Abstract
Methodological limitations have hampered the use of heavy water (2H2O), a convenient, universal biosynthetic label, for measuring protein synthesis. Analyses of 2H-labeled amino acids are sensitive to contamination; labeling of peptides has been measured for a few serum proteins, but this approach awaits full validation. Here we describe a method for quantifying protein synthesis by peptide mass spectrometry (MS) after 2H2O labeling, as applied to various proteins of the major histocompatibility complex (MHC). Human and murine antigen-presenting cells were cultured in medium containing 5% 2H2O; class I and class II MHC proteins were immunoprecipitated, bands were excised, and Ala-/Gly-rich, allele-specific tryptic peptides were identified by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Mass isotopomer distributions were quantified precisely by LC–MS and shifted markedly on 2H2O labeling. Experimental data agreed closely with models obtained by mass isotopomer distribution analysis (MIDA) and were consistent with contributions from Ala, Gly, and other amino acids to labeling. Estimates of fractional protein synthesis from peptides of the same protein were precise and internally consistent. The method was capable of discriminating between MHC isotypes and alleles, applicable to primary cells, and readily extendable to other proteins. It simplifies measurements of protein synthesis, enabling novel applications in physiology, in genotype/phenotype interactions, and potentially in kinetic proteomics.
Keywords: Peptide liquid chromatography/mass spectrometry, Heavy water labeling, Major histocompatibility complex proteins, Allelic polymorphism, Mass isotopomer distribution analysis, Protein synthesis
Protein synthesis has classically been measured by incorporation of radiolabeled amino acids, but this approach is difficult to use in animals and is too hazardous for routine human use. Moreover, turnover measurements by radiolabeling are sensitive to variable protein losses during sample preparation. Alternatively, labeling with 2H- or 13C-substituted essential amino acids, in cell culture (stable isotope labeling of amino acids in cell culture [SILAC])2 or in animals, has been used [1–4]. This approach is safe and the ratio of labeled to unlabeled species is quantified, so variable recovery has little impact on results. However, calculation of fractional protein synthesis requires an understanding of precursor/product relationships, which can be confounded by complexity in intracellular aminoacyl transfer RNA (tRNA) pools and by recycling of unlabeled amino acids. In vivo, complete replacement of endogenous amino acids takes several generations [2], and long-term experiments are expensive, due to the requirement of modified diets or infusion of the labeled precursor. The accuracy of the kinetic information obtained remains unclear.
An alternative approach for the analysis of long-lived proteins (t1/2 ≥ hours) employs stable isotope labeling of nonessential amino acids with heavy water (SINEW) [5–7]. This is an extension of well-established methods for measuring the synthesis of lipids and DNA [8,9]. Body water is routinely enriched to 5% heavy water (2H2O [deuterated water, D2O]) in rodents, and to 1–2% in humans, without perturbing normal physiology [8]. After 2H2O intake, 2H atoms rapidly equilibrate across body water pools. C—H bonds of free glycine and alanine [5–7,10], and of several other nonessential amino acids (NEAAs) [11], then approach isotopic equilibrium with body water. Thereafter, protein biosynthesis becomes the rate-limiting step for 2H incorporation (Fig. 1A). Body water turns over slowly [12], and a fixed enrichment is maintained indefinitely by regular oral 2H2O intake [8]. This enables extended labeling protocols for quantification of slow protein turnover rates. The analytical approach used in most studies to quantify label incorporation involves total hydrolysis of proteins of interest to single amino acids and quantification of 2H incorporation into NEAA (most often Ala) by gas chromatography–mass spectrometry (GC–MS) [5–7,13–15]. This approach is precise and sensitive, but it can easily be confounded by contamination with extraneous proteins or free amino acids; in practice, applications have been limited to relatively abundant, easily purified proteins. Analysis of 2H-labeled tryptic protein fragments [16,17] is less easily confounded, but studies have been limited to rodent serum albumin and contaminants of albumin preparations; methods for estimating fractional synthesis from experimental data are somewhat cumbersome, and validation of model assumptions underlying this methodology remains incomplete.
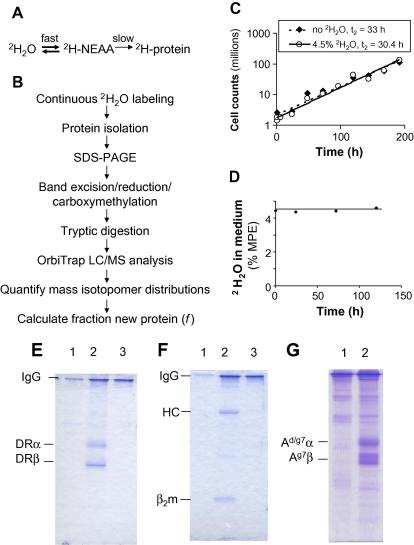
Fig. 1.
Application of SINEW with peptide LC–MS to cultured antigen presenting cell lines. (A) Biosynthetic labeling of nonessential amino acids of proteins from 2H2O. (B) Flow chart of experimental approach. (C) 2H2O labeling does not perturb cell growth. Growth kinetics of Priess cells are shown during 1 week of culture in medium with or without 4.5% 2H2O. Exponential curve fits and doubling times (t2) are shown. (D) Constant 2H2O enrichment in medium throughout the culture period (MPE, mole per cent excess over natural abundance) was verified by IRMS analysis of medium sampled at different times. (E–G) Protein isolation. HLA-DR (E), HLA-A/B/C (F), and H2-A (G) molecules were immunoprecipitated from host cell lines (Priess in panels E and F, M12.NOD in panel G) using appropriate mAbs: L243, W6/32, and OX-6, respectively (lanes 2). Lanes 1 represent control immunoprecipitates using irrelevant control antibody (Mac4 in panels E and F, MKD6 in panel G). Lanes 3 represent control immunoprecipitates from irrelevant cell lines (A20 cells in panels E and F). Samples were analyzed by nonreducing 12% SDS–PAGE and visualized by Coomassie blue staining. Positions of bands representing immunoprecipitating antibody, MHC class II α- and β-chains, MHC class I heavy chain (HC), and β2-microglobulin (β2m) are indicated. Under nonreducing conditions, the bands migrated slightly faster than the nominal molecular weights for each polypeptide (35 and 29 kDa for class II α- and β-chains, respectively, and 45 kDa for class I heavy chains; MW markers not shown).
Here we wished to examine the utility of the SINEW approach, combined with tryptic peptide analysis by liquid chromatography (LC)–MS (Fig. 1B), for quantifying the synthesis of major histocompatibility complex (MHC) proteins. These membrane glycoproteins, abundant on immune cells, present antigens to T lymphocytes of the adaptive immune system; their extensive genetic polymorphism is important in acute allogeneic transplant rejection and autoimmune disease susceptibility. The posttranslational regulation of their fate and function is of great interest in immunology and may be affected by polymorphism. Here we have developed a peptide LC–MS method for measuring the fractional synthesis of MHC protein allomorphs in cell culture by SINEW. The resultant innovations greatly simplify the analysis and enhance the versatility of this approach.
Materials and methods
Cell lines
The human Epstein–Barr virus (EBV)-transformed B cell line, Priess, was used to quantify synthesis of human leukocyte antigen (HLA)-DR. Priess cells are homozygous at the HLA-DR locus, expressing the DRB1∗0401 allele along with the nearly monomorphic DRA∗0101 gene product [18]. Monocyte-derived dendritic cells were differentiated with GM-CSF (50 ng/ml) and IL4 (1000 U/ml) for 7 days after immunomagnetic bead isolation of CD14+ monocytes from peripheral blood mononuclear cells. LCL721 cells were used to quantify the synthesis of HLA-B∗08 and B∗51 MHC class I heavy chain gene products [19]. A20 murine B lymphoma cells, derived from Balb/c mice (H-2d), or splenic B cells from Balb/c mice were used as a source of murine H2-Ad (AαdAβd heterodimer) MHC class II molecules [20]. Expression of MHC molecules was verified by immunofluorescent staining followed by flow cytometric analysis (data not shown).
For analysis of murine H2-Ag7 MHC class II molecules, M12.NOD cells that were derived from the M12.C3 cell line by transfection with a genomic construct coding for Ag7 β-chains (the Adα-chain, identical to that in the g7 haplotype, is endogenously expressed) were used. A 4.3-kbp BamHI–KpnI fragment, containing part of the 5′ flanking sequences of the Abb promoter [21], was cloned into pTCF. The entire Abg7 gene was subcloned from the cosmid NOD-1–3 [22] isolated from a NOD liver cosmid library [23] as an 11-kbp EcoRV fragment and subcloned into the ClaI site of pTCF [24] following the addition of ClaI linkers to each end. The 5′ end of the cloned Abg7 gene was extended by subcloning the 10-kbp KpnI–ClaI Abg7 fragment into the KpnI–ClaI site of the p 5′-Abb-TCF described above. For transfection, the extended Abg7–TCF plasmid was linearized with ClaI and added to M12.C3 cells as a calcium phosphate coprecipitate. Stable transfectants were selected with 0.8 mg/ml G418, and the expression of H2-Ag7 was confirmed by staining with OX-6 (AbD Serotec, Oxford, UK) and flow cytometric analysis.
Antibodies
L243 is a murine IgG2a monoclonal antibody (mAb) specific for the α-chain of assembled HLA-DR αβ heterodimers [25]. W6/32 is a murine IgG2a mAb specific for HLA-A, -B, and -C heavy chains assembled with β2-microglobulin [26]. MKD6 is an IgG2a mAb specific for H2-Ad [27]. OX-6 is an IgG1 mAb specific for a polymorphic determinant present on several H2-A alleles, including Ag7 [28]. Hybridoma supernatants were produced, and mAbs were isolated on protein A or protein G affinity columns (GE Biosciences), with low-pH elution; purified antibodies were buffer exchanged, quantified by absorbance at 280 nm, and stored at −20 °C in phosphate-buffered saline (PBS).
Cell culture and 2H2O labeling
Cells were maintained in suspension culture in a humidified CO2 incubator (37 °C) in RPMI 1640 medium (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM l-glutamine (Sigma), 1% penicillin/streptomycin, and 10 mM Hepes (Sigma). Exponentially growing cell suspensions (typically 105–106 cells/ml) were adjusted to approximately 5% 2H2O enrichment in medium by the addition of 99% (mol/mol) 2H2O (Cambridge Isotope Laboratories), which was adjusted to 0.9% (w/v) NaCl, sterile filtered, and (if needed) depleted of residual lipopolysaccharide on polymyxin B columns (Pierce) according to the manufacturer’s instructions. Cells were cultured in an incubator with 5% 2H2O in the humidifier dish, harvested at various times by centrifugation (350g, 10 min, 4 °C), counted by hemocytometer with trypan blue exclusion, washed in PBS, and stored at −80 °C. For long-term culture (>24 h), cells were counted and split daily with the addition of medium containing approximately 5% 2H2O. Labeling experiments were performed for at least five doublings to ensure nearly complete (≥97%) labeling. Medium and culture supernatants were analyzed for 2H2O enrichment using isotope ratio mass spectrometry (IRMS) after gravimetric dilution (MRC–HNR [Human Nutrition Research], Widdowson Laboratories, Fulbourn, Cambridge, UK) and equilibration with H2 gas [29].
MHC protein isolation
Immunoprecipitation was performed using modifications of established protocols [30]. Unlabeled and 2H2O-labeled cell pellets were lysed for 1 h at 4 °C in 1% CHAPS (Sigma–Aldrich) or 1% IGEPAL CA-630 (NP40) (Sigma) in 50 mM Tris (pH 8.0), 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), and protease inhibitors (Roche Complete Protease Inhibitors used per manufacturer’s instructions) at a ratio of up to 108 cells/ml lysis buffer. Nuclei and debris were removed by centrifugation (10,000g in a microcentrifuge, 30 min, 4 °C). After preclearing with protein A or protein G Sepharose (20 μl packed beads, Sigma–Aldrich), immunoprecipitation was performed using protein A or protein G Sepharose and 10–30 μg of the appropriate mAb (1 h to overnight, 4 °C). Negative controls included the last preclearing step, irrelevant control antibody, and precipitation from a cell lacking the protein of interest. Pellets were washed repeatedly in lysis buffer with 0.1% detergent. One-dimensional sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed on 12% acrylamide gels under nonreducing conditions. Gels were fixed and stained with Coomassie blue or silver, and bands were excised and stored at or below −20 °C.
Sample preparation and peptide MS
SDS–PAGE gel pieces were thawed and transferred into 96-well polymerase chain reaction (PCR) plates for automated sample preparation in a Mass Prep Station (Waters). Briefly, the gel bands were destained, reduced (dithiothreitol [DTT]), alkylated (iodoacetamide), and digested with trypsin overnight at 37 °C. Thereafter, 20 μl of supernatant was pipetted into a sample vial and loaded onto an autosampler for LC–tandem mass spectrometry (MS/MS) analysis.
All LC–MS/MS and LC–MS experiments were performed using an Eksigent NanoLC-1D Plus (Eksigent Technologies, Dublin, CA, USA) high-performance liquid chromatography (HPLC) system and an LTQ OrbiTrap mass spectrometer (ThermoFisher, Waltham, MA, USA). Peptides were separated by reverse-phase chromatography using a flow rate of 300 nl/min and an LC Packings PepMap 100 C18 column (75 μm i.d. × 150 mm, 3 μm particle size, Dionex, Sunnyvale, CA, USA). Peptides were loaded onto a precolumn (Dionex Acclaim PepMap 100 C18, 5 μm particle size, 100 Å pore size, 300 μm i.d. × 5 mm) from the autosampler with 0.1% formic acid for 5 min at a flow rate of 10 μl/min. The 10-port valve was then switched to allow elution of peptides from the precolumn onto the analytical column. Solvent A was 0.1% aqueous formic, acid and solvent B was acetonitrile + 0.1% formic acid. The gradient employed was 5–50% B in 40 min. The LC eluate was sprayed into the mass spectrometer by means of a New Objective nanospray source. For MS/MS experiments, all m/z values of eluting ions were measured in the OrbiTrap mass analyzer set at a resolution of 7500. Peptide ions with charge states of 2+ and 3+ were then isolated and fragmented in the LTQ linear ion trap by collision-induced dissociation, and MS/MS spectra were acquired.
For peptide identification, the data were processed using Bioworks Browser (version 3.3.1 SP1, Thermo Fisher Scientific). MS/MS data were converted to dta (text) files using the Sequest Batch Search tool (within Bioworks). The data files were converted to a single mgf file using an SSH script in the SSH Secure Shell Client program (version 3.2.9, build 283, SSH Communications). These combined files were then submitted to the Mascot search algorithm (Matrix Science, London, UK) and searched against the mouse or human NCBI database, as appropriate, using a fixed modification of carbamidomethyl (for cysteines) and a variable modification of oxidation (for methionines).
The peptide identifications from the Mascot search were matched to the M0 precursor peak in the chromatograms through careful scrutiny of the MS/MS data contained within the Mascot search result and the MS/MS spectra from the raw data. Once the MS/MS data for individual unlabeled peptides had been matched, the retention times of the peptides were established and this information was used for the subsequent integrations (see below). For quantification of mass isotopomer distributions, selected ion monitoring (SIM) experiments, in which limited m/z regions were exclusively scanned for specific unlabeled or labeled ions, were performed. Peak integration was performed using Xcalibur QualBrowser software (version 2.0, Thermo Fisher Scientific). Briefly, peaks were smoothed within QualBrowser (Boxcar, 7 points) to remove any spikes within the peaks before reconstructed ion chromatograms were generated for each mass isotopomer and the areas under the peaks were measured. The reconstructed ion chromatograms posed no difficulties with respect to baseline drift or variable peak shapes. Peak contamination was rarely seen and was readily detectable by visual inspection of chromatograms when present; the data shown were from uncontaminated chromatograms. Each peak was integrated across the same retention time range to ensure that the areas of the peaks were consistent for each mass isotopomer. Measured peak areas were copied into Microsoft Excel spreadsheets for further analysis. The same peak integration procedure was used for unlabeled, partially labeled, and fully labeled peptides.
Calculations
Cell growth
Log-transformed cumulative cell counts (cell density per milliliter × volume, corrected for removal of cell samples or dilution since the start of the experiment) were plotted as a function of time. Doubling times were calculated from the slope of the best-fitting straight line according to the relationship:
Fractional protein synthesis
Standard nomenclature for mass isotopomer distribution analysis (MIDA) calculations was used [31]. For each peptide, the mass species corresponding to the monoisotopic molecular weight was defined as M0. This species comprises only the lowest atomic weight isotopes of each element (i.e., all 12C/1H/16O/14N/32S). Replacement of 1H by 2H, replacement of 12C by 13C, and so on at any one position adds 1 Da, and the collection of these isotopomers (isotopic isomer) is referred to as the M1 mass isotopomer. Any combination of two of these substitutions, or replacement of 16O by 18O, adds 2 Da, yielding M2 and so on.
Integrated LC peak intensities (designated I) measured across the first-dimension MS mass envelope of the peptide of interest were converted to fractional molar abundances (designated A) by dividing the peak intensity of the mass isotopomer Mi (i = 0, 1, 2, …, N) by the sum of the peak intensities of all (N + 1) quantifiable mass isotopomers in the peptide mass envelope:
To measure the changes in mass isotopomer distributions over time during a continuous 2H2O labeling experiment, the factional molar abundance of each mass isotopomer at baseline was subtracted:
Replicate measurements of Ai and ΔAi were averaged, and standard deviations were calculated.
Fractional protein synthesis (f) was calculated from the fractional abundance of each mass isotopomer using the relationship
| (1) |
where represents the maximal shift in fractional molar abundance of the mass isotopomer from baseline, obtained empirically from fully labeled samples. Estimates of f from different mass isotopomers within one peptide were averaged, and standard deviations were calculated. Mass isotopomers with low values of gave substantially noisier estimates of f (see Results) and were excluded from the analysis.
Mass isotopomer distribution analysis
MIDA uses combinatorial probability algorithms to calculate the theoretical mass isotopomer distribution for any biomolecule both at baseline (i.e., with all isotopes present at their natural abundance) and after stable isotope labeling. MIDA algorithms have been described exhaustively elsewhere [31] and were used in a software implementation kindly provided by R.A. Neese and M.K. Hellerstein.
The expected baseline mass isotopomer distributions, Ai,baseline, were calculated from the elemental composition of each peptide, which was determined from its sequence and corrected for added protons (+1/charge acquired) and for carbamidomethylation of any Cys residues present. In addition, the mass isotopomer distributions of fully labeled peptides were modeled. In the simplest model, used throughout the main text, the measured 2H2O enrichment in medium was taken to represent the precursor pool enrichment (designated p in the standard nomenclature used in MIDA calculations [31]). Mass isotopomer distributions were then calculated, using MIDA software, for different numbers of labeling sites (designated n). Alternative models (see Supplementary material) invoking uniform label dilution were calculated with MIDA software using the indicated values of n and p. A nonuniform dilution model was assembled in two steps. First, the baseline mass isotopomer distribution and the contribution of the fully labeled hydrogens were modeled using MIDA software. Second, a spreadsheet program was used to adjust for the contribution of the partially labeled hydrogen atoms using Eq. (A3) of Ref. [31].
ΔAi values can be determined accurately only for those mass isotopomers that can be quantified in both the unlabeled and labeled samples [31]. To allow comparison of MIDA calculations to experiment, the predicted distributions were truncated to include only the (N + 1) mass isotopomers that were experimentally quantifiable and normalized to add up to 100%:
The truncation perturbs the linear relationship between measured mass isotopomer abundances and f [31]; however, this bias is negligible when the measured mass isotopomer distributions account for more than 90% of the total mass envelope [31], as was the case here (data not shown). The goodness of fit of the observed mass isotopomer distributions, Ai,obs was assessed by calculating root mean square deviations (RMSDs) from the truncated, normalized MIDA values:
where the summation was performed over all (N + 1) observed mass isotopomers.
Results
Approach and peptide identification
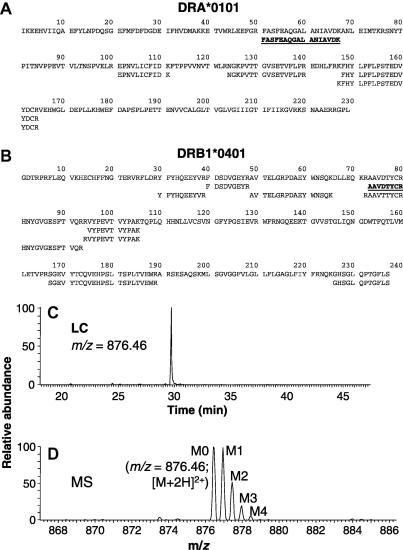
Initially, the synthesis of HLA-DR0401 molecules in Priess cells was analyzed using the SINEW approach with peptide LC–MS analysis (Fig. 1B). Cells grew indistinguishably in the presence and absence of 4.5% 2H2O in medium (Fig. 1C); the 2H2O enrichment remained constant throughout the culture period (Fig. 1D). HLA-DR αβ heterodimers were immunoprecipitated and visualized by Coomassie blue staining after SDS–PAGE analysis (Fig. 1E). To identify tryptic peptides suitable for kinetic analysis, unlabeled α- and β-chain bands were excised, digested with trypsin, and analyzed by LC–MS/MS for peptide sequencing. Matches to the DRα and DR0401β protein sequences were identified using the Mascot search algorithm and are shown in Fig. 2A and B. Peptides rich in Ala and/or Gly, two of the amino acids that are nearly fully labeled from 2H2O in mice [6], were identified in both chains (Table 1; underlined in Fig. 2A and B). The quantification of mass isotopomer distributions in the relevant m/z range is shown for the DRα (51–67) peptide in Fig. 2C and D. A single LC peak of the expected m/z ratio was observed for the lowest (M0) mass isotopomer of the peptide, comprising only the lowest atomic weight isotopes of each element (Fig. 2C). Even in the absence of 2H2O labeling, a series of mass isotopomers (M0–M4) was present at this retention time (Fig. 2D) owing to the presence of stable isotopes in nature (e.g., 1.09% 13C, 0.02% 2H). The individual mass peaks were separated by 0.5 m/z units due to the z = 2 charge state of this peptide. The identity of this peptide was confirmed by comparing its MS/MS spectrum, extracted from raw data, with the MS/MS spectrum embedded in the Mascot search results (data not shown).
Fig. 2.
Identification of peptides suitable for 2H2O labeling studies. (A and B) Identification of Ala-/Gly-containing HLA-DR4-derived tryptic peptides. DRα (A) and DRβ (B) bands excised from SDS gels of L243 immunoprecipitates from Priess cell extracts were reduced, carbamidomethylated, digested with trypsin, and analyzed by LC–MS/MS. Alignments of tryptic fragments to the sequence of their parent polypeptides are shown. Peptides selected for analysis are shown in bold type and underlined. (C and D) LC–MS analysis of mass isotopomer distributions for the DRα peptide, FASFEAQGALANIAVDK. (C) LC chromatogram of a DRα digest, with SIM for m/z = 876.45, corresponding to the M0 mass isotopomer of the doubly charged peptide. Integration of the principal LC peak for this mass isotopomer, and for higher order mass isotopomers, was used to quantify mass isotopomer distributions. (D) Mass spectrum for the LC peak of the intact DRα peptide showing a single, well-resolved set of mass isotopomers.
Table 1.
MHC protein-derived peptides used for analysis of 2H2O labeling.
| Cell | mAb | Polypeptidea | AAsb | Sequencec | m0/zd | ze |
|---|---|---|---|---|---|---|
| Priess | L243 | DRα | 51–67 | FASFEAQGALANIAVDKf | 876.46 | 2 |
| DR0401β | 73–80 | AAVDTYCR f | 478.22 | 2 | ||
| LCL721g | W6/32 | B8 | 132–145 | SWTAADTAAQITQR | 760.28 | 2i |
| B51 | 122–145 | DYIALNEDLSSWTAADTAAQITQR | 1327.48 | 2i | ||
| 317–338 | GGSYSQAASSDSAQGSDVSLTA | 1023.72 | 2 | |||
| A20 | MKD6 | Aα | 43–65 | LPEFGQLILFEPQGGLQNIAAEKh | 838.13 | 3 |
| M12.NOD | OX-6 | |||||
| A20 | MKD6 | Adβ | 73–80 | AEVDTACR | 461.21 | 2 |
| M12.NOD | OX-6 | Ag7β | 73–80 | AELDTACR | 468.22 | 2 |
Mascot searches identified multiple peptides from the indicated HLA or H-2 polypeptides following specific immunoprecipitation using cells and antibodies as shown. Peptides were selected on the basis of their Ala/Gly content and polymorphism.
Amino acid sequence numbering refers to the mature protein.
Amino acid sequences in single-letter code. Ala/Gly residues (whose side chain C—H bonds are known to be accessible to labeling from 2H2O) are underlined, and relevant polymorphisms are in bold.
Mass-to-charge ratios experimentally determined by LC–MS, where m0 mass represents the lowest mass isotopomer.
Charge determined by the spacing of peaks in the MS mass envelope.
The full set of HLA-DR0401-derived tryptic peptides is shown in Fig. 2A and B. No peptides were found to match the DRB4⁎0101-encoded DRw53 β-chain, which is coexpressed in the DR4 haplotype and pairs with DRα; thus, DRw53 might not have been coisolated in substantial amounts.
LCL721 is heterozygous in the MHC class I region; allele-specific HLA-B peptides were selected for analysis. Both polymorphisms within the peptide and adjacent polymorphisms affecting tryptic cleavage can be exploited; the latter is illustrated here by the Ser131 polymorphism in the B51 (122–145) peptide, which destroys a tryptic cleavage site present in B8.
The α-chains of Ad and Ag7 are identical; the peptide shown was found in MKD6 immunoprecipitates from A20 cells (Ad) and in OX-6 immunoprecipitates from M12.NOD (Ag7).
The z = 3 triply charged peptide species were also present, albeit at lower abundance.
Analysis of mass isotopomer distributions
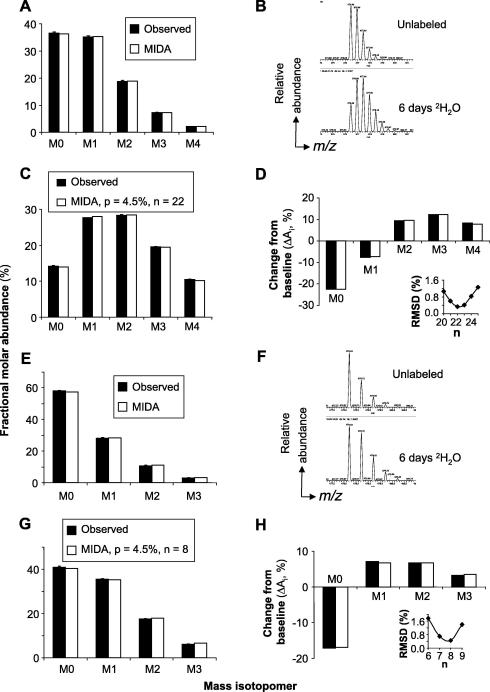
The LC peak of each of the mass isotopomers of the unlabeled DRα peptide was integrated, and its abundance was expressed as a percentage of the total abundance of the M0 to M4 mass isotopomers (Fig. 3A). Standard deviations (SDs) of six replicate injections ranged from 0.08% to 0.43% for the different mass isotopomers, indicating high analytical precision. MIDA allows baseline fractional molar abundances to be predicted from the elemental composition of the peptide; the combinatorial probability algorithms used in MIDA have been reviewed [31]. The data agreed with MIDA predictions (Fig. 3A) to within experimental error; the RMSD between MIDA prediction and experiment was 0.15% for the average of the replicates and ranged from 0.25% to 0.38% for the six individual determinations. The measured distributions were also reproducible between experiments (see Fig. S1a in Supplementary material). When input cell numbers were varied over two orders of magnitude, the abundance of the DR molecules recovered, and of the DRα peptide after LC–MS analysis, varied accordingly (Fig. S1b). The shape of the mass isotopomer distributions was maintained at lower analyte abundance, albeit with larger deviations from MIDA predictions (Fig. S1b). Thus, the mass isotopomer distributions of tryptic peptides are measured accurately and precisely by LC–MS, with only modest loss of precision at low analyte abundance; moreover, RMSD values represent a useful estimate of measurement accuracy.
Fig. 3.
Mass isotopomer distributions of HLA-DR4-derived tryptic peptides. (A–D) Analysis of DRα (51–67) peptide. (A) MIDA model and experimental quantification (means ± SDs of N = 6 replicate injections) of mass isotopomer distributions of the unlabeled DRα peptide. (B) Mass spectrum of the DRα peptide obtained before and after 6 days of culturing Priess cells in 4.5% 2H2O. (C) MIDA model and experimental quantification (means ± SDs, N = 5) of mass isotopomer distributions after 2H2O labeling. The model assumes that n = 22 labeling sites are accessible to label without dilution of the precursor pool enrichment (p) relative to the 2H2O enrichment in medium; the value of n was adjusted for optimal fit to the data. (D) The difference between the mass isotopomer distributions of the fully labeled and unlabeled DRα peptide is shown. The inset shows the RMSD of the experimental data from a series of models in which p was kept at 4.5% and n was varied. The lowest RMSD value was obtained at n = 22. (E–H) The same analysis was applied to the DR0401β (73–80) peptide: (E) unlabeled mass isotopomer distributions (means ± SDs, N = 5 replicates); (F) mass spectra of unlabeled versus extensively labeled peptide; (G) mass isotopomer distributions of labeled peptide (means ± SDs of N = 5 replicate analyses, RMSD = 0.45% vs. model with n = 8 labeling sites and p = 4.5%); (H) difference analysis and dependence of model fit on n (inset).
Next, the shifts in peptide mass isotopomer distributions were evaluated after growing Priess cells in 2H2O for approximately five doublings (Fig. 1C and D), so that approximately 97% of the cells, and thus at least 97% of protein, would incorporate 2H at 4.5% 2H2O enrichment. 2H2O labeling caused a visible shift in the mass isotopomer distribution of the tryptic DRα (51–67) peptide (Fig. 3B and C). The SD of the fractional molar abundances of the M0 to M4 mass isotopomers after five replicate injections ranged from 0.10% to 0.18% (Fig. 3C). By comparison, the changes in fractional mass isotopomer abundance on labeling were substantial, ranging from −22.5% for M0 to +12.3% for M3 (Fig. 3D).
MIDA algorithms were used to model the effects of 2H2O labeling on the mass isotopomer distributions [31]. The shape of the labeled distributions depends on the number of labeling sites (n) and the isotope enrichment in the precursor pool (p). During 2H2O labeling, 2H atoms enter the peptide individually at an unknown number of labeling sites within the NEAAs present; the precursor/product relationship depends on the amino acid composition of the peptide. The simplest model involves a fixed number (n) of biosynthetic labeling sites, at each of which 2H is biosynthetically incorporated from 2H2O without label dilution from competing sources (i.e., at p = 4.5% 2H enrichment in this experiment). This model gave excellent agreement with the labeled mass isotopomer distributions when the value of n was adjusted for optimal fit to the data (n = 22–23 in several independent determinations, RMSD = 0.2–0.3%) (Fig. 3C and D, and Table 2). The optimal value of n was well matched to the total number of C—H bonds [22] nominally attributable to the side chains of Ala (n = 4/residue) and Gly (n = 2). Thus, 2H incorporation into the DRα peptide under these culture conditions is almost entirely accounted for by labeling of Ala and Gly, although contributions from other amino acids, and label dilution in Ala and Gly, were not ruled out by our data.
Table 2.
Summary of MIDA calculations.
| Peptidea | n (Ala+Gly)b | Experiment | Source | Labeling (days) | 2H2O (p) c (%) | n (experimental)d | RMSD (%)e |
|---|---|---|---|---|---|---|---|
| DRα (51–67) | 22 | 1 | Priess | 6 | 4.5 | 22 | 0.33 |
| 2 | Priess | 6f | 4.5 | 22 | 0.33 | ||
| 8 | 4.5 | 23 | 0.20 | ||||
| 3 | DC | Nil | – | – | 2.02 | ||
| 3 | 4.9 | 22 | 0.75 | ||||
| DR0401β(73–80) | 8 | 1 | Priess | 6 | 4.5 | 8 | 0.54 |
| 2 | Priess | 6f | 4.5 | 8 | 0.45 | ||
| 8 | 4.5 | 9 | 0.53 | ||||
| B8 (132–145)g | 16 | 1 | LCL721 | Nil | – | – | 0.27 |
| 7 | 4.5 | 15 | 0.32 | ||||
| B51 (122–145)g | 20 | LCL721 | Nil | – | – | 0.67 | |
| 7 | 4.5 | 28 | 0.23 | ||||
| B51 (317–338) | 22 | 1 | LCL721 | Nil | – | – | 0.58 |
| 7 | 4.5 | 24 | 0.59 | ||||
| Aα (43–65) | 14 | 1 | A20 | Nil | – | – | 0.33 |
| 6 | 4.8 | 22 | 0.57 | ||||
| 2 | A20 | Nil | – | – | 0.26 | ||
| 5 | 5.6 | 24 | 0.87 | ||||
| 3 | M12.NOD | Nil | – | – | 0.22 | ||
| 5 | 5.0 | 26 | 0.17 | ||||
| 4 | M12.NOD | Nil | – | – | 0.26 | ||
| 5 | 5.6 | 24 | 0.87 | ||||
| 5 | Balb/c spl | Nil | – | – | 0.33 | ||
| Adβ (73-80) | 8 | 1 | A20 | Nil | – | – | 0.27 |
| 5 | 4.8 | 9 | 0.57 | ||||
| 2 | Balb/c spl | Nil | – | – | 0.27 | ||
| Ag7β (73–80) | 8 | 1 | M12.NOD | Nil | – | – | 1.73 |
| 6 | 5.0 | 11 | 0.29 |
See Table 1 for sequences and masses.
Number of C—H bonds in Ala and Gly residues of the peptide.
2H2O enrichment in medium (atom % excess over natural abundance) measured by IRMS.
MIDA calculations were performed using medium 2H2O enrichment to represent p and varying the number of labeling sites (n); the value of n that gave the best fit to the data is given.
RMSD values indicate the goodness of fit of the optimized MIDA model (per noted) to the data.
Average of five replicate analyses shown in Fig. 3. Statistics for the corresponding unlabeled DR mass isotopomer distributions are given in the text, Fig. 3, and Fig. S1 in the Supplementary material.
Data for the z = 2 charge state are given. Triply charged (z = 3) species gave similar best-fit values of n (14 for B8 peptide, 27 for B51) but gave greater RMSD values related to their lower abundance.
The DR0401β (73–80) peptide was also analyzed. Its unlabeled mass isotopomer distribution was reproducible, conforming to MIDA predictions (Figs. 3E and S1c). SDs ranged from 0.09% to 0.43% for different mass isotopomers; RMSD was 0.43% for the average of five replicates, and RMSDs for individual determinations ranged between 0.19% and 0.71%. For the labeled samples, the best model fit to the data was obtained assuming n = 8 or 9 labeling sites labeled at p = 4.5% 2H enrichment (Fig. 3F–H and Table 2). This labeling pattern is well matched to the eight labeling sites attributable to the two Ala residues in this peptide.
We wished to examine whether the MHC-derived peptides also incorporate the 2H label at exchangeable sites, such as solvent-exposed O—H and N—H bonds [32], which would confound the analysis of biosynthetic label incorporation at nonexchangeable C—H bonds. Thus, unlabeled Priess cells were lysed in 2H2O-enriched lysis buffer, immunoprecipitated, and washed in unenriched buffer. LC–MS analysis revealed no distortion of the unlabeled mass isotopomer distributions due to 2H2O exposure post lysis (Fig. S1d). Thus, label retention at exchangeable sites does not confound measurements of protein synthesis.
The analysis was extended to other MHC proteins. Human HLA-A, -B, and -C class I molecules were isolated from EBV-transformed B cells, and murine H2-Ag7 class II molecules were isolated from M12.NOD transfectants (Fig. 1F and G). H2-Ad molecules, isolated from A20 B lymphoma cells, were obtained in lower abundance and detected by silver staining (not shown). LC–MS/MS analyses identified suitable peptides within H2-Ad and H2-Ag7 α- and β-chains (Table 1; note that both alleles share the same α-chain). A mixture of HLA-B8 and -B51 heavy chain-derived peptides was identified in HLA class I immunoprecipitates from LCL721 cells, allowing the identification of Ala-/Gly-rich peptides that were specific for either allele (Table 1). The unlabeled mass isotopomer distributions of all peptides conformed to MIDA predictions (Table 2 and Fig. S2 in Supplementary material, left panels). The mass isotopomer distributions after extensive 2H2O labeling were substantially shifted and were modeled well by assuming a fixed number of labeling sites (n) that incorporate 2H at the measured body water enrichment (Table 2 and Fig. S2, right panels). For several peptides, the best-fit values of n were close to the number of C—H bonds attributable to Ala and Gly. For the B51 (122–145) and Aα (43–65) peptides, however, only 20 of 28 and 14 of approximately 24 labeling sites, respectively, were explained by Ala and Gly, implying contributions from other amino acids to labeling (Table 2).
Quantification of fractional protein synthesis
In metabolic labeling experiments, the enrichment in the isotope-labeled precursor pool is generally kept constant, so that newly synthesized molecules incorporate the label to the same extent regardless of when they are synthesized. In the SINEW approach, however, the NEAA tRNAs used for protein synthesis are labeled indirectly from 2H2O. At early time points, label equilibration might be incomplete, so that p or n might increase over time. This would confound the estimation of fractional protein synthesis (f) from 2H2O labeling data. Previous work has shown that C—H bonds of Ala and Gly precursors are rapidly labeled from 2H2O [5–7,10], but considerable label dilution has been noted in Pro [15] and data for 2H2O labeling of other NEAAs in cell culture are unavailable. Thus, the possibility of delayed label entry was a concern, particularly for peptides whose label incorporation patterns were only partially accounted for by their Ala and Gly content.
Therefore, we considered the behavior of mass isotopomer distributions during a labeling time course assuming that there is no change in p or n. If this assumption is correct, old molecules, which retain the baseline mass isotopomer distribution, will gradually be replaced (during protein turnover) or diluted (due to cell growth) with new molecules, all of which possess a mass isotopomer distribution identical to that of fully labeled molecules. Thus, the mass isotopomer distribution, Ai(t), of a partially turned-over protein pool (i.e., the fractional molar abundance, A, of mass isotopomer i at time t) is the weighted average of the unlabeled [Ai(0)] and fully labeled () mass isotopomer distributions; the weighting factor, f(t), represents fractional protein synthesis:
Thus, fractional protein synthesis, f(t), can be calculated as
| (1) |
Therefore, if there is no change in precursor/product relationships during the experiment, each mass isotopomer will change from its baseline to its fully labeled value at the same rate, characteristic of the rate of fractional protein synthesis, and will provide an independent estimate of f. In contrast, if the label equilibrates poorly during early time points, this will distort the mass isotopomer distributions of molecules synthesized early on; therefore, analysis of different mass isotopomers using Eq. (1) will yield systematically different estimates of f (see Fig. S3 in Supplementary material).
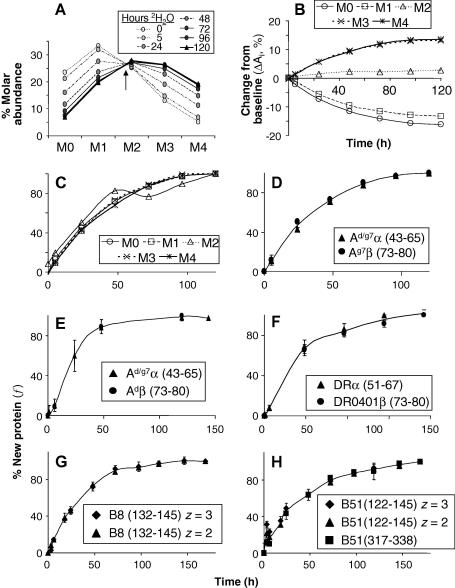
Accordingly, we examined whether the analysis of different mass isotopomers yields convergent estimates of f according to Eq. (1). Fig. 4A shows the mass isotopomer distributions of the H2-Aα (43–65) peptide from Ag7 molecules isolated after varying times of 2H2O labeling of M12.NOD cells. All intermediate distributions intersected at a common “iso-abundant” point near M2 (arrow). This observation was consistent with the intermediate distributions representing binary mixtures of unlabeled and fully labeled molecules (analogous to the isosbestic point found in absorbance spectra of binary reactant/product mixtures that is lost in more complex mixtures). Fig. 4B shows the abundance change of each mass isotopomer over time; values of f, calculated from each mass isotopomer using Eq. (1), are shown in Fig. 4C. All mass isotopomers gave convergent estimates of f. Estimates from M2, however, were less precise because M2 changed much less than the other mass isotopomers during the experiment, resulting in a bigger impact of analytical error on estimates of f. A similar time course was observed for the Ag7β (73–80) peptide, further validating this method of estimating f (Fig. 4D).
Fig. 4.
Measurement of fractional MHC protein synthesis. (A–D) Analysis of H2-Aα peptide obtained from Ag7 immunoprecipitates after varying times of continuous labeling of M12.NOD cells. (A) Mass isotopomer distributions after different times of labeling. Note the intersection of all mass spectra at an iso-abundant point near M2 (arrow). (B) Evolution of fractional mass isotopomer abundances from their baseline values as a function of labeling time. Each mass isotopomer shifted from its baseline value to its fully labeled abundance with the same kinetics, reflecting the rate of fractional protein synthesis. (C) Convergent estimates of fractional H2-Ag7 protein synthesis as a function of labeling time obtained from analysis of different mass isotopomers of the Aα peptide. (D) Indistinguishable estimates of fractional H2-Ag7 protein synthesis obtained from analysis of α- and β-chain peptides. Data were averaged for the different mass isotopomers within each peptide (excluding those that exhibited experimental noise due to small abundance shifts such as M2 in the α-chain peptide). Error bars represent SDs and in most instances are smaller than the symbols. Similar analyses were performed for α- and β-chain peptides of H2-Ad molecules isolated from A20 cells (E) and for HLA-DR4 molecules isolated from Priess cells (F). (G and H) The indicated peptides specific for the HLA-B8 (G) and HLA-B51 (H) alleles were analyzed after W6/32 immunoprecipitation from LCL721 cells. In panel G, two charge states of the same peptide were analyzed. In panel H, two peptides were analyzed, and one of these was present in two charge states, as indicated.
The labeling kinetics of H2-Ad from A20 cells (Fig. 4E), HLA-DR0401 from Priess cells (Fig. 4F), HLA-B8 (Fig. 4G), and HLA-B51 (Fig. 4H) from LCL721 were also analyzed. In each case, convergent estimates of f were obtained from different mass isotopomers within any one peptide. This result was consistent with the hypothesis that the precursor/product relationships for peptide labeling from 2H2O are effectively independent of time, justifying the use of Eq. (1) to calculate f from 2H2O-labeled peptide mass isotopomer distributions. The estimates of f were validated by comparison between different charge states of the same peptide, different peptides of the same polypeptide, or a peptide from a partner chain (Figs. 4D–H). The estimates were less precise for H2-Ad molecules; this was attributed to the low expression of these molecules in A20 cells, resulting in low MS abundance. The fractional synthesis rates measured in these experiments corresponded to half-lives on the order of approximately 1 day (Fig. 4D–H), with different contributions from cell growth and from replacement of turned-over proteins to fractional synthesis, depending on the MHC molecule and its cellular background (A. De Riva et al., manuscript in preparation).
Studies in primary APCs
This analytical approach also was feasible for primary antigen-presenting cells (APCs) cultured in vitro or obtained ex vivo. Unlabeled and fully labeled mass isotopomer distributions of the DRα peptide, obtained from immature human monocyte-derived dendritic cells, and the baseline mass isotopomer distributions of the Ad α and β peptides, obtained from Balb/c splenic B cells, closely conformed to MIDA predictions (Table 2). Thus, the SINEW approach will be applicable to primary APCs.
Discussion
Here we have used peptide LC–MS, in conjunction with the SINEW approach (Fig. 1B), to quantify the fractional synthesis of various MHC proteins. In the process of validating this method, we tested key underlying assumptions, explored the generality and versatility of the method, and greatly simplified the algorithms used to calculate fractional protein synthesis from experimental data. Importantly, the method is capable of distinguishing turnover rates of closely related MHC protein allotypes present in the same cell. Collectively, these improvements substantially broaden the utility of the SINEW approach.
Specificity is achieved at several steps in the procedure. MHC proteins of interest are enriched by immunoprecipitation followed by SDS–PAGE. However, contamination with the immunoprecipitating antibody, noncovalently associated or copurified proteins, and keratin remains detectable by LC–MS/MS (data not shown). Nonetheless, suitable peptides specific for the proteins of interest are found, and uncontaminated mass envelopes are obtained, as indicated by the close match to MIDA predictions. Unique peptide masses and retention times allow discrimination between peptides derived from MHC class I and class II allelic variants even when several MHC molecules are present. In Fig. 4G and H, we analyzed two alleles encoded at the HLA-B class I MHC locus, but HLA-A and -C were coisolated using the W6/32 antibody. Preliminary studies in another lymphoblastoid cell line (not shown) suggest that the entire class I haplotype will be resolvable provided that the HLA type is available. Thus, peptide LC–MS greatly improves analytical specificity in the SINEW approach compared with the analysis of 2H incorporation into protein-derived NEAAs after total hydrolysis. The tolerance of our method to complexity in the protein samples suggests that our approach may have applications in kinetic proteomics.
Measured mass isotopomer abundances must be both precise and accurate to enable determination of f by stable isotope labeling [31]. OrbiTrap MS analysis satisfies these criteria, comparing favorably with the performance of other MS modalities in previous studies [16,17]; the data are analytically precise (SDs = 0.2–0.3% for high-abundance samples) and accurate (abundance independent and consistent with MIDA predictions). Accuracy diminishes somewhat at lower analyte abundance; thus, the best results require minimization of sample losses throughout the procedure. Nonetheless, analysis of MHC molecules isolated from as few as 105 EBV-B cells was possible, comparing favorably with 35S radiolabeling. Substantial shifts in abundance are observed after complete 2H2O labeling on the order of −20% for M0 and +10% for other mass isotopomers in each peptide. Thus, estimation of f to within a few percentage points is possible, provided that mass isotopomers that show the greatest fractional abundance shifts during labeling are selected for analysis. Allowing for the contribution of cell growth to fractional synthesis measured by our approach, our measurements (Fig. 4 and data not shown) are compatible with the MHC protein half-lives measured previously by radiolabeling in immortalized B cell lines [30,33] and monocyte-derived dendritic cells [34].
The success of this approach relies on the identification of peptides that incorporate sufficient label from 2H2O. Prospective identification of such peptides from their amino acid sequence remains a complex challenge because the efficiency with which each amino acid incorporates the 2H label depends on the activity of complex biosynthetic pathways and on the abundance of unlabeled amino acids present in culture medium, and this may dilute any label incorporated via endogenous amino acid synthesis to an unknown extent. Previous work showed that C—H bonds in Ala and Gly are nearly fully equilibrated with 2H from 2H2O in vivo in rodents, contributing n = 4 and n = 2 labeling sites each, respectively, to peptides containing these amino acids [6,11]. We reasoned that these amino acids should be well labeled in vitro as well based on previous studies showing good labeling of protein-derived Ala in cell culture [13] and based on the lack of Ala and a modest amount of Gly present in RPMI medium. Accordingly, we preselected peptides on the basis of Ala/Gly content. In several of the peptides examined, Ala or Gly labeling explained most or all of the labeling sites inferred from MIDA calculations, but this did not rule out contributions from other amino acids that would have been expected from previous work [11]. Indeed, in other peptides, such as the Aα peptide, a substantial proportion of labeling sites must have arisen from labeling of other NEAAs. Similarly, in previous studies performed in vivo, some well-labeled peptides that were not rich in Ala/Gly were found [16,17]. A preliminary analysis of 21 peptides (including data in Table 1 and unpublished work) reinforces our impression that numerous amino acids contribute to labeling in our system (data not shown).
Counting of Ala and Gly residues, therefore, will often underpredict the extent of labeling and will miss some well-labeled peptides. However, most proteins contain some Ala-/Gly-rich peptides, and other well-labeled peptides may be identified empirically if needed, so this does not appear to be a serious limitation in practice. Moreover, we observed no peptides in which the apparent number of labeling sites (n) was markedly less than that attributable to Ala/Gly, suggesting that the counting of Ala and Gly provides a useful lower bound on the number of labeling sites that may be expected. Importantly, this lower bound is likely to apply in vivo in rodents, where Ala and Gly are known to be well labeled. A more complete and accurate set of rules for predicting highly labeled peptides from amino acid sequence will require refinement of predictive algorithms and validation in larger data sets.
Given the potential complexity of precursor/product relationships for 2H2O labeling of NEAAs, it is interesting that a simple MIDA model, in which a fixed number of labeling sites are assumed to be labeled at the 2H enrichment in medium water, accounts quantitatively for the observed labeling patterns, as was also found previously for selected rodent serum proteins [17]. However, more complex MIDA models, invoking label dilution in some or all of the NEAA precursor pools, fit the data equally well (see Fig. S4 in Supplementary material). Thus, neither the experimental data nor the MIDA models are sensitive to the biochemical details that govern labeling of peptides from 2H2O. Indeed, the calculation of fractional protein synthesis, using Eq. (1), makes no assumptions about the precursor/product relationships, as captured in MIDA calculations (values of n and p), nor does it assume any knowledge as to the metabolic source of the label (e.g., whether 2H is introduced via Ala, Gly, or any other amino acid). For this reason, the approach outlined here permits calculation of fractional protein synthesis despite our incomplete understanding of the biochemical details underlying the observed labeling patterns.
The use of Eq. (1) to calculate f represents an application of the rise-to-plateau approach [35]. This is a rigorous approach for determining fractional synthesis that is feasible when a stable isotope label can be administered continuously for extended periods, allowing empirical definition of plateau labeling. Labeling to plateau is possible in the SINEW approach both in cell culture and in living animals [6]. The only additional assumption inherent in Eq. (1) is that the precursor/product relationship remains constant on the time scale of the experiment, which was consistent with our empirical results. It should be noted that the assumption of a constant precursor/product relationship also underlies the MIDA-type algorithms used in previous work to calculate f [16,17], although this assumption was not previously tested experimentally. Moreover, our model calculations (see Figs. S3 and S5 in Supplementary material) show that minor violations of these assumptions (small fluctuations in values of n or p or delayed label equilibration from 2H2O into NEAA precursor pools) have only minimal impact on estimates of f. This argues for the robustness of our approach. Operationally, Eq. (1) greatly simplifies calculation of f and of the associated experimental error.
Our validation studies focused on cultured, immortalized B cell lines, but the method also is applicable in human monocyte-derived dendritic cells and murine splenic B cells. By extrapolation from our results, in vivo studies in rodents appear to be feasible, consistent with previous work on abundant serum proteins [16,17]. 2H2O enrichments of 5%, used in the cell culture studies presented here, are routinely used for extended continuous labeling of rodents in vivo; 2H2O label equilibration with Ala and Gly, which are present in the peptides we selected, is rapid and nearly complete in vivo [5–7,10]. Moreover, overall label incorporation in cell culture appears to be less complete than would be expected from the data of Commerford and coworkers [11] in mice (based on data in Table 1 and unpublished observations). This is likely due to competition with unlabeled exogenous amino acids in medium and serum. These considerations suggest that the values of n in vivo will not be the same as in cell culture but rather will generally be higher. This would pose no difficulty in calculating f from shifts in mass isotopomer distributions in vivo and may even make in vivo analyses more robust. However, the precursor/product relationships (best-fit values of n for the peptides of interest) would need to be redetermined in vivo because they cannot be simply extrapolated from results obtained in cell culture.
Further work will be required to examine whether the approach can be extended to human in vivo studies because lower 2H2O enrichments (1–2% in plasma) are routinely attained in humans and additional sources of label dilution may be present [6]. However, MIDA calculations predict that at 1–2% 2H2O enrichment, the shifts in mass isotopomer abundance for fully labeled HLA-B and -DR peptides would range from −4% to −13% in M0 and from +2% to +8% in the best higher order mass isotopomers. This may be sufficient to estimate f.
The approach can readily be extended to other proteins. A powerful feature of 2H2O labeling is the ability to combine studies of protein dynamics with analyses of lipid synthesis [36] and cell proliferation (measured as DNA synthesis, reviewed in Ref. [8]) using the same biosynthetic label. The use of peptide LC–MS in SINEW experiments promises to become a convenient and powerful tool for studies of protein synthesis and turnover in a variety of biomedically relevant in vitro settings and ultimately in kinetic proteomics. The ability to distinguish biosynthetic rates of protein allomorphs, in particular, will be an invaluable tool for dissecting the role of protein dynamics in genotype/phenotype relationships. Extension to in vivo settings appears to be feasible and would open up a wide range of potential applications in which synthesis and turnover of long-lived proteins may be measured in healthy human populations and animal models and their perturbation by diseases and therapeutic interventions may be quantified.
Acknowledgments
This work was funded by a project grant from Cambridge Arthritis Research Endeavour, the Stanley Elmore Senior Research Fellowship at Sidney Sussex College, and a Senior Research Fellowship from Arthritis Research UK (formerly Arthritis Research Campaign, 18543) to R.B. We thank Leslie Bluck and Marilena Leventi (MRC–Human Nutrition Research, Widdowson Laboratories) for IRMS analysis of 2H enrichments in medium, and we thank Jenny Phillips, Anne Cooke, J. S. Hill Gaston (Departments of Pathology and Medicine, University of Cambridge), and Brigitta Stockinger (Division of Molecular Immunology, MRC–National Institute for Medical Research) for providing cell lines and antibodies. Jane C. Goodall and Claudia Prevosto provided help with dendritic cell culture, and Michael J. Bacon and Svenja Hester provided technical assistance. Richard A. Neese and Marc K. Hellerstein (Department of Nutritional Sciences and Toxicology, University of California, Berkeley) kindly provided software for MIDA calculations.
Footnotes
Abbreviations used: SILAC, stable isotope labeling of amino acids in cell culture; tRNA, transfer RNA; SINEW, stable isotope labeling of nonessential amino acids with heavy water; 2H2O, heavy water; NEAA, nonessential amino acid; GC–MS, gas chromatography–mass spectrometry; LC, liquid chromatography; MHC, major histocompatibility complex; EBV, Epstein–Barr virus; HLA, human leukocyte antigen; mAb, monoclonal antibody; PBS, phosphate-buffered saline; FBS, fetal bovine serum; IRMS, isotope ratio mass spectrometry; EDTA, ethylenediaminetetraacetic acid; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; DTT, dithiothreitol; MS/MS, tandem mass spectrometry; HPLC, high-performance liquid chromatography; SIM, selected ion monitoring; MIDA, mass isotopomer distribution analysis; f, fractional protein synthesis; p, precursor pool enrichment; n, number of sites of label incorporation; RMSD, root mean square deviation; SD, standard deviation; APC, antigen-presenting cell.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ab.2010.04.018.
Appendix A. Supplementary data
References
- 1.Papageorgopoulos C., Caldwell K., Shackleton C., Schweingrubber H., Hellerstein M.K. Measuring protein synthesis by mass isotopomer distribution analysis (MIDA) Anal. Biochem. 1999;267:1–16. doi: 10.1006/abio.1998.2958. [DOI] [PubMed] [Google Scholar]
- 2.Kruger M., Moser M., Ussar S., Thievessen I., Luber C.A., Forner F., Schmidt S., Zanivan S., Fassler R., Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Harsha H.C., Molina H., Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat. Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 4.Beynon R.J., Pratt J.M. Metabolic labeling of proteins for proteomics. Mol. Cell. Proteomics. 2005;4:857–872. doi: 10.1074/mcp.R400010-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Dufner D.A., Bederman I.R., Brunengraber D.Z., Rachdaoui N., Ismail-Beigi F., Siegfried B.A., Kimball S.R., Previs S.F. Using 2H2O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1277–E1283. doi: 10.1152/ajpendo.00580.2004. [DOI] [PubMed] [Google Scholar]
- 6.Busch R., Kim Y.K., Neese R.A., Schade-Serin V., Collins M., Awada M., Gardner J.L., Beysen C., Marino M.E., Misell L.M., Hellerstein M.K. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim. Biophys. Acta. 2006;1760:730–744. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 7.Previs S.F., Fatica R., Chandramouli V., Alexander J.C., Brunengraber H., Landau B.R. Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end-stage renal disease. Am. J. Physiol. Endocrinol. Metab. 2004;286:E665–E672. doi: 10.1152/ajpendo.00271.2003. [DOI] [PubMed] [Google Scholar]
- 8.Busch R., Neese R.A., Awada M., Hayes G.M., Hellerstein M.K. Measurement of cell proliferation by heavy water labeling. Nat. Protoc. 2007;2:3045–3057. doi: 10.1038/nprot.2007.420. [DOI] [PubMed] [Google Scholar]
- 9.Turner S.M., Roy S., Sul H.S., Neese R.A., Murphy E.J., Samandi W., Roohk D.J., Hellerstein M.K. Dissociation between adipose tissue fluxes and lipogenic gene expression in ob/ob mice. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1101–E1109. doi: 10.1152/ajpendo.00309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belloto E., Diraison F., Basset A., Allain G., Abdallah P., Beylot M. Determination of protein replacement rates by deuterated water: validation of underlying assumptions. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1340–E1347. doi: 10.1152/ajpendo.00488.2006. [DOI] [PubMed] [Google Scholar]
- 11.Commerford S.L., Carsten A.L., Cronkite E.P. The distribution of tritium among the amino acids of proteins obtained from mice exposed to tritiated water. Radiat. Res. 1983;94:151–155. [PubMed] [Google Scholar]
- 12.Raman A., Schoeller D.A., Subar A.F., Troiano R.P., Schatzkin A., Harris T., Bauer D., Bingham S.A., Everhart J.E., Newman A.B., Tylavsky F.A. Water turnover in 458 American adults 40–79 yr of age. Am. J. Physiol. Renal Physiol. 2004;286:F394–F401. doi: 10.1152/ajprenal.00295.2003. [DOI] [PubMed] [Google Scholar]
- 13.Fanara P., Banerjee J., Hueck R.V., Harper M.R., Awada M., Turner H., Husted K.H., Brandt R., Hellerstein M.K. Stabilization of hyperdynamic microtubules is neuroprotective in amyotrophic lateral sclerosis. J. Biol. Chem. 2007;282:23465–23472. doi: 10.1074/jbc.M703434200. [DOI] [PubMed] [Google Scholar]
- 14.Lindwall G., Hsieh E.A., Misell L.M., Chai C.M., Turner S.M., Hellerstein M.K. Heavy water labeling of keratin as a non-invasive biomarker of skin turnover in vivo in rodents and humans. J. Invest. Dermatol. 2006;126:841–848. doi: 10.1038/sj.jid.5700189. [DOI] [PubMed] [Google Scholar]
- 15.Gardner J.L., Turner S.M., Bautista A., Lindwall G., Awada M., Hellerstein M.K. Measurement of liver collagen synthesis by heavy water labeling: effects of profibrotic toxicants and antifibrotic interventions. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1695–G1705. doi: 10.1152/ajpgi.00209.2006. [DOI] [PubMed] [Google Scholar]
- 16.Wang B., Sun G., Anderson D.R., Jia M., Previs S., Anderson V.E. Isotopologue distributions of peptide product ions by tandem mass spectrometry: quantitation of low levels of deuterium incorporation. Anal. Biochem. 2007;367:40–48. doi: 10.1016/j.ab.2007.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao G.G., Garg M., Lim S., Wong D., Go V.L., Lee W.N. Determination of protein synthesis in vivo using labeling from deuterated water and analysis of MALDI–TOF spectrum. J. Appl. Physiol. 2008;104:828–836. doi: 10.1152/japplphysiol.00976.2007. [DOI] [PubMed] [Google Scholar]
- 18.Brodsky F.M., Parham P., Barnstable C.J., Crumpton M.J., Bodmer W.F. Monoclonal antibodies for analysis of the HLA system. Immunol. Rev. 1979;47:3–61. doi: 10.1111/j.1600-065x.1979.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 19.Ohta N., Bach F.H. NO1: an HLA-DQw1-associated determinant present on loss mutants not expressing DQw1. Hum. Immunol. 1986;16:91–99. doi: 10.1016/0198-8859(86)90038-8. [DOI] [PubMed] [Google Scholar]
- 20.Glimcher L.H., Kim K.J., Green I., Paul W.E. Ia antigen-bearing B cell tumor lines can present protein antigen and alloantigen in a major histocompatibility complex-restricted fashion to antigen-reactive T cells. J. Exp. Med. 1982;155:445–459. doi: 10.1084/jem.155.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larhammar D., Hammerling U., Denaro M., Lund T., Flavell R.A., Rask L., Peterson P.A. Structure of the murine immune response I-A beta locus: sequence of the I-A beta gene and an adjacent β-chain second domain exon. Cell. 1983;34:179–188. doi: 10.1016/0092-8674(83)90148-4. [DOI] [PubMed] [Google Scholar]
- 22.Lund T., O’Reilly L., Hutchings P., Kanagawa O., Simpson E., Gravely R., Chandler P., Dyson J., Picard J.K., Edwards A., Kioussis D., Cooke A. Prevention of insulin-dependent diabetes mellitus in non-obese diabetic mice by transgenes encoding modified I-A β-chain or normal I-E α-chain. Nature. 1990;345:727–729. doi: 10.1038/345727a0. [DOI] [PubMed] [Google Scholar]
- 23.Lund T., Simpson E., Cooke A. Restriction fragment length polymorphisms in the major histocompatibility complex of the non-obese diabetic mouse. J. Autoimmun. 1990;3:289–298. doi: 10.1016/0896-8411(90)90147-k. [DOI] [PubMed] [Google Scholar]
- 24.Grosveld F.G., Lund T., Murray E.J., Mellor A.L., Dahl H.H., Flavell R.A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982;10:6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampson L.A., Levy R. Two populations of Ia-like molecules on a human B cell line. J. Immunol. 1980;125:293–299. [PubMed] [Google Scholar]
- 26.Barnstable C.J., Bodmer W.F., Brown G., Galfre G., Milstein C., Williams A.F., Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA, and other human cell surface antigens: new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 27.Watts T.H., Brian A.A., Kappler J.W., Marrack P., McConnell H.M. Antigen presentation by supported planar membranes containing affinity-purified I-Ad. Proc. Natl. Acad. Sci. USA. 1984;81:7564–7568. doi: 10.1073/pnas.81.23.7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reizis B., Eisenstein M., Bockova J., Konen-Waisman S., Mor F., Elias D., Cohen I.R. Molecular characterization of the diabetes-associated mouse MHC class II protein, I-Ag7. Int. Immunol. 1997;9:43–51. doi: 10.1093/intimm/9.1.43. [DOI] [PubMed] [Google Scholar]
- 29.Scrimgeour C.M., Rollo M.M., Mudambo S.M., Handley L.L., Prosser S.J. A simplified method for deuterium/hydrogen isotope ratio measurements on water samples of biological origin. Biol. Mass Spectrom. 1993;22:383–387. doi: 10.1002/bms.1200220704. [DOI] [PubMed] [Google Scholar]
- 30.Busch R., Doebele R.C., von Scheven E., Fahrni J., Mellins E.D. Aberrant intermolecular disulfide bonding in a mutant HLA-DM molecule: implications for assembly, maturation, and function. J. Immunol. 1998;160:734–743. [PubMed] [Google Scholar]
- 31.Hellerstein M.K., Neese R.A. Mass isotopomer distribution analysis at eight years: theoretical, analytic, and experimental considerations. Am. J. Physiol. 1999;276:E1146–E1170. doi: 10.1152/ajpendo.1999.276.6.E1146. [DOI] [PubMed] [Google Scholar]
- 32.Yan X., Watson J., Ho P.S., Deinzer M.L. Mass spectrometric approaches using electrospray ionization charge states and hydrogen–deuterium exchange for determining protein structures and their conformational changes. Mol. Cell. Proteomics. 2004;3:10–23. doi: 10.1074/mcp.R300010-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Lanzavecchia A., Reid P.A., Watts C. Irreversible association of peptides with class II MHC molecules in living cells. Nature. 1992;357:249–252. doi: 10.1038/357249a0. [DOI] [PubMed] [Google Scholar]
- 34.Cella M., Engering A., Pinet V., Pieters J., Lanzavecchia A. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature. 1997;388:782–787. doi: 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 35.Hellerstein M. Methods for measuring polymerisation biosynthesis: three general solutions to the problem of the “true precursor”. Diabetes Nutr. Metab. 2000;13:46–60. [PubMed] [Google Scholar]
- 36.Hellerstein M.K., Kletke C., Kaempfer S., Wu K., Shackleton C.H. Use of mass isotopomer distributions in secreted lipids to sample lipogenic acetyl-CoA pool in vivo in humans. Am. J. Physiol. 1991;261:E479–E486. doi: 10.1152/ajpendo.1991.261.4.E479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.