Abstract
Types A and B Niemann-Pick disease (NPD) result from the deficient activity of acid sphingomyelinase (ASM), due to mutations in the sphingomyelin phosphodiesterase 1 (SMPD1) gene. Here we report the identification, characterization and genotype/phenotype correlations of eight novel mutations in six unrelated NPD patients. These mutations included seven missense mutations: c.631T > C (p.W211R), c.757G > C (p.D253H), c.940G > A (p.V314M), c.1280A > G (p.H427R), c.1564A > G (p.N522S), c.1575G > C (p.Q525H) and c.1729A > G (p.H577R), and a novel frameshift mutation, c.1657delACCGCCT (fsT553). Each missense mutation was expressed in 293T or COS-7 cells; mutant enzymes p.W211R, p.D253H, p.H427R and p.H577R had <1% of expressed wild-type activity, whereas p.V314M, p.N522S and p.Q525H had 21.7%, 10.1% and 64% of expressed wild-type activity, respectively. The c.1564A > G mutation obliterated a known N-glycosylation site and its p.N522S mutant enzyme had ~10% of expressed wild-type activity. Western blot analysis revealed that each mutant protein was expressed at near wild-type amounts, despite their differences in residual activity. The novel seven-base deletion occurred at codon 553, leading to a premature truncation after residue 609. The expression studies predicted the clinical phenotypes of the six patients: two type A patients had genotypes with only type A alleles [c.631T > C (p.W211R), c.757G > C (p.D253H) and c.1729A > G (p.H577R)], and the other four type B disease patients had at least one neuroprotective mutant type B allele [c.940G > A (p.V314M), c.1280A > G (p.H427R), c.1564A > G (p.N522S) and c.1575G > C (p.Q525H)] that expressed >5% residual ASM activity. Thus, these new mutations provide novel genotype/phenotype correlations and further document the genetic heterogeneity in types A and B NPD.
INTRODUCTION
Types A and B Niemann-Pick disease (NPD) are autosomal recessive sphingolipidoses caused by mutations in the sphingomyelin phosphodiesterase 1 (SMPD1) gene that results in the deficient activity of lysosomal acid sphingomyelinase (ASM, E.C. 3.1.4.12). Type A NPD (MIM# 257200) is an infantile neurodegenerative disease characterized by massive hepatosplenomegaly, rapidly progressive psychomotor deterioration, and death in the first few years of life, whereas type B NPD (MIM# 607616) is a nonneurologic, visceral form with hepatosplenomegaly, pulmonary disease and survival into adolescence and/or adulthood. An intermediate phenotype, called type A/B, may include neurologic involvement, mild developmental delay, and cherry-red maculae, along with the type B visceral manifestations (1). Note that NPD type C (MIM# 257220) is a different disease caused by mutations in the NPC1 or NPC2 genes, which are involved in cholesterol transport (2).
The SMPD1 gene (MIM# 607608, GenBank# M81780.1) is composed of six exons and is located on chromosome 11p15.1–11p15.4 (1). The human ASM protein is synthesized as a 75-kDa, glycosylated prepolypeptide, which is converted to a precursor 72-kDa form. The precursor is subjected to two different processing events. A minor portion is cleaved in the endoplasmic reticulum–Golgi complex, yielding a 57-kDa form, and the majority is processed to a 70-kDa mature form (3). Six N-glycosylation sites exist in the protein, of which five are occupied (4).
Of the mutations in the SPMD1 gene causing types A and B NPD (see the Human Gene Mutation Database: http://www.hgmd.org/), only a few have been found to occur frequently in specific ethnic or demographic groups. Most mutations are “private,” occurring in one or a few families. For newly diagnosed infants and children, mutation analysis and subsequent expression studies can be used to predict the patient’s NPD subtype. Frameshift mutations due to splicing, small and large insertions and deletions and splicing defects typically have little or no residual ASM activity and are called type A alleles. Missense and other lesions (such as in-frame codon deletions and splicing mutations) that retain significant residual activity (>5% of in vitro–expressed wild-type activity) are neuroprotective and are called type B alleles (5). Inheritance of two type A alleles predicts a type A phenotype with a neurodegenerative disease course. In contrast, inheritance of one type B allele will be neuroprotective and is predictive for a type B phenotype, even if the other allele has a type A lesion.
Although many mutations have been reported in patients with the types A and B phenotypes, expression studies are the most accurate way to predict which missense mutations have residual enzyme activity and may be neuroprotective. To date, however, expression studies have been performed for only a small number of missense mutations (6,7). Here we present the clinical and molecular findings for six unrelated type A and B NPD patients. Seven novel missense mutations and one novel frameshift mutation were identified. The missense mutations were expressed in vitro, providing genotype/phenotype correlations for these patients that were consistent with their clinical phenotypes.
SUBJECTS AND METHODS
Subjects
Patients with NPD were admitted to the Mount Sinai General Clinical Research Center for evaluation. The diagnosis of each child was previously confirmed by the demonstration of reduced ASM activity in isolated leukocytes and/or cultured skin fibroblasts. Of the six patients in this study, two were identified as having significant, rapidly progressive neurologic symptomatology, consistent with type A or the intermediate type A/B phenotype. The other four patients were normal neurologically and classified as having the type B phenotype. The clinical findings in these patients are summarized in Table 1. All patients and/or their guardians provided written informed consent for the evaluation and genotype/phenotype studies, which were approved by the Mount Sinai Institutional Review Board.
Table 1.
Clinical findings in the NPD patients.
| Patient no./sex | Age at diagnosis, years | Current age, years | Heptosplenomegaly | Developmental delaya | Clinically based NPD type |
|---|---|---|---|---|---|
| 1/M | Infancy | 2 | Massive | + | A |
| 2/M | Infancy | 3 | Massive | +/− | A/B |
| 3/F | 2 | 6 | Moderate | − | B |
| 4/M | 9 | 12 | Mild | − | B |
| 5/F | 3 | 25 | Moderate | − | B |
| 6/M | 3 | 8 | Moderate | − | B |
Developmental delay was assessed by standard clinical evaluation and determined to be normal (+), abnormal (−), or intermediate (+/−).
DNA Extraction and Mutation Identification
Genomic DNA was extracted from the patients’ leukocytes or cultured fibroblasts by using standard protocols. Each exon, 50 flanking intronic nucleotides, 1750 nucleotides upstream of the first initiation codon, and 310 nucleotides into the 3′-untranslated region of the SMPD1 gene were amplified by use of polymerase chain reaction (PCR) as previously described (8). The PCR products were directly sequenced in the forward and reverse directions by using the Big Dye Terminator Cycle Sequencing v3.1 kit (Perkin Elmer Applied Biosystems, Foster City, CA, USA), and an ABI PRISM 3730xl DNA analyzer (Perkin Elmer Applied Biosystems), according to the manufacturer’s instructions.
SMPD1 nucleotides were numbered according to sequence RefSeq NM_000543.3, with +1 being the A of the ATG start codon. Similarily, the ATG start codon was designated +1 for amino acid numbering, according to the preprotein sequence NP_000534.3.
Vector Construction and Site-Directed Mutagenesis
All mutations were introduced into the wild-type full-length SMPD1 cDNA cloned in pBluescript by PCR-based site-directed mutagenesis by using the QuickChange II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s instructions. All constructs were resequenced to ensure that no spurious mutation had been introduced by the mutagenesis procedure. For protein expression, the mutant and wild-type cDNAs were subcloned into the pcDNA3.1(−) expression vector.
Cell Culture and Transient Transfection
COS-7 and 293T cells were grown in 100-mm tissue-culture dishes with Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, Poole, UK) supplemented with 10% fetal bovine serum and antibiotics (Gibco, Grand Island, NY, USA). For the transfection, 30 × 105 cells were split in antibiotic-free medium and transfected 24 h later (when cells were at 90% confluence) with 3 μg of wild-type or mutant plasmid, by using the FuGENE 6 transfection reagent (Roche, Indianapolis, IN, USA). As a negative control, we used cells transfected with an empty pcDNA3.1(−) vector, and 48 h after transfection, cells were harvested by scraping and centrifuging. Cell pellets were washed twice with phosphate buffered saline (PBS) and stored at − 80ºC before ASM activity determinations. After thawing, transfected cells were disrupted by five freeze-thaw cycles in Tris buffer, pH 7.0, containing 0.2% Igepal to prepare cell extracts. Approximately 10% of each cellular extract volume was used for Western blotting.
Enzymatic Analysis
ASM activity was measured in the transfected cells by using the BODIPY-conjugated C12 sphingomyelin (cat #D7711; Molecular Probes, Carlsbad, CA, USA) substrate as previously described (9). Protein concentrations were determined by the Lowry method. In every expression experiment, transfections were performed three times for the wild-type and each mutant construct. An empty vector was used as a negative control. The residual enzymatic activity of each mutant allele was expressed as a percentage of the mean expressed activity of the wild-type construct transfected in the same experiment. The ASM activity of the negative control was subtracted from the expressed activity.
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis and Western Blot Analysis
Protein extracts from transfected COS-7 cells (35 μg of protein/lane) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (12.5% polyacrylamide) and electrophoretically transferred onto nitrocellulose membranes. Each membrane was blocked with 5% nonfat milk in PBS, containing 0.2% Tween 20 (PBST). The blotted membranes were probed with polyclonal antihuman ASM antibody for 4 h in 3% nonfat milk in PBS with 0.1% Tween, washed four times with PBST, and then four times with PBS. Antirabbit IgG antibody (Sigma-Aldrich) was used as a secondary antibody, and the immunoreactive bands were detected by incubation of the membrane for 2 min in the following solution: 10 mL of 100 mmol/L Tris-HCl pH 9.0, 50 μL of 45 mmol/L p-coumaric acid, 50 μL of luminal, and 10 μL of 30% H2O2.
RESULTS
Mutation Analyses
SMPD1 mutation analyses were performed on genomic DNA isolated from the six unrelated NPD patients (Table 1). Ten mutations were identified, eight of which were novel mutations, including seven missense mutations and one seven-nucleotide deletion (c.1657delACCGCCT, designated fsT553) (Table 2). The novel missense mutations were: c.631T > C (p.W211R), c.757G > C (p.D253H), c.940G > A (p.V314M), c.1280A > G (p.H427R), c.1564A > G (p.N522S), c.1575G > C (p.Q525H), and c.1729A > G (p.H577R). The seven-nucleotide deletion occurred in codons 553 and 554 of exon 6, and caused a frameshift that introduced 57 different amino acids in residues 553 through 609 of the 631-residue wild-type enzyme, with premature truncation after residue 609. In patient 6 the second allele was not found by sequencing 1750 nucleotides upstream of the initiation ATG, all exons, at least 50 intronic nucleotides at each intron/exon boundary, and 310 nucleotides into the 3′-untranslated region.
Table 2a.
Novel SMPD1 mutations identified in the type A and B NPD patients.
| Mutationa | Exon/codon | Predicted amino acid substitution | |
|---|---|---|---|
| c.631T > C | 2/211 | p.W211R | Tryptophan → Arginine |
| c.757G > C | 2/253 | p.D253H | Aspartate → Histidine |
| c.940G > A | 2/312 | p.V314M | Valine → Methionine |
| c.1280A > G | 4/427 | p.H427R | Histidine → Arginine |
| c.1564A > G | 6/522 | p.N522S | Asparagine → Serine |
| c.1575G > C | 6/525 | p.Q525H | Glutamine → Histidine |
| c.1729A > G | 6/577 | p.G577R | Histidine → Arginine |
| c.1657del7 | 6/553 | p.fsT553 | deletion of ACCGCCT |
Numbered according to the first in-frame ATG in reference sequence NM_000543.3.
Table 2b.
Comparison of the partial nucleotide and predicted amino-acid sequences of the wild-type and deletion mutation c.1657del7.a
The seven-base deletion (boxed in the wild-type sequence) occurred after nucleotide 1657 in the SMPD1 gene.
Indicates premature termination.
Transient Expression and Western Blot Analysis of Wild-Type and Mutant ASM Enzymes
To evaluate the functional effect of the novel missense mutations on ASM activity, we performed transient expression studies. The wild-type SMPD1 cDNA and/or cDNAs bearing each mutation were inserted into the pcDNA3.1(−) vector and transiently transfected into 293T or COS-7 cells. As a negative control, cells were transfected with an empty pcDNA vector. After transfection, cell extracts were assayed for ASM activity by using BODIPY-conjugated C12 sphingomyelin as substrate (Table 3). Mutant enzymes p.V314M, p.N522S and p.Q525H each had significant residual ASM activity in vitro (21.7%, 10.1% and 63.9% of expressed wild-type activity, respectively), whereas p.W211R, p.D253H, p.H427R and p.H577R had less than 1% of expressed wild-type activity.
Table 3.
Transient expression of novel SMPD1 missense mutations in 293-T cells.
| Construct/mutation | Mean ASM activity, U/mg protein | % Wild-type activity | Stability,a % initial activity | Predicted phenotype |
|---|---|---|---|---|
| ASM (wild type) | 546 | 100 | 55.9 | WT |
| W211R | 2.34 | 0.43 | NDb | A |
| D253H | 0.13 | 0.02 | NDb | A |
| V314M | 118 | 21.7 | 18.2 | B |
| H427R | 0.10 | 0.02 | NDb | A |
| N522S | 55.1 | 10.1 | 45 | B |
| Q525H | 349 | 63.9 | 19.9 | B |
| H577R | 2.66 | 0.49 | NDb | A |
For stability studies, 24 h after transfection cell extracts were prepared and incubated at 55°C, pH 7.0, for 30 min prior to enzyme assay.
ND: thermostability was not determined for these alleles, because the expressed activities were already at the lower limit of detectability.
Heat-inactivation studies were performed to determine whether the three mutations expressing residual ASM activity were thermolabile. As shown in Table 3, after 30 min at 55°C, pH 7.0, the wild-type and p.N522S mutant enzymes retained ~56% and ~45% of initial activity, respectively, whereas the p.V314M and p.Q525H mutant enzymes were thermolabile, having 18.2% and 19.9% of initial activity, respectively.
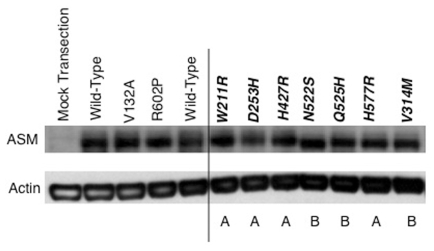
Western blot analysis was performed to determine the relative stability of the mutant enzymes (Figure 1). Notably, despite differences in residual ASM activity, each of the mutations expressed essentially wild-type amounts of the mutant ASM polypeptide, which migrated to a position of ~72 kDa, with the exception of the p.N522S enzyme, which migrated to a slightly lower position (~69–70 kDa), a finding consistent with the loss of its N-glycosylation site (4).
Figure 1.
Western blot analysis of wild-type and mutant ASM enzymes following transient expression in COS-7 cells. Each of the seven novel missense mutations was introduced into the full-length SMPD1 cDNA. Below each lane the predicted phenotype of the mutations (based on transient expression data in COS-7 or 293T cells; see Table 3) is shown. As controls, the wild-type cDNA also was used, along with mutant constructs representing two known mutations (V132A and R602P). β-actin was used as a loading control. The mock transfection included the empty pcDNA vector. The immunoblot is representative of three independent experiments.
DISCUSSION
By performing mutation analysis of six unrelated NPD patients, we identified eight novel SMPD1 gene lesions; seven of these were missense mutations and the eighth was a seven-nucleotide deletion in exon 6 that caused a frameshift mutation inserting 57 different amino acids from residues 553 through 609, followed by premature termination of the 631-residue wild-type enzyme.
The identification and characterization of these mutations provided genotype/phenotype correlations that were consistent with the clinical findings in each of these patients. Two patients had a type A phenotype (Table 1). Patient 1, who was homozygous for the c.631T > C (p.W211R) mutation that expressed less than 0.5% of wild-type activity, was a severely affected male with type A disease (massive hepatosplenomegaly and severe psychomotor retardation). Patient 2 was heteroallelic for missense mutations c.757G > C (p.D253H) and c.1729A > G (p.H577R), both mutant enzymes having little, if any residual activity. Of note, patient 2 was 3 years old when NPD was diagnosed, and clinically had an intermediate A/B phenotype. The expression data would indicate a more severe type A phenotype.
The other four patients had type B disease. Patient 3 had missense mutation c.940G > A (p.V314M) and the previously reported deletion, 573delT, which resulted in early termination of the ASM polypeptide (10). The fact that the p.V314M mutant enzyme had significant residual activity when expressed provided sufficient neuroprotective activity in this clinically diagnosed 6-year-old patient, consistent with her type B phenotype. Patient 4 had the novel missense mutation, c.1280A > G (p.H427R), and the previously reported c.689G > A (p.R230H) mutation (8). Although the p.H427R mutant enzyme had little to no residual activity when expressed in vitro, the previously reported p.R230H enzyme had sufficient activity to account for the type B phenotype (7). Patient 5 had the novel N-glycoslyation mutation, c.1564A > G (p.N522S), and the novel seven-base deletion in exon 6, c.1657delACCGCCT (fsT553). The p.N522S mutation occurred at one of the five functional N-glycoslyation sites in ASM, and previous site-directed mutagenesis at this site (introducing a glycine instead of the asparagine residue) resulted in ~5% residual ASM activity in vitro (4). These findings are consistent with the expression data reported here, which demonstrate that the p.N522S mutant protein had ~10% residual ASM activity in vitro. Patient 5 had type B NPD with moderate hepato splenomegaly and no neurological involvement (Table 4). Because the fsT553 frameshift mutation presumably does not express residual ASM activity, these data indicate that the ~10% in vitro activity of the p.N522S enzyme was neuroprotective in this patient. In patient 6, only one lesion was identified, the missense mutation c.1575G > C (p.Q525H), which had the highest expressed activity in vitro (~64% of expressed wild-type activity), but was thermolabile, which resulted in ~20% of its initially expressed activity after 15 minutes at 55°C, pH 7.0. The other mutant allele in this patient was not detected, indicating that it likely resulted from within an intron that was not included in our PCR and sequencing reactions, or resulted from an intronic lesion that deleted a whole exon or exons, created a cryptic splice site or was due to deletion of the entire gene, and so on.
Table 4.
Genotype/phenotype correlations in NPD patients.a
| Patient no. | Mutant allele 1/expression phenotype | Mutant allele 2/expression phenotype | Predicted/known phenotype |
|---|---|---|---|
| 1 | W211R/A | W211R/A | A/A |
| 2 | D253H/A | H577R/A | A/A/B |
| 3 | W314M/B | 573delT/A | B/B |
| 4 | H427R/A | R230Hb/B | B/B |
| 5 | N522S/B | 1657del7/A | B/B |
| 6 | Q525H/B | Not determined | ?/B |
To determine whether the mutations identified resulted in reduced levels of the ASM polypeptide, we carried out Western blot analysis following transient expression in COS-7 cells. Of note, each of the seven novel missense mutations expressed near normal levels of the ASM polypeptide, although four of the seven mutant proteins (p.W211R, p.D253H, p.H427R and p.H577R) had little, if any, residual ASM activity in vitro. These findings are consistent with other reported results showing that near-normal levels of mutant ASM were present in cells from type A NPD patients (11). As expected, each of the three missense mutations expressing residual ASM activity also had near-normal levels of their respective mutant enzyme proteins. However, results of heat inactivation studies indicated that two of these proteins (pV314M and pQ525H) were less stable than the wild-type enzyme.
In summary, eight new mutations causing types A and B NPD in six unrelated patients were identified and characterized by in vitro expression assays. These findings predicted the clinical phenotypes of these six patients. These results further underscore the genetic heterogeneity of the mutations causing types A and B NPD, and provide additional information for predicting the clinical phenotypes in newly diagnosed infants and children with this disease.
ACKNOWLEDGMENTS
The authors thank the staff of the Clinical Research Center at Mount Sinai for their assistance with the care of our NPD patients.
This work was supported in part by NIH grant 5 R01 HD28607 and by a grant UL1RR029887 from the National Center for Research Resources, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
DISCLOSURE
EH Schuchman is an inventor on patents licensed to Genzyme, Luminex and Ambion for the diagnosis and/or treatment of NPD. He and Mount Sinai could benefit financially from royalties obtained from these licenses. EH Schuchman also received a research grant from Genzyme for studies of NPD.
Online address: http://www.molmed.org
REFERENCES
- 1.Schuchman EH. The pathogenesis and treatment of acid sphingomyelinase-deficient Niemann-Pick disease. J. Inherit. Metab. Dis. 2007;30:654–63. doi: 10.1007/s10545-007-0632-9. [DOI] [PubMed] [Google Scholar]
- 2.Patterson MC, et al. Niemann-Pick disease type C: a lipid trafficking disorder. In: Scriver CR, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. 8th ed. McGraw-Hill; New York: 2001. pp. 3611–34. [Google Scholar]
- 3.Ferlinz K, Hurwitz R, Vielhaber G, Suzuki K, Sandhoff K. Occurrence of two molecular forms of human acid sphingomyelinase. Biochem. J. 1994;301:855–62. doi: 10.1042/bj3010855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlinz K, et al. Functional characterization of the N-glycosylation sites of human acid sphingomyelinase by site-directed mutagenesis. Eur. J. Biochem. 1997;243:511–7. doi: 10.1111/j.1432-1033.1997.511_1a.x. [DOI] [PubMed] [Google Scholar]
- 5.Levran O, Desnick RJ, Schuchman EH. Niemann-Pick type B disease: identification of a single codon deletion in the acid sphingomyelinase gene and genotype/phenotype correlations in type A and B patients. J. Clin. Invest. 1991;88:806–10. doi: 10.1172/JCI115380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dardis A, et al. Functional in vitro characterization of 14 SMPD1 mutations identified in Italian patients affected by Niemann Pick Type B disease. Hum. Mutat. 2005;26:164. doi: 10.1002/humu.9353. [DOI] [PubMed] [Google Scholar]
- 7.Pavlu-Pereira H, et al. Acid sphingomyelinase deficiency. Phenotype variability with prevalence of intermediate phenotype in a series of twenty-five Czech and Slovak patients: a multi-approach study. J. Inherit. Metab. Dis. 2005;28:203–27. doi: 10.1007/s10545-005-5671-5. [DOI] [PubMed] [Google Scholar]
- 8.Simonaro CM, Desnick RJ, McGovern MM, Wasserstein MP, Schuchman EH. The demographics and distribution of type B Niemann-Pick disease: novel mutations lead to new genotype/phenotype correlations. Am. J. Hum. Genet. 2002;71:1413–9. doi: 10.1086/345074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, Chen F, Dagan A, Gatt S, Schuchman EH. A fluorescence-based, high-performance liquid chromatographic assay to determine acid sphingomyelinase activity and diagnose types A and B Niemann-Pick disease. Anal. Biochem. 2003;314:116–20. doi: 10.1016/s0003-2697(02)00629-2. [DOI] [PubMed] [Google Scholar]
- 10.Ricci V, et al. Screening of 25 Italian patients with Niemann-Pick A reveals fourteen new mutations, one common and thirteen private, in SMPD1. Hum. Mutat. 2004;24:105. doi: 10.1002/humu.9258. [DOI] [PubMed] [Google Scholar]
- 11.Jones I, He X, Katouzian F, Darroch PI, Schuchman EH. Characterization of common SMPD1 mutations causing types A and B Niemann-Pick disease and generation of mutation-specific mouse models. Mol. Genet. Metab. 2008;95:152–62. doi: 10.1016/j.ymgme.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]