Abstract
We observed that human rhabdomyosarcoma (RMS) cells highly express a tissue factor (TF) that promotes thrombin formation, which indirectly and directly affects RMS progression. First, we found that thrombin activates platelets to generate microvesicles (PMVs), which transfer to RMS cells’ α2β3 integrin and increase their adhesiveness to endothelial cells. Accordingly, RMS cells covered with PMVs showed higher metastatic potential after intravenous injection into immunodeficient mice. Furthermore, PMVs activate mitogen-activated protein kinase (MAPK)p42/44 and AKT to chemoattract RMS cells. We also found that RMS cells express functional protease-activated receptor-1 (PAR1) and PAR3 and respond to thrombin stimulation by MAPKp42/44 and MAPKp38 phosphorylation. To our surprise, thrombin did not affect RMS proliferation or survival; it inhibited RMS cells’ chemotactic and adhesive properties. However, when PAR1-specific agonist thrombin receptor-activating peptide (TRAP)6 was employed, which does not activate PAR3, selective PAR1 stimulation enhanced RMS proliferation. To learn more on the role of PAR1 and PAR3 antagonism in RMS proliferation and metastasis, we knocked down both receptors by employing a short hairpin (sh)RNA strategy. We found that while thrombin does not affect growth of PAR1−/− cells, it stimulated proliferation of PAR3−/− cells. More importantly, PAR3−/− cells, in contrast to PAR1−/− ones, formed larger tumors in immunodeficient mice. We conclude that thrombin is a novel; underappreciated modulator of RMS metastasis and that we have identified a novel role for PAR3 in thrombin signaling.
Keywords: rhabdomyosarcomas, thrombin, microvesicles, metastases, chemotaxis
INTRODUCTION
RMS is the most common soft-tissue sarcoma of adolescence and childhood, accounting for 5% of all malignant tumors in patients under 15 years of age. Most RMS tumors originate in the head and neck region, urogenital tract, and extremities (1–3). It is well known that RMS cells, particularly alveolar (A)RMS, can infiltrate the bone marrow (BM) and, because they can resemble hematologic blasts, may sometimes be misdiagnosed as acute leukemia cells (1–3).
There are two major histological subtypes of RMS: the alveolar (A)RMS and embryonal (E)RMS (4–10). Clinical evidence indicates that ARMS is more aggressive and has a significantly worse outcome than ERMS. Genetic characterization of RMS has identified markers that show excellent correlation with histological subtype. Specifically, ARMS is characterized by the translocation t(2;13)(q35;q14) in 70% of cases or the variant t(1;13)(p36;q14) in a smaller percentage of cases. These translocations disrupt the paired box-containing (PAX)3 and PAX7 genes on chromosomes 2 and 1, respectively, and the forkhead in RMS (FKHR) gene on chromosome 13. As such, they generate PAX3-FKHR and PAX7-FKHR fusion genes. These fusion genes encode the fusion proteins PAX3-FKHR and PAX7-FKHR, which are believed to act in cell survival and deregulation of the cell cycle in ARMS cells (4–10).
The major clinical problem of RMS is its ability to metastasize and infiltrate various organs. Several chemokines [e.g., stromal-derived factor-1 (SDF-1), interferon-inducible T-cell alpha chemoattractant (I-TAC)], growth factors [e.g., hepatocyte growth factor (HGF), insulin growth factor-1 (IGF-1)] and cytokines [e.g., Leukemia inhibitory factor (LIF)] are involved in the spread and metastasis of RMS (11–14). Interestingly, several reports also link RMS spread to sub-acute intravascular coagulation, which, in some cases, may even lead to a hemorrhagic tendency as a result of consumption coagulopathy (15–17). In particular, coagulopathy appears to be a common feature of ARMS. Because the association of cancer dissemination and thrombosis is a well-known phenomenon for various cancer types (e.g., pancreatic, brain, lung, breast, prostate, and breast) (18–26), we became interested in whether the coagulation system may also modulate the metastatic behavior of human RMS cells.
We report that human RMS cells lines express a TF on their surface that activates conversion of pro-thrombin to thrombin, which may affect several aspects of cancer metastasis (27, 28). First, it may indirectly influence tumor growth by releasing pro-metastatic circular membrane fragments from activated platelets called plated-derived microvesicles (PMVs). As we reported for lung cancer (29) and breast cancer cells (30), PMVs may stimulate tumor cells and transfer integrin receptors to their surface, which increase their pro-metastatic properties. Thrombin possesses proteolytic activity and may also directly stimulate tumor cells by interacting with PARs. PARs are unique members of the G protein-coupled receptor (GPCR) family activated primarily by proteases. Protease cleavage exposes a tethered ligand at the N-terminus that subsequently binds to a conserved region in the second extracellular loop, which leads to intracellular signaling. There are currently 4 known PARs: PAR1; PAR3; and PAR4, which are activated primarily by thrombin; and PAR2, which is activatedby trypsin (31–35). However, PAR2 is not cleaved by thrombin it plays a role in signaling by engaging PAR1 in response to thrombin (36, 37).
It has been reported that thrombin may increase chemotaxis, adhesion, proliferation, and invasiveness of several tumor cell types (38–42). For example, blocking endogenous thrombin by an infusion of hirudin reduces metastasis of experimental lung cancer up to 400 times in mice (38). Nevertheless, the molecular mechanisms responsible for all these effects may vary with tumor type. Generally, because of the pleiotropic effects of thrombin, e.g., activation of platelets, stimulation of angiogenesis, and direct stimulation of tumor cells via PARs, thrombin-tumor cell interactions are not well understood.
In the current report, we provide novel evidence that RMS-expressed TF promotes the generation of thrombin, which regulates pro-metastatic properties of RMS cells indirectly (by PMV generation) and directly (by stimulation of PARs). Thus, thrombin is a novel, underappreciated, pro-metastatic factor for these cells and modulation of blood coagulation cascade (BCC) may become an important therapeutic strategy to prevent RMS progression.
MATERIALS AND METHODS
Cell lines
We used human RMS cell lines (gift of Dr. Frederic Barr, University of Pennsylvania, Philadelphia) comprising ARMS lines (RH2, RH4, RH28, RH30, and CW9019) and ERMS lines (RH18, RD, and SMS-CTR), all established at St. Jude Hospital. RMS cells used for experiments were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich, St. Louis, MO) and supplemented with 100 IU/ml penicillin, 10 μg/ml streptomycin, and 50 μg/ml neomycin (Life Technologies, Inc., Grand Island, NY) in the presence of 10% heat-inactivated fetal bovine serum (FBS; Life Technologies).. Cells were cultured in a humidified 5% CO2 atmosphere at 37°C at an initial cell density of 2.5 × 104 cells/flask (Corning, Cambridge, MA) and the media were changed every 48 hours.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). Messenger (m)RNA (0.5μg) was reverse-transcribed with 500 U of Moloney murine leukemia virus (MoMLV)-RT. The resulting complementary (c)DNA fragments were amplified using 5 U of Thermus aquaticus (Taq) polymerase. Primer sequences for human PAR1 were forward primer 5′ – GTG GCC GCC TGC TTC AG – 3′ and reverse primer 5′ – CAG CAG CAT AAG CTC GTG CAT – 3′; for human PAR3 were forward 5′ – GTT GCC CAC TTT TTG TCA GAG T – 3′ and reverse primer 5′ – TCC AAA TAC CCA GTT GTT CCC – 3′; for human PAR4 were forward 5′ – TGA GCA ACA TGG TAA AAC CCC – 3′ and reverse primer 5′ – TGC CAC AAT GCC TGG TTC A – 3′; for human tissue factor were forward 5′-AAC CCA AAC CCG TCA ATC AAG -3′; and reverse primer 5′-TCC TTC ACA ATC TCG TCG GTG -3′; and for human β-actin were forward 5′ – GGA AAT CGT GCG TGA CAT TAA GG – 3′ and reverse primer 5′ – CTG ATC CAC ATC TGC TGG AAG GT – 3′.
Detection of TF
RMS cells were detached from culture dishes by employing Cell Stripper (BD Becton Dickinson PharMingen, San Diego, CA), a non-enzymatic cell de-attachment solution, and were washed two times in phosphate-buffered saline (PBS) before lysing for 10 minutes on ice in M-Per lysing buffer (Pierce, Rockford, IL) containing protease and phosphatase inhibitor cocktails (Sigma, Milwaukee, WI). Subsequently, the extracted proteins were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and the fractionated proteins were transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, NH) as previously described. Presence of human TF protein was detected using commercial rabbit and horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (Ig)G as secondary antibodies (Abs; Santa Cruz Biotech., Santa Cruz, CA). Equal loading in the lanes was evaluated by stripping the blots and reprobing with mouse mAb against β-actin (Sigma-Aldrich, St. Louis, MO) and horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (Ig)G as secondary antibodies (Abs; Santa Cruz Biotech., Santa Cruz, CA). The membranes were developed with an electrochemiluminescent (ECL) reagent (Amersham Life Sciences, Little Chalfont, GBR), dried, and subsequently exposed to HyperFilm (Amersham Life Sciences).
Measurement of TF activity
RMS cells were detached from culture dishes by employing Cell Stripper (BD Becton Dickinson PharMingen) and were washed two times in PBS. One million RMS cells were lysed and solubilized with 15 mM octyl-β-D-glucopyranoside at 37°C for 15 minutes. Fresh cell lysates were assayed using the AssaySense Human Tissue Factor Chromogenic Activity Assay Kit (Assaypro, St. Charles, MO) according to the manufacturer’s instructions. Briefly, cell lysates were supplemented with coagulation factor VII and X (FVII and FX) and incubated for 30 minutes at 37°C. Next, FXa substrate was added to the mixture and absorbance was read at 405nm. TF activity was evaluated using a standard curve based on standards supplemented by the manufacturer.
Fluorescence-activated cell sorting (FACS) analysis
The expression of PAR1 protein on RMS cell lines was evaluated by FACS. RMS cells were detached from culture dishes by employing Cell Stripper (BD Becton Dickinson PharMingen). The PAR1 and PAR3 antigen was detected with phycoerythrin (PE)-conjugated monoclonal (m)Abs (Becton Dickinson PharMingen). Samples stained with appropriate isotype controls (Becton Dickinson PharMingen) were examined in parallel.
Signal transduction studies
RMS cell lines were kept in RPMI medium containing low levels of bovine serum albumin (BSA; 0.5%) to render them quiescent and were divided and stimulated with optimal doses of thrombin (1U/ml) for 2, 5, and 15 minutes at 37°C before lysing for 10 minutes on ice in M-Per lysing buffer (Pierce) containing protease and phosphatase inhibitor cocktails (Sigma). Subsequently, the extracted proteins were separated on a 10% SDS-PAGE and the fractionated proteins were transferred to a nitrocellulose membrane (Schleicher & Schuell) as previously described. Phosphorylation of the intracellular kinases, 44/42 MAPK (Thr 202/Tyr 204), AKT (Ser 473), and p38 MAPK proteins was detected using commercial mouse phospho-specific mAb (p44/42) or rabbit phospho-specific polyclonal Abs for each of the remainder (all from New England Biolabs, Beverly, MA) with HRP-conjugated goat anti-mouse IgG or goat anti-rabbit IgG as secondary Abs (Santa Cruz Biotech) as described (13). Equal loading in the lanes was evaluated by stripping the blots and re-probing with appropriate mAbs: p42/44 anti-MAPK Ab clone #9102 (New England Biolabs). The membranes were developed with an ECL reagent (Amersham Life Sciences), dried, and subsequently exposed to HyperFilm (Amersham Life Sciences).
Chemotaxis assay
The 8-μm pore polycarbonate membranes were covered with 50 μL of 0.5% gelatin. RMS cells were detached from culture dishes by employing Cell Stripper (BD Becton Dickinson PharMingen) and were washed in RPMI 1640, resuspended in RPMI 1640 with 0.5% BSA, and seeded at a density of 3 × 104 in 120 μL into the upper chambers of Transwell inserts (Costar Transwell, Corning Costar, Corning, NY). The lower chambers were filled with 0.5% BSA RPMI 1640 (control) or conditioned media (CM) and CM together with thrombin, TRAP6, PAR2, or PAR4 agonists. After 24 hours, the inserts were removed from the Transwells. Cells remaining in the upper chambers were scraped off with cotton wool and cells that had transmigrated were stained by Hema 3 (Protocol, Fisher Scientific, Pittsburgh, PA) and counted either on the lower side of the membranes or on the bottom of the Transwells.
Adhesion to human umbilical vein endothelial cells (HUVECs)
RMS cells were labeled before assay with the fluorescent dye calcein-AM for 30 minutes in serum-free media. Next, cells were incubated with control media, CM harvested from human BM-derived fibroblasts (BM-CM), and CM with thrombin. Cells were added for 15 minutes (105 cells/well) to the 96-well plates covered by HUVECs, which had been pretreated for 16 hours with tumor necrosis factor (TNF)-alpha (5ng/mL). After the non-adherent cells had been discarded, cells that adhered to the HUVECs were evaluated using a fluorescent microscope.
Time lapse monitoring of the locomotion of individual cells
The images of human RMS migrating on plastic at 37°C were evaluated with an inverted microscope using phase contrast optics. Analysis of cell migration began 18 hours after cell seeding. The locomotion images were recorded with a charge-coupled device (CCD) camera. CW9019, RD, RH5, RH28, and RH30 cells were plated to Corning flasks at a density of 104 cells/cm2 and were mock-treated or prestimulated by SDF-1 (300 ng/mL) or by SDF-1 (300 ng/mL) together with thrombin (5U/mL) for 60 minutes before recording. The cell trajectories were constructed from 48 subsequent cell centroid positions recorded for 240 minutes at 5-minute intervals. The cell trajectories were presented in circular diagrams (43) and the lengths of cell tracks were calculated in under the conditions addition to the final displacement and cell tracks (20) were recorded.
The following parameters characterizing cell locomotion were computed for each cell using procedures written in the Mathematica language, including: (1) total length of cell trajectory (in micrometers); (2) whether the trajectory was a sequence of N straight-line segments, each corresponding to cell centroid translocation within one time interval between two successive images; (3) total length of the final displacement of the cell from the starting point to the final position, i.e., distance between the first and last points of the cell track (in micrometers); (4) average speed of cell locomotion defined as total length of cell trajectory/time of recording; and (5) the ratio of cell displacement length to cell trajectory length, called the coefficient of movement efficiency (CME). (44–46) shRNA. The peptide (p)RNA-U6.1/Neo plasmid carrying shRNA for PAR1 and PAR3 was engineered by annealing the single strand oligonucleotides 5′ – GAT CCC GCA TGT ACG CCT CTA TCT TGT TCA AGA GAC AAG ATA GAG GCG TAC ATG TTT TTT CCA AA - 3′ and 5′-AGC TTT TGG AAA AAA CAT GTA CGC CTC TAT CTT GTC TCT TGA ACA AGA TAG AGG CGT ACA TGC GG – 3′ (for PAR1) and 5′ –GAT CCC GCA CTT GCG AGT CCT CAT CTT TCA AGA GAA GAT GAG GAC TCG CAA GTG TTT TTT CCA AA - 3′ and 5′ – AGC TTT TGG AAA AAA CAC TTG CGA GTC CTC ATC TTC TCT TGA AAG ATG AGG ACT CGC AAG TGC GG – 3′ (for PAR3) and inserting the double strand oligonucleotide in the pRNA-U6.1/Neo plasmid (GenScript Co, Piscataway, NJ). RMS cells were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Isolation of PMVs
Human peripheral blood (PB) platelets were isolated from healthy volunteer donors who had given informed consent; the protocols used were approved by the institutional review board of the University of Louisville. The platelets were activated by thrombin (0.1 U/mL) and collagen (4 g/mL; Sigma-Aldrich) for 30 minutes at 37°C by stirring and were centrifuged twice at 2,000g for 15 minutes at 4°C. The PMV-enriched supernatants were collected and centrifuged at 24,000g for 1 hour at 4°C. The pellets were washed and resuspended in N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer, pH 7.4. The PMVs were characterized by staining with PE-conjugated anti-human Abs against IIb3 (Coulter-Immunotech, Marseille, FRA), P-selectin (CD62; Becton Dickinson, San Jose, CA), and CXC chemokine receptor 4 (CXCR4; Becton Dickinson Pharmingen). PMVs were fixed in 1% paraformaldehyde prior to FACS analysis using the FACScan (Becton Dickinson). Concentrations of both PMVs were estimated by Bradford assay.
Detection of platelet-specific receptors on human RMS
To demonstrate the presence of PMVs on the surface of target cells, RMS cancer cells incubated with PMVs were stained with PE-anti-CD41 Ab (Coulter-Immunotech) and analyzed by FACS. As isotype controls, we employed PE-goat-anti-mouse Abs.
In vivo model of metastasis of RMS cells
Five- to six-week-old male severe combined immunodeficient (SCID)-Beige inbred mice (National Cancer Institute, Bethesda, MD) were injected intravenously with 5 × 106 RMS cells that had been incubated with PMVs, thrombin, or a combination of both. Before injection, cells were washed in PBS. After 48 hours, mice were euthanized and their lungs were collected. The presence of RMS cells, i.e., murine-human chimerism, was evaluated as the difference in the level of human-satellite. DNA was amplified in the extracts isolated from the BM-derived- and lung-derived cells using real-time PCR. Briefly, DNA was isolated using the QIAamp DNA Mini kit (Qiagen). Detection of human-satellite and murine β-actin DNA levels was accomplished using real-time PCR and an ABI Prism 7000 Sequence Detection System. A 25-μL reaction mixture contained 12.5 μL SYBR Green PCR Master Mix, 300 ng DNA template, 5′-GGG ATA ATT TCA GCT GAC TAA ACA G-3′, 5′-TTT CGT TTA GTT AGG TGC AGT TAT C-3′, and 5′-AAA CGT CCA CTT GCA GAT TCT AG-3′ primers for the satellite and 5′-GGA TGC AGA AGG AGA TCA CTG-3′ forward and 5′-CGA TCC ACA CGG AGT ACT TG-3′ reverse primers for the β-actin. Ct was determined as described before (13). The number of human cells present in murine lungs (degree of chimerism) was calculated from the standard curve obtained by mixing different numbers of human cells with a constant number of murine cells.
Cell proliferation
Cells were plated in culture flasks at an initial density of 104 cells/cm2. In some experiments, cells were cultured in the presence or absence thrombin or TRAPs. The cell number was calculated 72 hours after culture initiation. At the indicated time points, cells were harvested from the culture flasks by trypsinization and the number of cells was determined using a Bürker hemocytometer (American Optical Corp., Buffalo, NY).
Statistical Analysis
All results are presented as mean ± standard error (SEM). Statistical analysis of the data was performed using the nonparametric Mann-Whitney test, with p<0.05 considered significant.
RESULTS
Human RMS cell lines express TF and activate coagulation cascade
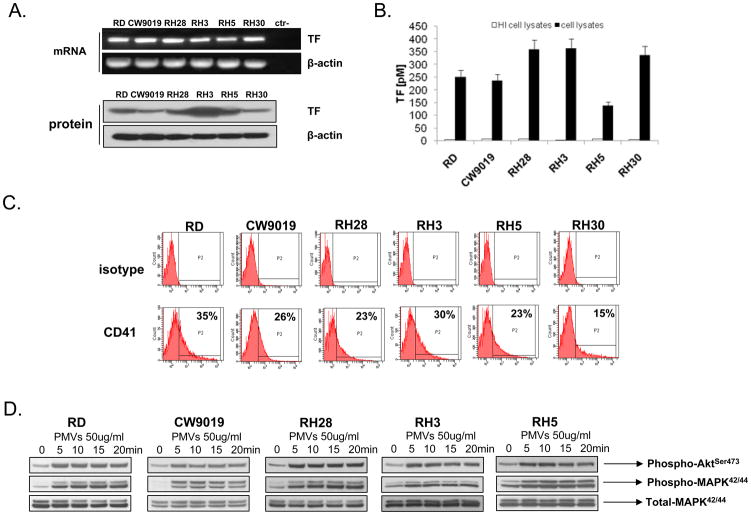
TF activates blood coagulation cascade (BCC) via the extrinsic pathway by converting pro-thrombin into thrombin. We report here for the first time that human RMS cell lines express TF at the mRNA and protein levels (Fig. 1A and Supplementary Figure 1). Expression of TF on RMS cells lines was constitutive and did not increase during hypoxia or upon stimulation by TNF-α or lipopolysaccharide (LPS; data not shown). More importantly, we noticed that TF present on the surface of the RMS cells triggers BCC, as demonstrated by employing the TF activity assay (Fig. 1 panel B). Based on this observation and our previous work (29, 30), we hypothesized that TF-triggered activation of BCC and thrombin release may affect several aspects of RMS cell biology that are related to tumor progression and metastasis and focused on this issue in our current paper.
Figure 1. Human RMS cells express TF and activate coagulation cascade that generates PMVs.
Panel A: Human RMS cells express TF at the mRNA and protein levels. The experiment was repeated three times with similar results. A representative study is shown. Panel B: TF expressed on human RMS cell lines has the ability to initiate coagulation cascade, as shown by TF activity assay. The experiment was repeated three times with similar results. A representative study is shown. Panel C: RMS cells were incubated with PMVs, then washed and stained with anti-α2β3 (CD41)-specific Ab. As shown, PMVs transfer CD41 from platelets to RMS cells. The experiment was repeated three times with similar results. A representative study is shown. Panel D: PMVs activate AKT and MAPKp42/44 in human RMS cells in a time-dependent manner. The experiment was repeated three times with similar results. A representative study is shown.
Thrombin activated by RMS cells leads to generation of prometastatic PMVs
We reported that thrombin may affect metastatic potential of cancer cells indirectly by releasing from activated platelets circular membrane fragments called platelet-derived microvesicles (PMVs) (29, 30). Accordingly, we reported in our previous work that TF expressed on the surface of lung cancer (29) and breast cancer cells (30) activates via thrombin blood platelets to release PMVs which subsequently promote the metastatic potential of human cancer cells by transferring platelet-derived adhesion molecules to their surfaces. In addition PMV may directly stimulate tumor cell growth via proteins and active lipids expressed on their surfaces (29, 30).
Here, we evidenced by FACS analysis (Fig. 1 panel C) that, similarly to human lung and breast cancer cells, PMVs transfer to RMS cells’ α2β3 integrin (CD41), which is crucial for interaction of platelets with the endothelium. Transfer of these adhesion molecules to RMS cells may increase their metastatic properties. Furthermore, similarly as we reported for human lung and breast cancer cells (29, 30) PMVs directly stimulated in RMS cells AKT and MAPKp42/44 (Fig. 1 panel D).
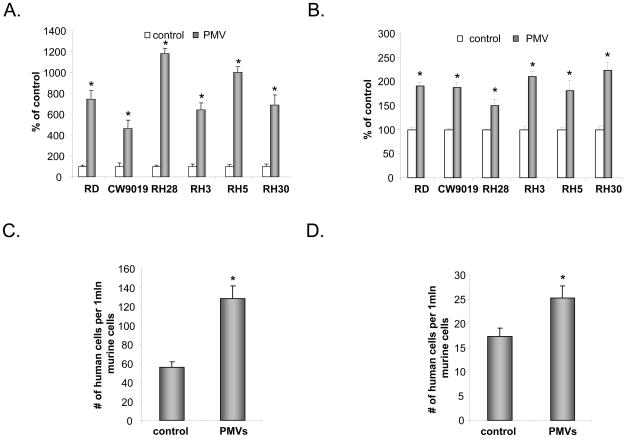
Furthermore, PMVs also chemoattract human RMS cells and increase their adhesion to endothelial cells (Fig. 2 panels A and B, respectively). More important, in an in vivo assay, we noticed that selected RMS cell line (CW9019 and RH30) covered by PMVs possess increased seeding efficacy into BM after intravenous injection into SCID mice (Fig. 2 panels C and D). The seeding efficiency for RMS cells in murine BM was evaluated by real-time PCR by employing detection of human α-satellite gene and the number of human cells present in murine BM (degree of chimerism) was calculated from the standard curve obtained by mixing different numbers of human cells with a constant number of murine cells as described (13).
Figure 2. PMV mediates chemotaxis and adhesion of RMS cells.
Panels A and B: PMV induces chemotaxis (Panel A) and adhesion (Panel B) of RMS cells. Data from four separate experiments are pooled together * p < 0.01. CW9019 (Panel C) or RH30 (Panel D) cells were pre-incubated with PMVs or medium alone and were then injected intravenously into immunodeficient mice. Forty-eight hours later, the seeding efficiency for RMS cells in murine BM was evaluated by real-time PCR by detection of human α-satellite gene. The number of human cells present in murine BM (degree of chimerism) was calculated from the standard curve obtained by mixing different numbers of human cells with a constant number of murine cells as described (13). Data from four separate experiments are pooled together * p < 0.01.
Thus, these data strongly suggests thrombin may affect metastatic potential of RMS cells indirectly through PMVs relased from activated platelets.
RMS cells express functional thrombin receptors (PARs)
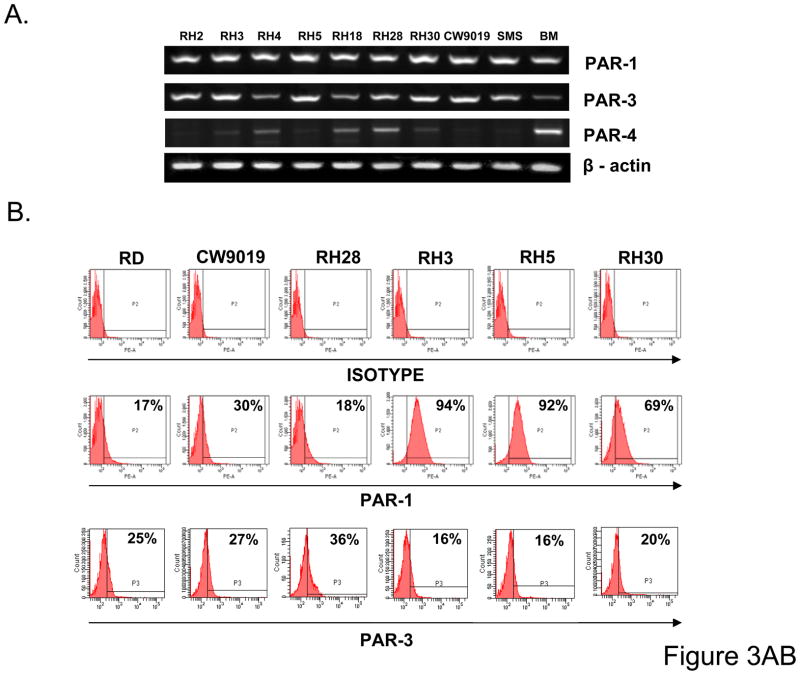
Next we become interested if RMS cells express thrombin-binding PARs -1, -3, and -4. In fact, we noticed that all 9 RMS cell lines employed in our studies express mRNA for PAR1 and PAR3 and 3 out of 9 cell lines (RH4, RH18, and RH28) express PAR4 mRNA (Fig. 3 panel A) at very low levels. More importantly, we confirmed by FACS analysis that the main thrombin receptors, PAR1 and PAR3, are expressed at the protein level on the surface of RMS cells (Fig. 3 panel B).
Figure 3. RMS cells express functional thrombin receptors.
Panel A: Expression of PAR1, -3, and -4 at the mRNA level in human RMS cells. The experiment was repeated two times with similar results. A representative study is shown. Panel B: Flow cytometry analysis of PAR1 on human RMS cell lines. The experiment was repeated three times with similar results. A representative study is shown. Phosphorylation of MAPK p42/44, AKT, and MAPK p38 in selected human RMS cell lines stimulated by thrombin (1U/ml) (Panel C) or PAR1-agonist TRAP6 (50uM) (Panel D) for 2, 5, 10, and 15 minutes. The experiment was repeated three times with similar results. A representative study is shown.
To demonstrate that these receptors are functional, we stimulated human RMS cells by thrombin and noticed activation of MAPKp42/44 in 5 out of 9 cell lines (RD, CW9019, RH28, RH3, and RH5) and activation of p38 in CW9019 and RH3 cells (Fig. 3 panel C). Interestingly, stimulation of RMS cells by thrombin did not affect activation of AKT kinase in all these cell lines. Subsequently, all cell lines that expressed functional PAR1 and PAR3 receptors (RD2, CW9019, RH28, RH3 and RH5) were employed for further studies. In parallel, RH30 cells that express non-functional PAR1 and PAR3 were employed as negative controls.
Because thrombin may activate three receptors, i.e., PARs -1, -3, and -4, in the next step we tried to address which one of these receptors is crucial for thrombin signaling. It is known that thrombin activates PARs by cleaving the N-terminal fragment of the receptor and unmasking the signaling peptide, which subsequently binds to the receptor and triggers signaling (47). To determinate which PAR in RMS cells is involved in thrombin signaling, we used synthetic peptides that mimic unmasked peptides and that are PAR1-and PAR4-specific agonists.
First, we noticed similar pattern of intracellular signaling in RMS cells stimulated by thrombin and TRAP6, a specific PAR1 agonist (Fig. 3 panel C and D respectively). At the same time however, we did not observe any activation of intracellular signaling after stimulation of three RMS cell lines that expressed PAR4 mRNA (Fig. 2 panel A) after exposure to the PAR4-specific agonist (data not shown). This suggests that PAR4 is not involved in thrombin signaling in RMS cells. Because activation of PAR3 by thrombin unleashes the peptide, which does not bind PAR3 (44), we were not able to use a specific peptide to activate this receptor. However, these data clearly demonstrate that PAR1 and PAR3 are responsible for thrombin signaling in RMS cells.
Thrombin inhibits RMS chemotaxis
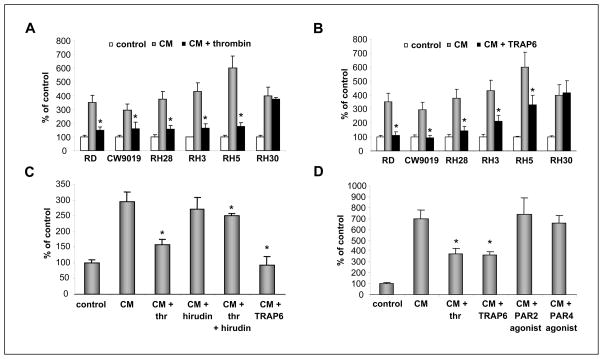
We reported that human RMS cells are strongly chemoattracted by BM-CM, which are sources of several RMS chemoattractants. These include SDF-1 and HGF/scatter factor (SF), which are both factors responsible for BM infiltration by RMS cells (11, 12). Therefore, in our chemotaxis experiments, we choose to employ BM-CM as a source of chemoattractants. We noticed that both thrombin (Fig. 4 panel A) as well as the selective PAR1 stimulatory peptide TRAP6 inhibit chemotaxis of RMS cells (Fig. 4 panel B), except for RH30 cells, which do not express functional PARs as mentioned and served as negative control. We also learned that inhibition of chemotaxis by thrombin was dose dependent. To support this, we observed ~50% inhibition of chemotaxis at concentrations of 0.5U/ml and almost complete inhibition at doses of 5U/ml (Supplementary Fig. 2 panel A). Furthermore, a similar effect was observed regardless of whether thrombin was added to the lower chamber together with BM-CM or if cells were exposed during the chemotaxis assay to thrombin that was added to the upper chamber (Supplementary Fig. 2 panel B).
Figure 4. Thrombin regulates the pro-metastatic potential of RMS cells.
Thrombin (Panel A) and TRAP6 (Panel B) decrease the chemotactic response of human RMS cells to BM-CM. Data from four separate experiments are pooled together * p < 0.001. Panel C: Hirudin reverses an inhibitory effect of thrombin but not TRAP6 on the chemotactic response of RMS cells to BM-CM. Data from three separate experiments are pooled together * p < 0.01. Panel D: Thrombin and TRAP6, but not PAR2- and PAR4-agonist, inhibit the chemotactic response of human RMS cells to BM-CM. Data from four separate experiments are pooled together * p < 0.01.
Next, to check the specificity of thrombin in the inhibition of RMS chemotaxis, we employed hirudin, which is a specific thrombin inhibitor. Figure 4 panel C shows that hirudin prevented thrombin-dependent inhibition of chemotaxis of CW9019 cells that respond robustly to stimulation by both ligands (Figure 3 panel C and D), but did not have any effect when PAR1 agonist TRAP6 was employed (Fig. 4 panel C).
We also found by employing CW9019 cells that thrombin-mediated inhibition of chemotaxis was thrombin-specific and not seen when thrombin was replaced by PAR2 or PAR4 agonists (Fig. 4 panel D). Overall, these data suggest that on RMS cells, PAR1 mediates inhibition of a chemotactic response of human RMS after exposure to thrombin.
Thrombin inhibits SDF-1-induced locomotion of RMS cells
We reported that SDF-1, one of major chemoattractants for RMS cells present in BM-CM (11), induces the locomotion of these cells. To address this issue, we studied movement on polystyrene dishes of two human ERMS cell lines (RD and CW9019) and three ARMS cell lines (RH28, RH5, and RH30) using time-lapse monitoring of the movement of individual cells. Figure 5 shows the trajectories of RMS cell migration in the absence (left panel) or presence (middle panel) of SDF-1 and (right panel) SDF-1 + thrombin in the culture media. Analyses of these trajectories and mean values as well as standard errors for the parameters of cell locomotion are summarized in Table 1.
Figure 5. Effect of thrombin on the SDF-1-induced motility of RMS cell lines.
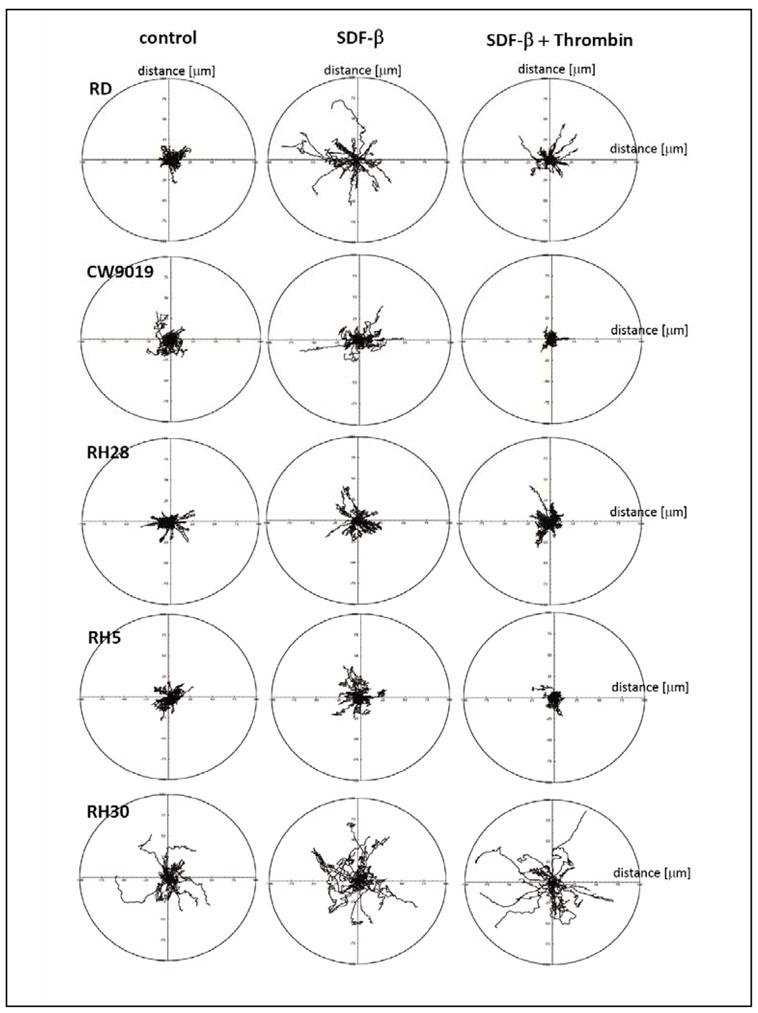
The composite trajectories of RD, CW9019, RH28, RH5, and RH30 cells migrating in the absence (left panel) or presence (middle panel) of SDF-1 and (right panel) SDF-1 + thrombin in the culture media are shown in circular diagrams drawn with the initial point of each trajectory at the origin of the plot. Data from 4 separate experiments are pooled together.
Table 1.
Parameters characterizing the movement of rhabdomyosarcoma RD, RH5, CW9019, RH28, and RH30 cells moving in control medium in the presence of SDF-1 (300 ng/mL), and in the presence of SDF-1 (300 ng/mL) with thrombin (5U/mL).
| Cell line | TLcT | AScL | TLcD | AVcD | CME |
|---|---|---|---|---|---|
| RD | |||||
| Control | 102.39 ± 16.57 | 0.43 ± 0.06 | 18.10 ± 14.49 | 0.07 ± 0.06 | 0.19 ± 0.17 |
| SDF | 112.97 ± 21.16* | 0.47 ± 0.08* | 32.77 ± 24.47* | 0.13 ± 0.10* | 0.30 ± 0.2* |
| SDF+THROMBIN | 106.25 ± 16.31 | 0.44 ± 0.06 | 11.81 ± 7.76§ | 0.04 ± 0.03§ | 0.11 ± 0.08§ |
| RH5 | |||||
| Control | 113.91 ± 33.10 | 0.47 ± 0.13 | 15.33 ± 6.98 | 0.06 ± 0.03 | 0.15 ± 0.10 |
| SDF | 111.31 ± 19.12 | 0.46 ± 0.07 | 18.14 ± 11.19 | 0.07 ± 0.04 | 0.16 ± 0.10 |
| SDF+THROMBIN | 127.33 ± 18.40§ | 0.53 ± 0.07§ | 6.67 ± 4.91*§ | 0.03 ± 0.02*§ | 0.05 ± 0.04*§ |
| RH28 | |||||
| Control | 102.45 ± 26.71 | 0.42 ± 0.11 | 12.95 ± 8.82 | 0.05 ± 0.03 | 0.14 ± 0.10 |
| SDF | 107.24 ± 19.52 | 0.44 ± 0.08 | 18.45 ± 10.50* | 0.08 ± 0.04* | 0.17 ± 0.12 |
| SDF+THROMBIN | 120.31 ±23.03*§ | 0.5 ± 0.09*§ | 16.26 ± 10.66 | 0.07 ± 0.04 | 0.15 ± 0.13 |
| RH30 | |||||
| Control | 108.81 ± 12.18 | 0.45 ± 0.05 | 26.93 ± 17.62 | 0.11 ± 0.07 | 0.24 ± 0.16 |
| SDF | 149.00 ± 20.00* | 0.62 ± 0.08* | 38.13 ± 20.04* | 0.17 ± 0.08* | 0.31 ± 0.05 |
| SDF+THROMBIN | 122.99 ±17.18*§ | 0.51 ± 0.07*§ | 45.98 ± 24.75* | 0.18 ± 0.10* | 0.37 ± 0.19* |
| CW9019 | |||||
| Control | 122.21 ± 27.87 | 0.50 ± 0.11 | 14.09 ± 9.65 | 0.06 ± 0.04 | 0.12 ± 0.10 |
| SDF | 105.96 ± 15.98* | 0.44 ± 0.06* | 21.05 ± 17.03* | 0.08 ± 0.07* | 0.21 ± 0.19* |
| SDF+THROMBIN | 105.27 ± 20.24* | 0.44 ± 0.08* | 6.32 ± 6.21*§ | 0.02 ± 0.02*§ | 0.06 ± 0.07*§ |
TLcT - Total length of cell trajectory [μm].
AScL - Average speed of cell locomotion [μm/min] is defined as the total length of the cell trajectory/time of recording (240 minutes).
TLcD - Total length of cell displacement [μm].
AVcD - Average velocity of cell displacement [μm/min] is defined as the length of the final cell displacement/time of recording.
CME - Coefficient of movement efficiency is the ratio of cell displacement to cell trajectory length. CME would equal 1 for a cell moving persistently along one straight line in one direction and 0 for a random movement.
Note: Values are given as the mean ± SEM,
statistically significant vs. control,
vs. SDF at P<.05 (Mann-Whitney test).
Analysis of individual cell trajectories showed that SDF-1 stimulated motility of RD, CW9019, RH 30, and RH5 cells, but not RH28 cells. Exposure of RD, CW9019, and RH5 cells to thrombin caused a statistically significant decrease in the average SDF-1-induced speed of locomotion (AScL) as well as ~3-fold decrease in the total length of cell displacement (TLcD) as compared with SDF-1-stimulated cells (Table 1). Motile activity of RH30 not expressing functional PARs did not change in the presence of thrombin + SDF-1 in the comparison to locomotion stimulated by SDF-1 alone. It is worth noting that because the migrating RMS cells made tortuous tracks (CME), their final displacement may be much smaller than the total length of their trajectories.
Thrombin inhibits RMS adhesion to endothelium
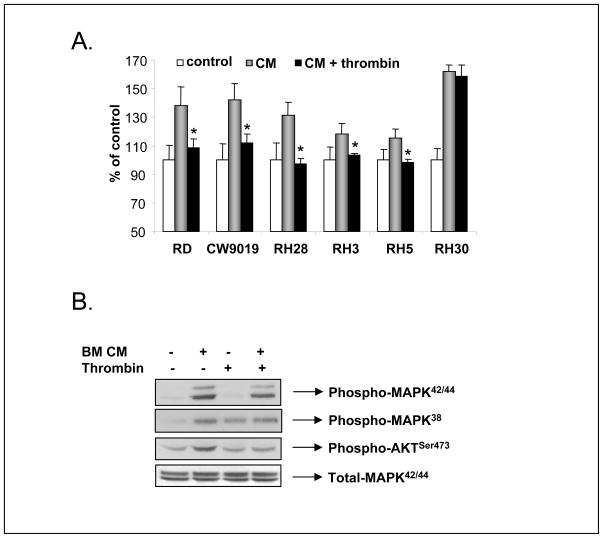
One of the important steps in the metastatic process is adhesion of tumor cells circulating in the blood stream or lymph vessels to the endothelium. In our experiments, we employed HUVECs pretreated by TNF-α as a substrate for RMS cell adhesion. Prior to the adhesion assay, RMS cells were pre-stimulated with BM-CM for 15 minutes. Figure 6 panel A shows that thrombin inhibited adhesion of RMS cells except in RH30 cells, which as mentioned above lack functional PARs.
Figure 6. Thrombin regulates the adhesive potential of RMS cells.
Panel A: Thrombin decreases the BM-CM induced adhesive response of human RMS cells. Data from four separate experiments are pooled together * p < 0.001. Panel B: Thrombin inhibits phosphorylation of AKT, but not MAPKp42/44 induced by BM-CM in human RMS cells (RH5). The experiment was repeated three times with similar results. A representative study is shown.
Interestingly, we also observed negative effect of thrombin on PMVs-induced chemotaxis and adhesion (Supplementary Figure 3 A and B) as well as PMVs-mediated in vivo spread of RMS cells injected into SCID mice (Supplementary Figure 3 C and D). Thus, our data show that thrombin-PAR-1, -3 axis in RMS cells, in contrast to other tumors inhibits metastatic potential of malignant cells.
These observations imply that thrombin may interfere with important signaling pathways involved in cell migration and adhesion. In fact Figure 4 panel B shows that stimulation of RH5 cells, which show the highest chemotactic responsiveness (Fig. 4 panels A and B), by BM-CM activates MAPKp42/44, MAPKp38, and AKT. However, pre-stimulation of these cells by thrombin (2 U/ml) before exposure to BM-CM prevented AKT activation.
Effect of PAR1 and PAR3 on RMS cell proliferation
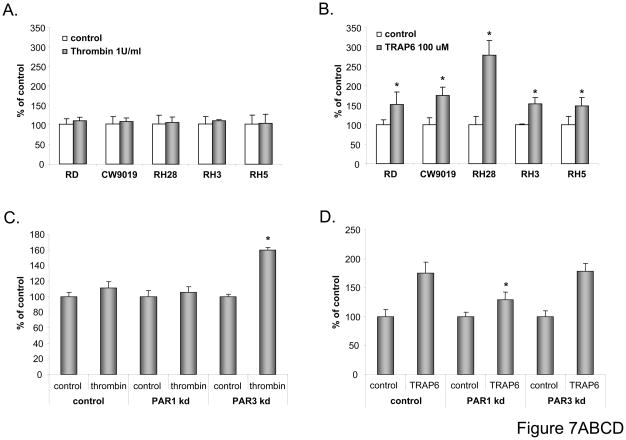
To evaluate whether thrombin may stimulate tumor growth similarly as described for other cancer cell lines (49), we exposed human RMS cell lines to thrombin (1U/mL). When cells were counted 72 hours later, no effect on cell proliferation was noticed (Fig. 7 panel A). To our surprise, however, when we employed TRAP6 for stimulation, which activates PAR1 selectively, we noticed increased proliferation of RMS cells (Fig. 7 panel B).
Figure 7. Effect of thrombin and thrombin receptors on proliferation of RMS cells in vitro and in vivo.
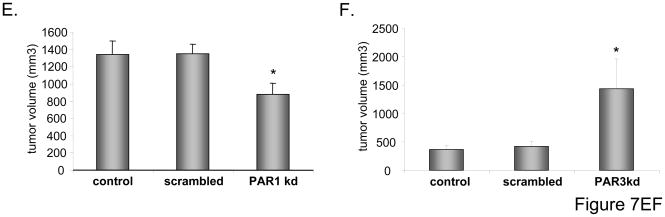
Panel A: RMS cells were cultured in the presence or absence of thrombin (1u/mL). After 72 hours, the number of cells was evaluated by hematocytometer. Panel B: RMS cells were cultured in the presence or absence of TRAP6 (100uM). After 72 hours, the number of cells was evaluated by hematocytometer. Panels C and D: CW9019, CW9019 PAR1−/−, and -PAR3−/− cells were cultured in the presence of thrombin (Panel C) or TRAP6 (Panel D). Numbers of cells were evaluated 72 hours after stimulation. Data from four separate experiments are pooled together * p < 0.001. Panel E: CW9019, CW9019 scrambled, or CW9019 PAR1−/− cells were inoculated into the hind limb muscles of SCID-Beige inbred mice. Tumors were measured after 6 weeks. Panel F: CW9019, CW9019 scrambled, or CW9019 PAR3−/− cells were inoculated into the hind limb muscles of SCID-Beige inbred mice. Tumors were measured after 3 weeks. Data from four separate experiments (6 mice/group each) are pooled together * p < 0.01.
This indicated that thrombin affects RMS proliferation by activating opposite responses from PAR1 and PAR3. To test this hypothesis we knocked down (−/−) PAR1 by employing shRNA [~80% as assessed by real-time quantitative (RQ)-PCR] and PAR3 (~90% as assessed by RQ-PCR) in CW9019 cells (Supplementary Fig. 4 panels A and B, respectively). Subsequently, PAR1−/− or PAR3−/− cells were stimulated by thrombin (Fig. 7 panel C) or TRAP6 (Fig. 7 panel D). As expected, while PAR3knock-down resulted in an increase of RMS cell proliferation after stimulation by thrombin (Fig. 7 panel C), PAR3−/− cells did not affect the response of RMS cells to TRAP6 (Fig. 7 panel D).
Interestingly, we also found that basic chemotaxis of CW9010 cells to BM-CM was less affected in PAR1−/− cells after exposure to thrombin or TRAP6 (Supplementary Fig. 5). These data overall support that activation of PAR1 in wild type cells is responsible for inhibition of chemotaxis in response to thrombin and that this phenomenon may be related to a negative effect of thrombin on AKT activation (Fig. 6 panel B).
Pleiotropic effect of PAR on tumor growth in vivo
Finally, to learn more on the roles of PAR1 and PAR3 during in vivo tumor growth, we intramuscularly inoculated SCID-Beige mice with CW9019 RMS cells and PAR1−/− (Fig. 7 panel E) or PAR3−/− (Fig. 7 panel F) cells. Three weeks later (PAR3−/−) and six weeks later (PAR1−/−), mice were sacrificed and tumor diameters were measured. We noticed a significant decrease in tumor growth in CW9010 cells where PAR1 was down-regulated (Fig. 7 panel E) and, at the same time, enhanced tumor formation in cells with PAR3−/− (Fig. 7 panel F).
Overall these in vivo data corroborate our in vitro results and suggest novel mechanism that activation of PAR3 by thrombin inhibits the pro-proliferative effect of the thrombin-PAR1 axis (Fig. 8).
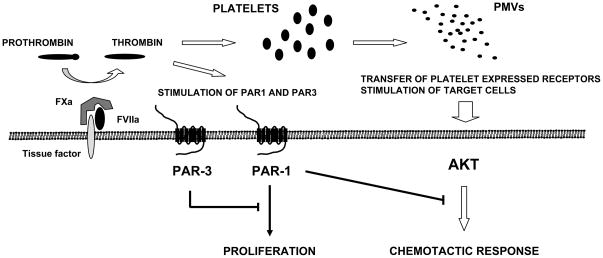
Figure 8. Pro-metastatic consequences of TF expression by RMS cells.
TF expressed on surface of RMS cells activates conversion of prothrombin to thrombin, which: i) activates platelets that release pro-metastatic PMVs; and ii) directly interacts with PAR1 and PAR3 on RMS cells. A negative regulatory effect of PAR3 on PAR1 is indicated.
DISCUSSION
RMS is the most common soft-tissue sarcoma of adolescents and children and frequently infiltrates the BM to the degree that it mimics acute lymphoblastic leukemia (1–3). The prognosis is poor in particular for the more aggressive and metastatic ARMS type (4–11). In addition to rapid tumor expansion, distant metastases and BM involvement are major problems for successful RMS therapy, making support of chemotherapy by new and efficient anti-metastatic treatment strategies necessary.
In our previous reports, we focused on a role of chemokines (SDF-1, I-TAC, and interleukin-8), HGF (11, 12, 50), and LIF (13) in RMS metastasis. In this current work, we are interested in the role of BCC in RMS spread. To justify this, activation of BCC was demonstrated as having an important function in the progression of several tumors (38–42, 51); more importantly, disseminated intravascular coagulation has been reported in several cases of RMS (15, 16).
We found that human RMS cells express functional TF on their surface. It is well known that TF may affect several aspects of tumor metastasis by promoting conversion of pro-thrombin to thrombin. TF-triggers activation of BCC and released in this process thrombin: i) activates platelets that release pro-metastatic PMVs; ii) stimulates tumor cells directly via PARs; iii) cleaves fibrinogen to cause fibrin formation; and iv) activates complement protein component 5 (C5), which releases anaphylatoxin C5a and increases vascular permeability. All these processes collectively enhance tumor metastasis.
In our previous work, we demonstrated that thrombin generates PMVs by stimulating and activating blood platelets. These PMVs increase the pro-metastatic properties of human lung (29) and breast cancer (30) cells. Accordingly, PMVs may directly stimulate tumor cells and increase their proliferation on one hand; on the other, they may transfer several platelet-associated receptors to the tumor cells and make them more adhesive to the endothelium. In the current study, we noticed that PMVs activate phosphorylation of MAPK p42/44 and AKT and transfer α2β3 integrin (CD41) to the surface of RMS cells, similarly to lung (29) or breast cancer cells (30). This results in a chemotactic response of RMS cells to the PMV gradient and increased adhesiveness to fibronectin. Thus, PMVs may modulate metastatic behavior of RMS cells similarly as with other types of human cancer (29, 30).
Next, thrombin as a proteolytic enzyme may activate cells via PARs. So far, four PARs were identified (PAR1, -2, -3, -4), three of which are known to be activated by the N-terminal cleavage by thrombin (PAR1, -3, -4). The expression of PARs may differ with the tumor type (38–42). In our studies, we noticed human RMS cells express functional thrombin PAR1 and PAR3 and that stimulation of RMS by thrombin induces phosphorylation of MAPKp42/44 and MAPKp38, but not AKT, in these cells. Moreover, we noticed that thrombin inhibits AKT activation in RMS cells exposed to BM-CM.
Furthermore and to our surprise, direct stimulation of RMS cells by thrombin negatively affected their chemotactic and adhesive properties in response to BM-CM that we employed as a source of several chemoattractants (e.g., SDF-1, HGF) that mediate BM infiltration by RMS cells (11–14). This negative effect of thrombin on RMS chemotaxis may support D’Amico et al. data who proposed intra-tumor injection of thrombin as potential antimetastatic therapeutic strategy (53). Moreover, in contrast to other tumors (49), thrombin did not affect proliferation of our RMS cell lines. Unexpectedly, however, when PAR1-specific agonist TRAP6 was employed, which does not activate PAR3, it increased RMS proliferation. This suggested a potentially inhibitory influence of PAR3 on potentially pro-proliferate PAR1 signaling in RMS cells.
To learn more about the roles of PAR1 and PAR3 in RMS proliferation and metastasis, we knocked down both receptors employing a shRNA strategy. As expected we found that while thrombin does not affect growth of PAR1−/− cells, it stimulated proliferation of PAR3−/− cells. More importantly, PAR3−/− cells in contrast to PAR1−/− formed larger tumors in immunodeficient mice. These data demonstrate a novel role of PAR3 as a negative regulator of PAR1 signaling (Fig. 8). To support this notion, PAR1 may dimerize in endothelial cells with non-signaling PAR3; this binding and dimerization of both receptors potentializes thrombin signaling through PAR1 (52). In our RMS model, this PAR1-PAR3 interaction, however, shows a negative or inhibitory effect. Further studies are required to identify the proteins involved and clarify the molecular basis of this negative crosstalk.
In conclusion, we provide evidence for the first time that RMS-expressed TF activates pro-thrombin and that thrombin is a novel, underappreciated, pleiotropic, and pro-metastatic factor for these cells (Fig. 7). First, platelets activated in proximity to the tumor by thrombin release PMVs that chemoattract RMS cells and transfer to their surface several platelet-expressed receptors and adhesion molecules that are crucial for adhesion interaction with the endothelium. Second, thrombin may also directly interact with PAR1 and PAR3 on RMS cells. Activation of RMS cells by thrombin strongly inhibits their chemotactic and adhesive properties. Thus, by decreasing the responsiveness of RMS cells to local chemoattractants and decreasing adhesiveness of RMS cells, thrombin probably promotes the release of malignant cells from the primary tumor into circulation. Consequently, RMS cells that are covered by PMVs egress into circulation and respond to chemoattractants in distant organs for metastasis. Finally, we provide novel evidence for a PAR3-negative regulatory loop controlling the thrombin-PAR1 axis in RMS cells. This molecular basis of this phenomenon is currently under investigation in our laboratory.
Supplementary Material
Expression of tissue factor in human RMS cells at RNA level by real time PCR.
Panel A: Thrombin inhibits chemotactic activity of CW9019 cells to BM-CM in a dose-dependent manner. Panel B: Thrombin inhibits chemotactic activity of CW9019 cells to BM-CM when added to upper-, lower-, or both chambers. Data from three separate experiments are pooled together * p < 0.001.
Panels A and B: Thrombin reduces PMV-induced chemotaxis (Panel A) and adhesion (Panel B) of RMS cells. Data from four separate experiments are pooled together * p < 0.01. CW9019 (Panel C) or RH30 (Panel D) cells were pre-incubated with thrombin, PMVs, or a combination of both and were then injected intravenously into immunodeficient mice. Forty-eight hours later, the seeding efficiency for RMS cells in murine BM was evaluated by real-time PCR. Data from four separate experiments are pooled together * p < 0.01.
Selected clones with stable transfection of shRNA targeted against PAR1 (CW9019 PAR1−/− - Panel A) and PAR3 (CW9019 PAR3−/− - Panel B). Expressions of PAR1 (Panel A) and PAR3 (Panel B) were evaluated by real time-PCR. Data from three separate experiments are pooled together * p < 0.001.
Inhibition of chemotactic activity of CW9019 cells to BM-CM by thrombin is PAR1- (Panel A) but not PAR3-dependent (Panel B), as shown using PAR1−/− and PAR3−/− cells. Data from three separate experiments are pooled together * p < 0.001.
Acknowledgments
Supported by NIH grant R01 CA106281, NIH R01 DK074720, the Henry M. & Stella M. Hoenig Endowment to MZR.
The abbreviations used are
- ARMS
alveolar rhabdomyosarcoma
- BSA
bovine serum albumin
- BCC
blood coagulation cascade
- CXCR4
CXC chemokine receptor 4
- ERMS
embryonal rhabdomyosarcoma
- HGF
hepatocyte growth factor
- IGF-1
insulin growth factor-1
- LIF
Leukemia Inhibitory factor
- HUVEC
human umbilical vein endothelial cell
- PAR
protease-activated receptor
- PMV
platelet-derived microvesicle
- RMS
rhabdomyosarcoma
- SDF-1
stromal-derived factor 1
- TF
tissue factor
References
- 1.Ruymann FB, Newton WA, Ragab AH, Donaldson MH, Foulkes M. Bone marrow metastases at diagnosis in children and adolescents with rhabdomyosarcoma. Cancer. 1984;53:368–373. doi: 10.1002/1097-0142(19840115)53:2<368::aid-cncr2820530233>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Dickman PS, Tsokos M, Triche TJ. Biology of rhabdomyosarcoma: cell culture, xenografts and animal models. In: Maurer HM, Ruymann FB, Pochedly C, editors. Rhabdomyosarcoma and Related Tumours in Children and Adolescents. Boca Raton, FL: CRC Press; 1991. pp. 49–88. [Google Scholar]
- 3.Sandberg AA, Stone JF, Czarnecki L, Cohen JD. Hematologic masquerade of rhabdomyosarcoma. Am J Hematol. 2001;68:51–57. doi: 10.1002/ajh.1148. [DOI] [PubMed] [Google Scholar]
- 4.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 5.Anderson MJ, Shelton GD, Cavenee KW, Arden KC. Embryonic expression of the tumour-associated PAX3-FKHR fusion protein interferes with the developmental functions of PAX3. Proc Natl Acad Sci U S A. 2001;98:1589–1594. doi: 10.1073/pnas.98.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 8.Collins MH, Zhao H, Womer RB, Barr FG. Proliferative and apoptotic differences between alveolar rhabdomyosarcoma subtypes: a comparative study of tumors containing PAX3-FKHR gene fusions. Med Pediatr Oncol. 2000;37:83–89. doi: 10.1002/mpo.1174. [DOI] [PubMed] [Google Scholar]
- 9.Bennicelli JL, Advani S, Schafer BW, Barr FG. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–4356. doi: 10.1038/sj.onc.1202812. [DOI] [PubMed] [Google Scholar]
- 10.Kelly KM, Womer RB, Barr F. 3-FKHR and PAX7-FKHR fusions in rhabdomyosarcoma. J Pediatr Hematol Oncol. 1998;20:517–518. doi: 10.1097/00043426-199809000-00027. [DOI] [PubMed] [Google Scholar]
- 11.Libura J, Drukala J, Majka M, Tomescu O, Navenot JM, Kucia M, et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood. 2002;100:2597–606. doi: 10.1182/blood-2002-01-0031. [DOI] [PubMed] [Google Scholar]
- 12.Jankowski K, Kucia M, Wysoczynski M, Reca R, Zhao D, Trzyna E, et al. Both hepatocyte growth factor (HGF) and stromal-derived factor-1 regulate the metastatic behavior of human rhabdomyosarcoma cells, but only HGF enhances their resistance to radiochemotherapy. Cancer Res. 2003;63:7926–35. [PubMed] [Google Scholar]
- 13.Wysoczynski M, Miekus K, Jankowski K, Wanzeck J, Bertolone S, Janowska-Wieczorek A, et al. Leukemia inhibitory factor: a newly identified metastatic factor in rhabdomyosarcomas. Cancer Res. 2007;67:2131–40. doi: 10.1158/0008-5472.CAN-06-1021. [DOI] [PubMed] [Google Scholar]
- 14.Grymula K, Tarnowski M, Wysoczynski M, Drukala J, Barr FG, Ratajczak J, Kucia M, Ratajczak MZ. Overlapping and Distinct Role of CXCR7-SDF-1/ITAC and CXCR4-SDF-1 Axes in Regulating Metastatic Behavior of Human Rhabdomyosarcomas. Int J Cancer. 2010 doi: 10.1002/ijc.25245. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiegl M, Weltermann A, Stindl R, Fonatsch C, Lechner K, Gisslinger H. Massive disseminated intravascular coagulation and hyperfibrinolysis in alveolar rhabdomyosarcoma: case report and review of the literature. Ann Hematol. 1999;78:335–8. doi: 10.1007/s002770050525. [DOI] [PubMed] [Google Scholar]
- 16.Sills RH, Stockman JA, 3rd, Miller ML, Stuart MJ. Consumptive coagulopathy. A complication of therapy of solid tumors in childhood. Am J Dis Child. 1978;132:870–2. [PubMed] [Google Scholar]
- 17.Semino A, Danova M, Perlini S, Palladini G, Riccardi A, Perfetti V. Unusual manifestations of disseminated neoplasia at presentation: right-sided heart failure due to a massive cardiac metastasis and autoimmune thrombocytopenia in pleomorphic rhabdomyosarcoma of the adult. Am J Clin Oncol. 2006;29:102–3. doi: 10.1097/01.coc.0000160067.48292.70. [DOI] [PubMed] [Google Scholar]
- 18.Baron JA, Gridley G, Weiderpass E, Nyren O, Linet M. Venous thromboembolism and cancer. Lancet. 1998;351:1077–1080. doi: 10.1016/S0140-6736(97)10018-6. [DOI] [PubMed] [Google Scholar]
- 19.Gerber DE, Grossman SA, Streiff MB. Management of Venous Thromboembolism in Patients With Primary and Metastatic Brain Tumors. J Clin Oncol. 2006;24:1310–1318. doi: 10.1200/JCO.2005.04.6656. [DOI] [PubMed] [Google Scholar]
- 20.Falanga A, Zacharski L. Deep vein thrombosis in cancer: the scale of the problem and approaches to management. Ann Oncol. 2005;16:696–701. doi: 10.1093/annonc/mdi165. [DOI] [PubMed] [Google Scholar]
- 21.Hilson AJW, Sorensen HT, MellemkjAer L, Olsen JH. Cancer and Venous Thromboembolism. New Eng J Med. 1998;339:703–703. [Google Scholar]
- 22.Blom JW, Osanto S, Rosendaal FR. The risk of a venous thrombotic event in lung cancer patients: higher risk for adenocarcinoma than squamous cell carcinoma. J Thromb Haemost. 2004;2:1760–5. doi: 10.1111/j.1538-7836.2004.00928.x. [DOI] [PubMed] [Google Scholar]
- 23.Caine GJ, Lip GY, Stonelake PS, Ryan P, Blann AD. Platelet activation, coagulation and angiogenesis in breast and prostate carcinoma. Thromb Haemost. 2004;92:185–90. doi: 10.1160/TH03-11-0679. [DOI] [PubMed] [Google Scholar]
- 24.Chew HK, Davies AM, Wun T, Harvey D, Zhou H, White RH. The incidence of venous thromboembolism among patients with primary lung cancer. J Thromb Haemost. 2008;6:601–608. doi: 10.1111/j.1538-7836.2008.02908.x. [DOI] [PubMed] [Google Scholar]
- 25.Wun T, Chew HK, Zhou H, Harvey D, White RH. Cause of death among patients with local or regional stage cancer of the breast, colon and lung who develop venous thromboembolism (VTE) Blood. 2006;108:433a. [Google Scholar]
- 26.Sgouros J, Maraveyas A. Excess premature (3-month) mortality in advanced pancreatic cancer could be related to fatal vascular thromboembolic events. A hypothesis based on a systematic review of phase III chemotherapy studies in advanced pancreatic cancer. Acta Oncol. 2007:1–10. doi: 10.1080/02841860701687267. [DOI] [PubMed] [Google Scholar]
- 27.Kasthuri RS, Taubman MB, Mackman N. Role of tissue factor in cancer. J Clin Oncol. 2009;27:4834–8. doi: 10.1200/JCO.2009.22.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rak J, Milsom C, Magnus N, Yu J. Tissue factor in tumour progression. Best Pract Res Clin Haematol. 2009;22:71–83. doi: 10.1016/j.beha.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Janowska-Wieczorek A, Wysoczynski M, Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J, et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–60. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 30.Janowska-Wieczorek A, Marquez-Curtis LA, Wysoczynski M, Ratajczak MZ. Enhancing effect of platelet-derived microvesicles on the invasive potential of breast cancer cells. Transfusion. 2006;46:1199–209. doi: 10.1111/j.1537-2995.2006.00871.x. [DOI] [PubMed] [Google Scholar]
- 31.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–64. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 32.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–68. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 33.Ishihara H, Connolly AJ, Zeng D, Kahn ML, Zheng YW, Timmons C, et al. Protease-activated receptor 3 is a second thrombin receptor in humans. Nature. 1997;386:502–6. doi: 10.1038/386502a0. [DOI] [PubMed] [Google Scholar]
- 34.Xu WF, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, et al. Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci U S A. 1998;95:6642–6. doi: 10.1073/pnas.95.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molino M, Barnathan ES, Numerof R, Clark J, Dreyer M, Cumashi A, et al. Interactions of mast cell tryptase with thrombin receptors and PAR-2. J Biol Chem. 1997;272:4043–9. doi: 10.1074/jbc.272.7.4043. [DOI] [PubMed] [Google Scholar]
- 36.Kawabata A, Saifeddine M, Al-Ani B, Leblond L, Hollenberg MD. Evaluation of proteinase-activated receptor-1 (PAR1) agonists and antagonists using a cultured cell receptor desensitization assay: activation of PAR2 by PAR1-targeted ligands. J Pharmacol Exp Ther. 1999;288:358–70. [PubMed] [Google Scholar]
- 37.Al-Ani B, Saifeddine M, Kawabata A, Renaux B, Mokashi S, et al. Proteinase-activated receptor 2 (PAR(2)): development of a ligand-binding assay correlating with activation of PAR(2) by PAR(1)- and PAR(2)-derived peptide ligands. J Pharmacol Exp Ther. 1999;290:753–60. [PubMed] [Google Scholar]
- 38.Hu L, Lee L, Campbell W, Perez-Soler R, Karpatkin S. Role of endogenous thrombin in tumor implantation, seeding and spontaneous metastasis. Blood. 2004;104:2746–2751. doi: 10.1182/blood-2004-03-1047. [DOI] [PubMed] [Google Scholar]
- 39.Klepfish Greco M, Karpatkin S. Thrombin stimulates melanoma tumor-cell binding to endothelial cells and subendothelial matrix. Int J Cancer. 1993;53:978–982. doi: 10.1002/ijc.2910530620. [DOI] [PubMed] [Google Scholar]
- 40.Nierodzik M, Kajumo F, Karpatkin S. Effect of thrombin treatment of tumor cells on adhesion of tumor cells to platelets in vitro and metastasis in vivo. Cancer Res. 1992;52:3267–3272. [PubMed] [Google Scholar]
- 41.Nierodzik M, Chen K, Takeshita K, Li J, Huang Y, Feng X, et al. Protease-activated receptor 1 (PAR-1) is required and rate-limiting for thrombin-enhanced experimental pulmonary metastasis. Blood. 1998;92:3694–3700. [PubMed] [Google Scholar]
- 42.Shi X, Gangadharan B, Brass L, Ruf W, Mueller B. Protease-activated receptors (PAR1 and PAR2) contribute to tumor cell motility and metastasis, Mol. Cancer Res. 2004;2:395–402. [PubMed] [Google Scholar]
- 43.Hu L, Roth JM, Brooks P, Ibrahim S, Karpatkin S. Twist is required for thrombin-induced tumor angiogenesis and growth. Cancer Res. 2008;68:4296–302. doi: 10.1158/0008-5472.CAN-08-0067. [DOI] [PubMed] [Google Scholar]
- 44.Gruler H, Nucitelli R. Neural creast cell galvanotaxis: new data and a novel approach to the analysis of both galvanotaxis and chemotaxis. Cell Motil Cytoskeleton. 1991;19:121–133. doi: 10.1002/cm.970190207. [DOI] [PubMed] [Google Scholar]
- 45.Friedl P, Noble PB, Zanker KS. Lymphocyte locomotion in three-dimensional collagen gels. Comparison of three quantitative methods for analysing cell trajectories. J Immunol Methods. 1993;165:157–165. doi: 10.1016/0022-1759(93)90341-4. [DOI] [PubMed] [Google Scholar]
- 46.Korohoda W, Drukała J, Sroka J, Madeja Z. Isolation, spreading, locomotion on various substrata, and the effect of hypotonicity on locomotion of fish keratinocytes. Biochem Cell Biol. 1997;75:277–286. [PubMed] [Google Scholar]
- 47.Miekus K, Czernik M, Sroka J, Czyz J, Madeja Z. Contact stimulation of prostate cancer cell migration: the role of gap junctional coupling and migration stimulated by heterotypic cell-to-cell contacts in determination of the metastatic phenotype of Dunning rat prostate cancer cells. Biol Cell. 2005;97:893–903. doi: 10.1042/BC20040129. [DOI] [PubMed] [Google Scholar]
- 48.Coughlin SR. How the protease thrombin talks to cells. Proc Natl Acad Sci U S A. 1999;96:11023–7. doi: 10.1073/pnas.96.20.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hansen KK, Saifeddine M, Hollenberg MD. Tethered ligand-derived peptides of proteinase-activated receptor 3 (PAR3) activate PAR1 and PAR2 in Jurkat T cells. Immunology. 2004;112:183–90. doi: 10.1111/j.1365-2567.2004.01870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wysoczynski M, Shin DM, Kucia M, Ratajczak MZ. Selective upregulation of interleukin-8 by human rhabdomyosarcomas in response to hypoxia: therapeutic implications. Int J Cancer. 2009;126:371–381. doi: 10.1002/ijc.24732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu L, Ibrahim S, Liu C, Skaar J, Pagano M, Karpatkin S. Thrombin induces tumor cell cycle activation and spontaneous growth by down-regulation of p27Kip1, in association with the up-regulation of Skp2 and MiR-222. Cancer Res. 2009;69:3374–81. doi: 10.1158/0008-5472.CAN-08-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLaughlin JN, Patterson MM, Malik AB. Protease-activated receptor-3 (PAR3) regulates PAR1 signaling by receptor dimerization. Proc Natl Acad Sci U S A. 2007;104:5662–7. doi: 10.1073/pnas.0700763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Amico AV, Nelson AC, Babayan RK. Thrombin: implications for intratumor therapy against metastasis. J Cancer Res Clin Oncol. 1988;114:129–32. doi: 10.1007/BF00417825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of tissue factor in human RMS cells at RNA level by real time PCR.
Panel A: Thrombin inhibits chemotactic activity of CW9019 cells to BM-CM in a dose-dependent manner. Panel B: Thrombin inhibits chemotactic activity of CW9019 cells to BM-CM when added to upper-, lower-, or both chambers. Data from three separate experiments are pooled together * p < 0.001.
Panels A and B: Thrombin reduces PMV-induced chemotaxis (Panel A) and adhesion (Panel B) of RMS cells. Data from four separate experiments are pooled together * p < 0.01. CW9019 (Panel C) or RH30 (Panel D) cells were pre-incubated with thrombin, PMVs, or a combination of both and were then injected intravenously into immunodeficient mice. Forty-eight hours later, the seeding efficiency for RMS cells in murine BM was evaluated by real-time PCR. Data from four separate experiments are pooled together * p < 0.01.
Selected clones with stable transfection of shRNA targeted against PAR1 (CW9019 PAR1−/− - Panel A) and PAR3 (CW9019 PAR3−/− - Panel B). Expressions of PAR1 (Panel A) and PAR3 (Panel B) were evaluated by real time-PCR. Data from three separate experiments are pooled together * p < 0.001.
Inhibition of chemotactic activity of CW9019 cells to BM-CM by thrombin is PAR1- (Panel A) but not PAR3-dependent (Panel B), as shown using PAR1−/− and PAR3−/− cells. Data from three separate experiments are pooled together * p < 0.001.