrest or apoptosis. Accordingly, p53 is frequently found in an inactivated, mutated form in many types of cancer (36–37). In this study, we found that the ectopic expression of SPRR3 downregulated p53 expression. Although the molecular factors regulating SPRR3 in colorectal cancer remain to be elucidated, these results lead us to propose that the effect of SPRR3 in promoting colorectal tumorigenesis is associated with the degradation of p53 caused by AKT activation.
To determine whether SPRR3 can be used as a prognostic or diagnostic marker in colorectal cancer, we are currently conducting an analysis of the biological and pathological characteristics of colorectal cancer. The results of the current study indicate that SPRR3 promotes cell proliferation, and that it may be a candidate biomarker for colorectal cancer.
Supplemental Data
ACKNOWLEDGMENTS
We thank SY Rha for generously providing CCD841 cells (Yonsei University, Seoul. Korea) and MR Wang and KY Choi for kindly providing SPRR3 and CTNNB1 plasmids (Peking Union Medical College and Chinese Academy of Medical Science, Beijing, China, and Yonsei University, Seoul. Korea). This work was supported by a grant from the Basic Research Program of the Korea Science and Engineering Foundation (R01-2006-000-10021-0), the Korean Health 21 Research and Development Project, Ministry of Health, Welfare and Family Affairs, Republic of Korea (A062254), and the education and research fund from the Asan Institute for Life Science, Seoul, Korea (2006-069).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Shin HR, et al. National cancer incidence for the year 2002 in Korea. Cancer Res. Treat. 2007;39:139–49. doi: 10.4143/crt.2007.39.4.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR. Colorectal cancer: a multipathway disease. Crit. Rev. Oncog. 2006;12:273–87. doi: 10.1615/critrevoncog.v12.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 4.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat. Rev. Cancer. 2008;8:415–24. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 5.Barrier A, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene. 2005;24:6155–64. doi: 10.1038/sj.onc.1208984. [DOI] [PubMed] [Google Scholar]
- 6.Bianchini M, et al. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int. J. Oncol. 2006;29:83–94. [PubMed] [Google Scholar]
- 7.Fan J, et al. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer. 2008;113:266–75. doi: 10.1002/cncr.23551. [DOI] [PubMed] [Google Scholar]
- 8.Kim JC, et al. Gene expression profiling: canonical molecular changes and clinicopathological features in sporadic colorectal cancers. World J. Gastroenterol. 2008;14:6662–72. doi: 10.3748/wjg.14.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tesfaigzi J, Th’ng J, Hotchkiss JA, Harkema JR, Wright PS. A small proline-rich protein, SPRR1, is upregulated early during tobacco smoke-induced squamous metaplasia in rat nasal epithelia. Am. J. Respir. Cell Mol. Biol. 1996;14:478–86. doi: 10.1165/ajrcmb.14.5.8624253. [DOI] [PubMed] [Google Scholar]
- 10.Pradervand S, et al. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. EMBO J. 2004;23:4517–25. doi: 10.1038/sj.emboj.7600454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracken S, Byrne G, Kelly J, Jackson J, Feighery C. Altered gene expression in highly purified enterocytes from patients with active coeliac disease. BMC Genomics. 2008;9:377–90. doi: 10.1186/1471-2164-9-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pyle AL, et al. Biomechanical stress induces novel arterial intima-enriched genes: implications for vascular adaptation to stress. Cardiovasc. Pathol. 2009;2010;19:e13–20. doi: 10.1016/j.carpath.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham JM, et al. Esophagin cDNA cloning and characterization: a tissue-specific member of the small proline-rich protein family that is not expressed in esophageal tumors. Cell Growth Differ. 1996;7:855–60. [PubMed] [Google Scholar]
- 14.Chen BS, et al. Decreased expression of SPRR3 in Chinese human oesophageal cancer. Carcinogenesis. 2000;21:2147–50. doi: 10.1093/carcin/21.12.2147. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Exogenous expression of Esophagin/SPRR3 attenuates the tumorigenicity of esophageal squamous cell carcinoma cells via promoting apoptosis. Int. J. Cancer. 2008;122:260–6. doi: 10.1002/ijc.23104. [DOI] [PubMed] [Google Scholar]
- 16.Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–88. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 17.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001;3:973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 18.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J. Biol. Chem. 2002;277:21843–50. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 19.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JC, et al. Establishment and biological characterization of human colorectal carcinoma cell lines expressing carcinoembryonic antigen. J. Korean Cancer Assoc. 1996;28:63–71. [Google Scholar]
- 21.Kimchi ET, et al. Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65:3146–54. doi: 10.1158/0008-5472.CAN-04-2490. [DOI] [PubMed] [Google Scholar]
- 22.Zlobec I, Lugli A. Prognostic and predictive factors in colorectal cancer. J. Clin. Pathol. 2008;61:561–9. doi: 10.1136/jcp.2007.054858. [DOI] [PubMed] [Google Scholar]
- 23.Lim SB, et al. Prognostic significance of lymphovascular invasion in sporadic colorectal cancer. Dis. Colon Rectum. 2009;53:377–84. doi: 10.1007/DCR.0b013e3181cf8ae5. [DOI] [PubMed] [Google Scholar]
- 24.Dong X, Seelan RS, Qian C, Mai M, Liu W. Genomic structure, chromosome mapping and expression analysis of the human AXIN2 gene. Cytogenet. Cell Genet. 2001;93:26–8. doi: 10.1159/000056942. [DOI] [PubMed] [Google Scholar]
- 25.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 26.Araki Y, et al. Regulation of cyclooxygenase-2 expression by the Wnt and ras pathways. Cancer Res. 2003;63:728–34. [PubMed] [Google Scholar]
- 27.Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci. 2008;99:631–7. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Testa JR, Bellacosa A. AKT plays a central role in tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10983–5. doi: 10.1073/pnas.211430998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCubrey JA, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv. Enzyme Regul. 2006;46:49–79. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–9. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 31.Zhou BP, et al. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 2001;3:245–52. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 32.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:339–45. [PubMed] [Google Scholar]
- 33.Pyle AL, et al. Regulation of the atheroma-enriched protein, SPRR3, in vascular smooth muscle cells through cyclic strain is dependent on integrin alpha1beta1/collagen interaction. Am. J. Pathol. 2008;173:1577–88. doi: 10.2353/ajpath.2008.080042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troussard AA, et al. Preferential dependence of breast cancer cells versus normal cells on integrin-linked kinase for protein kinase B/Akt activation and cell survival. Cancer Res. 2006;66:393–403. doi: 10.1158/0008-5472.CAN-05-2304. [DOI] [PubMed] [Google Scholar]
- 35.Loufrani L, et al. Key role of alpha(1)beta(1)-integrin in the activation of PI3-kinase-Akt by flow (shear stress) in resistance arteries. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H1906–13. doi: 10.1152/ajpheart.00966.2006. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008;7:979–87. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 37.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–65. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.