Abstract
Background
Over 150 000 Malawians have started antiretroviral therapy (ART), in which first-line therapy is stavudine/lamivudine/nevirapine. We evaluated drug resistance patterns among patients failing first-line ART on the basis of clinical or immunological criteria in Lilongwe and Blantyre, Malawi.
Methods
Patients meeting the definition of ART failure (new or progressive stage 4 condition, CD4 cell count decline more than 30%, CD4 cell count less than that before treatment) from January 2006 to July 2007 were evaluated. Among those with HIV RNA of more than 1000 copies/ml, genotyping was performed. For complex genotype patterns, phenotyping was performed.
Results
Ninety-six confirmed ART failure patients were identified. Median (interquartile range) CD4 cell count, log10 HIV-1 RNA, and duration on ART were 68 cells/μl (23–174), 4.72 copies/ml (4.26–5.16), and 36.5 months (26.6–49.8), respectively. Ninety-three percent of samples had nonnucleoside reverse transcriptase inhibitor mutations, and 81% had the M184V mutation. The most frequent pattern included M184V and nonnucleoside reverse transcriptase inhibitor mutations along with at least one thymidine analog mutation (56%). Twenty-three percent of patients acquired the K70E or K65R mutations associated with tenofovir resistance; 17% of the patients had pan-nucleoside resistance that corresponded to K65R or K70E and additional resistance mutations, most commonly the 151 complex. Emergence of the K65R and K70E mutations was associated with CD4 cell count of less than 100 cells/μl (odds ratio 6.1) and inversely with the use of zidovudine (odds ratio 0.18). Phenotypic susceptibility data indicated that the nucleoside reverse transcriptase inhibitor backbone with the highest activity for subsequent therapy was zidovudine/lamivudine/tenofovir, followed by lamivudine/tenofovir, and then abacavir/didanosine.
Conclusion
When clinical and CD4 cell count criteria are used to monitor first-line ART failure, extensive nucleoside reverse transcriptase inhibitor and nonnucleoside reverse transcriptase inhibitor resistance emerges, with most patients having resistance profiles that markedly compromise the activity of second-line ART.
Keywords: Africa, antiretroviral failure, public health approach, resistance, resource-limited setting
Introduction
In the Malawi antiretroviral treatment program, over 150 000 patients have initiated first-line antiretroviral therapy (ART) since 2001, the majority starting after ART became free of change in 2004 [1,2]. Given the limited resources, the program relies on a public health approach for initiation of ART using clinical staging and rarely uses CD4 cell counts as a basis for ART initiation [3]. Similarly, identification of ART failure is based primarily on clinical criteria in most settings or immunological criteria when CD4 cell counts are available. HIV-1 RNA levels are not routinely monitored [3].
The first-line regimen in Malawi is stavudine (d4T), lamivudine (3TC), and nevirapine (NVP). In the event of toxicity, one can substitute zidovudine (ZDV) for d4Tor efavirenz (EFV) for NVP. The current second-line regimen is ZDV, 3TC, tenofovir (TDF), and lopinavir/ritonavir (LPV/r), consistent with 2006 WHO ART guidelines [4]. A more recent WHO document on second-line treatment suggests using TDF/3TC or abacavir (ABC)/didanosine (DDI) for patients failing first-line treatment with a ZDV or d4T and 3TC backbone [5].
Early virological failure on a nucleoside reverse transcriptase inhibitor (NRTI) and nonnucleoside reverse transcriptase inhibitor (NNRTI) regimen is associated with emergence of the M184V mutation and an NNRTI resistance mutation in approximately 50–75% of patients in resource-rich settings [6–8]. Continuation of a failing regimen may be associated with more complex mutation patterns, as has been observed in several studies in developing countries [9–13]. The number and pattern of resistance mutations may depend on the exact components of the regimen, HIV-1 subtype [14], and the duration of failure. Substantial NRTI resistance may occur, making empiric selection of second-line NRTI difficult.
Given that the Malawian program relies on clinical or immunological criteria for changing ART, accumulation of multiple NRTI mutations is expected. We sought to describe the degree of resistance among patients with virologically confirmed first-line ART failure and determine the optimal empiric NRTI backbone for a second-line regimen given the resistance patterns found.
Methods
Study setting
The study was conducted in Malawi within the national ART program. Kamuzu Central Hospital (KCH) is a tertiary referral hospital for the central region of Malawi located in Lilongwe. The Lighthouse Trust clinic is the primary ART clinic for KCH. The Queen Elizabeth Central Hospital (QECH) is the tertiary referral hospital for the southern region of Malawi, located in Blantyre. The ART clinic of QECH is coordinated by the Malawi College of Medicine. The University of North Carolina (UNC) Project and the John Hopkins Project are research and care centers serving as partners for the Lighthouse clinic and QECH, respectively.
Study population
HIV-positive patients (age >13 years) who initiated ART with d4T/3TC/NVP within the Malawi national ART program and were suspected of failure on the basis of clinical (new or progressive stage 4 condition) or immunological (CD4 cell count decrease >30%, CD4 cell count less than the pretreatment value) findings were screened from January 2006 to June 2007. Those patients who were confirmed as treatment failures on the basis of the above criteria and who had HIV-1 RNA of more than 1000 copies/ml on a sample obtained at the time of failure formed the primary study population for resistance testing.
Procedures
Human experimentation guidelines of the US Department of Health and Human Services, those of the Malawi National Health Sciences Research Committee and the UNC at Chapel Hill and Heath Insurance Portability and Accountability Act regulations were followed in the conduct of this study. For all patients, basic demographics, ART treatment history, and previous trends in CD4 cell count were obtained. Blood samples for viral load, CD4 cell count, and plasma storage were obtained at the initial assessment visit for treatment failure prior to the initiation of second-line treatment. Genotyping was performed on all samples, and phenotyping was performed only on samples with more than one NRTI mutation.
Laboratory evaluations
Specimens were processed in either the UNC Project laboratory in Lilongwe or the Johns Hopkins laboratory in Blantyre. HIV-1 RNA levels were measured by the Roche Amplicor HIV-1 RNA Monitor kit (version 1.5; Roche Molecular Diagnostics, Pleasanton, California, USA). CD4 cell counts were measured using flow cytometry by either the Becton Dickinson FACSCount system (Becton Dickinson, Mountain View, California, USA) or the EPICS-MCL Beckman Coulter Pan-Leuco Gating method (Beckman Coulter, Brea, California, USA). Plasma was stored at −80°C until shipment to the USA. HIV genotype testing was performed by the UNC Center for AIDS Research Virology Core using the TruGene HIV-1 Genotyping Kit (Siemens Medical Solutions Diagnostics, Tarrytown, New York, USA). Drug susceptibility was assessed by the PhenoSenseHIV assay at Monogram Biosciences, Inc (South San Francisco, California, USA).
Analysis
All statistical analyses were performed using Stata 8.2 (Stata Corporation, College Station, Texas, USA). Simple descriptive statistics included means, medians, and proportions, as appropriate. Student’s t test, Wilcoxon rank-sum, and chi-square methods were applied as required.
For genotype analysis, mutations were generally categorized according to the International AIDS Society-USA recommendations [15]. Samples with M184V, M184I, and M184V/I were considered to have 3TC and emtricitabine (FTC) resistance. NNRTI mutations included K103N, Y181C, Y181I, G190A, G190S, V108I, Y188L, V106M, P225H, and K103NS. Additionally, K101E and G190E were also included as NNRTI mutations [16,17]. NRTI mutations included K65R and K70E (associated with TDF resistance), thymidine analog mutations (TAMs) M41L, D67N, K70R, L210W, T215Y, T215F, K219Q, and K219E, and multinucleoside mutations, including the 69 insertion complex and the 151 complex [15]. Virus with 69 insertion or Q151M complex with K65R or K70E were considered pan-nucleoside resistant by genotype. Mixtures that included a resistance mutation were considered resistant.
Factors associated with the emergence of K65R and K70E, pan-nucleoside resistance mutations (69 insertion, Q151M complex with K65R and K70E), and the presence of three or more TAM mutations were evaluated using logistic regression. Factors considered in the models included sex, type of identification of failure (clinical vs. not clinical), clinic location, ZDV use, HIV-1 RNA at time of failure identification, and CD4 cell count at time of failure identification.
For phenotype analysis, for those NRTIs that have both upper and lower clinical cut-offs in the Monogram assay (TDF, 4.0 and 1.4, DDI, 2.2 and 1.3, ABC, 6.5 and 4.5), we performed one analysis using the lower cut-off and a second analysis whereby we categorized the virus as resistant or sensitive on the basis of the upper cut-off. All drugs were considered to be partially active if their fold change in 50% inhibitory concentration, was between the upper and lower limit cut-offs. For 3TC(3.5), FTC(3.5), d4T(1.7), ZDV(1.9), EFV(3.0), and NVP(4.5), a single lower cut-off was used. We evaluated whether three potential second-line NRTI backbones, ZDV/3TC/TDF, FTC/TDF, and DDI/ABC, would be predicted to have activity against the individual viral variant.
Results
Over the 18-month period, 203 patients had suspected treatment failure on the basis of clinical and immunological criteria and had viral load performed. Of these, 88 patients were suppressed (HIV RNA <400 copies/ml), six had HIV RNA between 400–1000 copies/ml, three had previous protease inhibitor exposure, and 10 had insufficient stored sample. The 96 remaining patients served as the basis for this evaluation. Among these 96 patients, most were identified by CD4 cell count decline (87) or new or progressive WHO stage 4 conditions (16) or both. Ninety-two patients received d4T/3TC/NVP as initial therapy, and four patients had received ZDV/3TC dual therapy prior to initiation of d4T/3TC/NVP. Thirty patients had ZDV substituted for d4T, and nine had EFV substituted for NVP because of toxicity. The median CD4 cell count, HIV RNA, and time on ART were 68 cells/μl, 52 374 copies/ml [interquartile range (IQR) 16913–138259], and 36.5 months (range 8–127 months), and 50% were women (Table 1). Patients who were on ZDV at the time of failure evaluation had longer durations of ART treatment (48.6 vs. 34.7 months, P <0.001) with a median of 27 months (IQR 10–42) of d4T use prior to switch and a median ZDV treatment duration of 14.9 months (IQR 5.4–23).
Table 1.
Characteristics of clinical/immunological failure patients with confirmed virological failure (HIV-1 RNA >1000 copies/ml) (n = 96).
| Variable | Median or percentage |
|---|---|
| Age (years) | 38 (IQR 31–45) |
| CD4 cell count (cells/μl) | 68 (IQR 23–174) |
| <50 | 44 |
| 51–200 | 35 |
| >200 | 21 |
| Log10 transformed HIV RNA (copies/ml) | 4.72 (IQR 4.26–5.16) |
| <10 000 | 16 |
| 10 001–100 000 | 54 |
| >100 000 | 30 |
| Time on ART (months) | 36.5 (IQR 26.6–49.8) |
| 0–24 | 20 |
| >24–36 | 29 |
| >36–48 | 22 |
| >48 | 29 |
| Clinical failure | 17 |
| Immunological failure | 91 |
| Lilongwe | 60 |
| Blantyre | 40 |
ART, antiretroviral therapy; IQR, interquartile range.
Of the 96 samples, two did not amplify, leaving 94 samples that could be evaluated by genotyping. All samples were subtype C. The median number of resistance mutations (any class) was five (IQR 3–7) with a range of 0–11. Five samples had no mutations identified (Table 2). NNRTI mutations occurred in 93% of the samples. Two patients had HIV with only NNRTI mutations. The median number of NNRTI mutations was two (range 0–3), and of those with at least one NNRTI mutation (n =87), 39% had one, 52% had two, and 8% had three mutations. The most frequent NNRTI mutations were Y181C (55%), G190A (30%), K103N (28%), K101E (15%), Y188L (8%), V106M (7%), Y181I (6%), K103N/S (2%), P225H (1%), G190S (1%), and G190E (1%). Among patients with exposure to NVP but not to EFV (n =85), the most common mutations were Y181C (55%) and G190A (26%). Among those with exposure to both NVP and EFV (n =9), K103N (33%) and G190A (33%) mutations were the most common. Y181C was significantly more common among NVP-only-exposed patients (55 vs. 11%, P =0.012).
Table 2.
Patterns of mutations seen among patients with antiretroviral therapy failure in Malawi (n = 94). Full NRTI and NNRTI resistance profiles available online on journal website.
| Resistance patterna | n |
|---|---|
| Wildtype virus (no mutations identified) | 5 |
| M184 only | 0 |
| NNRTI mutations only | 2 |
| M184V and NNRTI mutations only | 15 |
| NNRTI mutations ± 184V-containing virus and additional mutations | |
| TAM-containing virus | 53 |
| K65R or K70E | 22 |
| K65R/K70E and TAM mutations | 7 |
| Q151M complex | 18 |
| Multi-nucleoside mutation combinations | |
| Q151M complex and K65R/K70E mutations | 15 |
| 69 insertion | 1 |
| Multi-nucleoside (Q151 and K65R/K70E or 69 insertion) | 16 |
Categories are not mutually exclusive. NNRTI, nonnucleoside reverse transcriptase inhibitor; TAM, thymidine analog mutations.
The M184Vor M184I mutation was present in virus from 77 patients (81%), although never as the only mutation. Fifteen patients (16%) had virus with M184V and NNRTI mutations only.
The most common mutation pattern was M184V and NNRTI mutations with one or more TAMs, which occurred in 56% of patients. Of the patients with TAM-containing virus (n =53), 28% had one, 28% had two, and 44% had three or more TAMs. The most frequent TAMs were T215 F/Y (73%), D67N (53%), K70R (36%), M41L (36%), K219 Q/E (23%), and L210W (23%). Patients who had ever received ZDV had more TAMs (1.8 vs. 1.2 mutations, P =0.055) and were more likely to have at least three TAMs (19% for d4T vs. 36% for ZDV, P =0.09). In spite of no known exposure to TDF, 23% (22/94) of patients acquired virus with the K70E (n =4) or K65R mutations (n =18). Eighteen patients developed virus with the Q151M mutation, and in 15 patients, the Q151M mutation was associated with either K65R or K70E. One patient developed a T69 insertion. In total, 16 patients had pan-nucleoside-resistant virus, on the basis of the presence of the K65R or K70E mutation in association with the Q151M complex or the presence of a 69 insertion. Compared with those with ZDV exposure, the emergence of the K65R, Q151M, and K70E mutations was more common in those exposed only to d4T: K65R (24 vs. 7%, P =0.05), K70E (6 vs. 0%, P =0.183), and Q151M (24 vs. 7%, P =0.05).
No patient with a CD4 cell count more than 200 cells/μl or HIV-1 RNA less than 10 000 copies/ml had pan-nucleoside-resistant virus. One-third of patients with HIV-1 RNA less than 10 000 copies/ml had TAMs. Only two of 30 patients who had ever received ZDV developed either K65R/K70E or pan-nucleoside resistance mutations.
Univariate and multivariate analyses were performed to look for associations with the emergence of pan-nucleoside resistance, at least three TAMS, or either K65R or K70E. A CD4 cell count of less than 100 cells/μl was strongly associated with emergence of K65R or K70E, pan-nucleoside resistance, and the emergence of three or more TAMs (Table 3). However, associations with ZDVuse were not consistently in the same direction. ZDV use was strongly protective for the emergence of K65R or K70E. ZDV use was protective for the emergence of pan-nucleoside resistance in the univariate analysis resistance, but the trend was no longer significant in the multivariate analysis [odds ratio (OR) 0.13, 95% confidence interval (CI) 0.02–1.07]. Conversely, ZDV was associated with an increased risk for emergence of three or more TAMs in the multivariate analysis (OR 3.4, 95% CI 1.7–10.97).
Table 3.
Univariate and multivariate logistic modeling of three potential outcomes for emergence of resistance among patients failing antiretroviral therapy based on clinical or immunological criteria in Malawi.
| Outcome of interest | At least three TAMs |
K65R or K70 E tenofovir-associated mutations |
Pan-nucleoside resistance Q151M and tenofovir mutations or 69 insertion |
|||
|---|---|---|---|---|---|---|
| Univariate | Multivariatea | Univariate | Multivariatea | Univariate | Multivariatea | |
| CD4 cell count (cells/μl) | ||||||
| >100 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 100 or less | 3.9 (1.2–12.6) | 5.57 (1.41–22.02) | 5.4 (1.5–19.7) | 6.1 (1.47–25.0) | 12.2 (1.5–97.0) | 9.9 (1.2–84.0) |
| ZDV use | ||||||
| No | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Yes | 2.3 (0.85–6.05) | 3.4 (1.07–10.97) | 0.18 (0.04–0.81) | 0.18 (0.04–0.94) | 0.12 (0.015–0.93) | 0.13 (0.02–1.07) |
Data are given as odds ratio (95% confidence interval). Outcomes include having at least three TAMs, the development of tenofovir-associated mutations (K65R or K70E) or expected pan-nucleoside resistance mutation (Q151M with K65R or K70E mutations or the 69 insertion). ART, antiretroviral therapy; TAM, thymidine analog mutations; ZDV, zidovudine.
Adjusted for HIV-1 RNA (copies/ml), clinical failure, clinic site, and sex. Duration of ART was not considered because of colinearity with the use of zidovudine.
Phenotypic drug susceptibility was determined for 70 samples with complex genotypes. Given the selection criteria, nearly all samples had high-level NNRTI resistance (96% for NVP and 83% for EFV) and 3TC resistance (97%). Substantial phenotypic resistance was demonstrated for most other NRTI agents (Fig. 1). The majority of viruses (61%) were fully susceptible to TDF, whereas fewer were fully susceptible to ZDV (37%), d4T (37%), ABC (33%), or DDI (1%). Partial susceptibility of the virus isolates was noted for TDF (33%), DDI (29%), and ABC (20%). Consistent with the genotypic findings, those with ZDV exposure were more likely to retain full susceptibility to TDF (82 vs. 56%, P =0.05).
Fig. 1. Fold change in 50% inhibitory concentration for individual nonnucleoside reverse transcriptase inhibitor and nucleoside reverse transcriptase inhibitor agents.
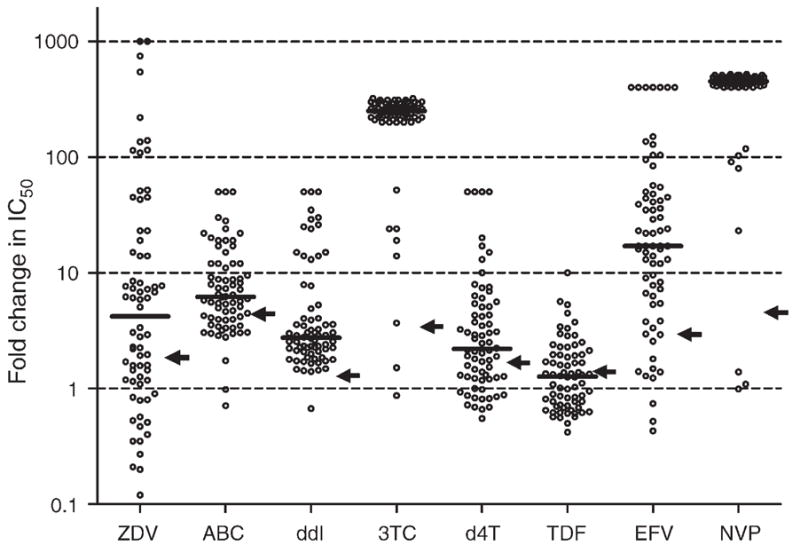
The bold horizontal line for each drug represents the median fold change and the arrows indicate the lower clinical cut-off or the biological cut-off, as applicable. Points clustered together at high fold-change values represent viruses with high-level resistance; an arbitrary fold-change value was assigned on the basis of the ratio between the highest drug concentration tested in the assay and the reference virus IC50 in that assay run. 3TC, lamivudine; ABC, abacavir; d4T, stavudine; DDI, didanosine; EFV, efavirenz; IC50, 50% maximal inhibitory concentration; NVP, nevirapine; TDF, tenofovir; ZDV, zidovudine.
We evaluated the expected activity of NRTI combinations recommended by the WHO considering only the phenotype results (Table 4) (n =70). Three NRTI combinations were evaluated, including ZDV/3TC/TDF, FTC/TDF, and DDI/ABC, taking advantage of the clinical cut-off available for TDF, ABC, and DDI (Table 4a–c). Table 5 includes an overall assessment of activity, including only fully sensitive NRTI or sensitive NRTI and those with partial susceptibility. Overall, the three-NRTI combination provided the greatest likelihood of one or two active NRTIs in either analysis, although 30 and 6% would have no active NRTI option depending on consideration of partial susceptibility (Table 5). Notably, the ABC/DDI combination performed poorly in either assessment, with neither NRTI active in 67% in the more stringent analysis and 46% when partial susceptibility was considered.
Table 4.
Resistance profiles according to combinations chosen.
| (a) Tenofovir | Zidovudinea |
(b) Tenofovir | Emtricitabine |
(c) Abacavir | Didanosine |
||||
|---|---|---|---|---|---|---|---|---|---|
| S (%) | RS (%) | S (%) | RS (%) | S (%) | P (%) | R (%) | |||
| S (%) | 29 | 33 | S (%) | 3 | 59 | S (%) | 1 | 21 | 10 |
| P (%) | 9 | 24 | P (%) | 0 | 33 | P (%) | 0 | 7 | 14 |
| R (%) | 0 | 6 | R (%) | 0 | 6 | R (%) | 0 | 0 | 46 |
(a–c) Bivariate comparisons including NRTI with clinical cut-offs. (a) Tenofovir/zidovudine, (b) Emtricitabine/tenofovir. (c) Abacavir/didanosine. n =70. NRTI, nucleoside reverse transcriptase inhibitor; P, partial susceptibility (between lower and upper cut-off); R, resistant (over upper cut-off); RS, reduced susceptibility for drugs with only one cut-off established; S, sensitive (below lower cut-off).
Addition of lamivudine does not modify results compared with zidovudine/tenofovir only.
Table 5.
Overall nucleoside reverse transcriptase inhibitor combination activity including either fully sensitive agents or agents with full sensitivity plus partial susceptibility.
| Drug combination | Sensitive only |
Sensitive and partial susceptibility |
||||
|---|---|---|---|---|---|---|
| Phenotyped specimens (n =70) | Two active NRTIs | One active NRTI | 0 active NRTI | Two active NRTIs | One active NRTI | 0 active NRTI |
| Emtricitabine/tenofovir | 3% | 59% | 39% | 3% | 91% | 6% |
| Abacavir/didanosine | 1% | 31% | 67% | 29% | 24% | 46% |
| Zidovudine/lamivudine/tenofovira | 29% | 42% | 30% | 38% | 56% | 6% |
NRTI, nucleoside reverse transcriptase inhibitor.
Addition of lamivudine does not modify results compared with zidovudine/tenofovir only.
Discussion
In this study, we confirmed that a high level of NRTI resistance could accumulate when ART failure is identified by clinical and immunological criteria. Our novel and most ominous finding, based on genotyping, was that 17% of patients would not be expected to have any active NRTI agents remaining. The use of d4T in subtype C virus does not follow predictable patterns in the emergence of mutations and appears to be promiscuous in its ability to select for K65R and K70E (associated with TDF resistance), multiple TAM pathways, Q151M, and T69 insertions [14,18].
D4T use was associated with the emergence of K65R and K70E mutations in 30% of patients. Although initially described to emerge at low frequency in patients receiving TDF-containing regimens [6,7], these mutations are now being identified in resource-poor settings where d4T/3TC/NVP is the first-line regimen [9,10,13,19]. ZDV use was independently associated with the lack of emergence of these mutations, consistent with observations that K65R increases ZDV sensitivity and TAMs and K65R appear to be antagonistic [20–22]. Although in subtype B virus, both thymidine analogs, d4T and ZDV, select for similar TAMs, our data demonstrate that d4T-based initial therapy in patients infected with subtype C virus selects for a broader array of mutations, including the K65R mutation, which may be because of differences in the nucleotide sequence of subtype C virus in this region of reverse transcriptase [14,23]. ZDV use, in contrast, was associated with the emergence of more than three TAMs. Higher levels of viral replication and lower CD4 cell counts, possibly related to longer duration of virological failure, are associated with K65R and other non-TAM NRTI mutations [24].
The higher proportion of extensive NRTI resistance in our survey, compared with other studies, likely reflects a longer duration of failure of our patients. High-level resistance to NRTI agents has been uncommonly reported after 12 months of d4T/3TC/NVP usage [19,25]. Compared with data from South Africa or Thailand [9,10], our patients had longer duration on ART (36.5 vs. 10.8 or 19 months), lower median CD4 cell count (median 68 vs. 161.5 or 174 cells/μl), and higher HIV-1 RNA levels (4.72 vs. 4.29 or 4.0 log10 copies/ml) at the time of failure, suggesting that patients in our cohort had likely been failing on therapy longer or had higher baseline levels of replication than these other cohorts with access to virological monitoring. Additionally, we found the highest levels of resistance, as defined by at least three TAMs, K65R or 70E, or pan-nucleoside resistance, among those with CD4 cell count less than 100 cells/μl. However, other than these indirect measures, we have no means of knowing the exact amount of time these patients were failing at the time of evaluation.
D4T/3TC/NVP serves as the first-line regimen for the majority of sub-Saharan Africa because of its low cost, its generic fixed-dose combination tablet, and the fact that it does not require laboratory monitoring of side effects as treatment is being initiated. However, long-term toxicities such as lactic acidosis, peripheral neuropathy, and lipoatrophy have raised concerns about long-term safety. Our data demonstrate that d4T may also have negative effects with respect to the emergence of resistance. To minimize the long-term side effects of d4T, a proposed switch to ZDV-based therapy after 6 months once the patient is stabilized and the chances for anemia are lower has been suggested [26]. Given our observation that ZDV decreased the odds of TDF and pan-NRTI resistance, this same strategy may also prove useful in preventing the emergence of pan-NRTI-resistant virus and preserving the use of TDF for second-line therapy. However, although ZDV use is associated with decreased odds of pan-nucleoside resistance, its use is associated with higher rates of virus with more than three TAMs compromising other NRTI activity and also limiting NRTI choices for second-line treatment.
Currently, WHO recommends FTC/TDF or ABC/DDI as the NRTI backbone of second-line ART regimens to be used in resource-limited settings [4,5]. On the basis of the complexity of assessing partial activity to ABC, DDI, and TDF using genotype analysis, we explored our more complex genotypes with phenotypic analysis [27]. Our findings suggest that neither drug would be fully active in these combinations in 67% of patients for ABC/DDI and 39% for FTC/TDF. The addition of ZDV, as in the Malawi ART program, to 3TC and TDF is supported by our findings and increases the likelihood of having two active NRTIs in the regimen, particularly if this determination is based on the lower cut points of the phenotypic assay. Many country programs, including that in Malawi, now use LPV/r for second-line regimens [3]. Data on the response to second-line treatment, stratified by the presence of broad nucleoside resistance, will be critical for guideline development and future research. Success of second-line therapy may be heavily dependent on the LPV/r component, which has shown substantial but less than optimal activity as initial monotherapy [28]. Outcomes of second-line therapy in developing world settings that have more virological monitoring and different viral subtypes cannot be extrapolated across all developing world settings as the activity of the nucleoside component may vary substantially. Nucleoside analog may only add cost and toxicity. Low success rates of second-line therapy in some settings raise the question of whether new classes of drugs may be needed if viral monitoring is not available. Paradoxically, the absence of virological monitoring may thwart the public health approach, that is, use of a single regimen for most patients, for second and later lines of therapy. Although recent modeling has suggested that virological monitoring would offer little additional benefit in terms of clinical outcomes, it is not clear that the potential for 15–25% prevalence of multi-NRTI or TDF resistance or both, and unknown subsequent clinical outcomes with limited active agents could be accounted for with certainty [29].
The fact that we did not find any pan-nucleoside resistance in patients with HIV-1 RNA less than 10 000 copies/ml, similar to other studies [24], supports the WHO guidelines of a virological trigger of 10 000 copies/ml for changing therapy. However, when clinical and immunological means are used to identify failures, as in our study, only 16% of the patients fell into this category. The strength of our study is that we have the largest description of resistance in a government program that uses clinical and immunological monitoring to define ART failure and limits second-line treatment to central hospitals. Additionally, we have included phenotype data to support the interpretation of the complex NRTI resistance patterns. All patients who were initiated on second-line treatment at these clinics during the 18-month study recruitment period were included. However, we acknowledge that many patients who were failing virologically may not have been included in this study that identified patients only through clinical and immunological means, and that patients failing by these criteria could be missed in busy public health-based clinics. Therefore, the actual number of patients with resistant virus is likely to be higher. However, if rapid progressors were preferentially selected through our screening procedures, the complex resistance patterns may have been overrepresented. As with all resistance studies, the genotype determined at the time of sampling may not reflect archived virus with other resistance patterns. For patients on ZDV, some mutations apparently selected by d4T (K65R, K70E) may have diminished to levels below the limit of detection, resulting in an underestimate in the prevalence of these mutations. As the phenotype cut-offs have been developed using patients primarily infected with subtype B, these cut-offs may not apply to patients infected with other subtypes, but recent evidence suggests no significant difference between subtype B and C [30]. However, this could influence our findings on expected drug susceptibility.
In summary, the high rate of broad NRTI resistance among patients on the most common first-line regimen in the developing world, monitored using clinical and immunological definitions of ART failure, strongly supports the need for improvement in monitoring of ART failure, in resource-poor settings. In lieu of virological monitoring, failing on ZDVappears to allow more options in terms of future treatment and argues for its consideration for broader rollout in favor of d4T-based regimens. In settings where clinical monitoring is used to detect ART failure and d4T-based regimens serve as the first line, the use of a ZDV/3TC/TDF backbone demonstrates the best chance for activity of second-line ART.
Acknowledgments
We are grateful for the funding from the National AIDS Commission of Malawi. We are very appreciative of the support from Monogram Biosciences, particularly Sunny Choe and the Clinical Reference Laboratory, for the phenotype analyses. We would like to thank the staff of the ART clinic of QECH and the Lighthouse clinic, the laboratory staff at UNC Project and Johns Hopkins Project and University of North Carolina CFAR Clinical and Virology Core Laboratory (P30 AI50410), UNC Project management, and the HIV Unit of the Ministry of Health. We dedicate this work to the late Dr George Joaki, who until his untimely death, served as the UNC project laboratory manager.
Mina C. Hosseinipour, Johnstone Kumwenda, Joep J.G. van Oosterhout, Sam Phiri, Joseph Eron and Ralf Weigel conceived the design and implemented of the study. Julie A.E. Nelson, Debbie Kamwendo, Neil Parkin and Susan A. Fiscus supported the laboratory analysis. Mina C. Hosseinipour wrote the first draft and performed the statistical analysis. All authors participated in the interpretation of data and critical review and revision of the manuscript.
References
- 1.Harries AD, Schouten EJ, Libamba E. Scaling up antiretroviral treatment in resource-poor settings. Lancet. 2006;367:1870–1872. doi: 10.1016/S0140-6736(06)68809-0. [DOI] [PubMed] [Google Scholar]
- 2.HIV Unit. ART quarterly report for December 2007. Ministry of Health; Malawi: 2007. [Google Scholar]
- 3.National AIDS Commission and Malawi Ministry of Health and Population. [Accessed January 2009];Treatment of AIDS: guidelines for the use of antiretroviral therapy in Malawi. (2). 2006 April; http://www.hivunitmohmw.org/Main/AntiretroviralTherapy.
- 4.WHO. Recommendations for a public health approach. 2006. Geneva: WHO; 2006. Antiretroviral therapy for HIV infection in adults and adolescents in resource-limited settings: toward universal access. [Google Scholar]
- 5.WHO. Report of a WHO Working Group Meeting. Geneva: WHO; 2007. Prioritizing second line antiretroviral drugs for adults and adolescents: a public health approach. [Google Scholar]
- 6.Margot NA, Lu B, Miller MD Study 903 Team. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in Study 903. HIV Med. 2006;7:442–450. doi: 10.1111/j.1468-1293.2006.00404.x. [DOI] [PubMed] [Google Scholar]
- 7.Gallant JE, DeJesus E, Arribas JR, Pozniak AL, Gazzard B, Campo RE, et al. Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine and efavirenz for HIV. N Engl J Med. 2006;354:251–260. doi: 10.1056/NEJMoa051871. [DOI] [PubMed] [Google Scholar]
- 8.Gulick R, Ribaudo H for the AIDS Clinical Trials Group Study A5095 Team. Triple nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med. 2004;350:1850–1861. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]
- 9.Marconi VC, Sunpath H, Lu Z, Gordon M, Koranteng-Apeagyei K, Hampton J, et al. South Africa Resistance Cohort Study Team. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008;46:1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Piyavong B, Chumpathat N, Chantratita W. Options for the second antiretroviral regimen for HIV-infected patients whose initial regimen of a fixed-dose combination of stavudine, lamivudine, and nevirapine fails. Clin Infect Dis. 2007;44:447–452. doi: 10.1086/510745. [DOI] [PubMed] [Google Scholar]
- 11.DART Virology Group and Trial Team. Virological response to a triple nucleoside/nucleotide analogue regime over 48 weeks in HIV-1 infected adults in Africa. AIDS. 2006;20:1291–1299. doi: 10.1097/01.aids.0000233572.59522.45. [DOI] [PubMed] [Google Scholar]
- 12.Sung H, Jung Y, Kang M, Baie I, Chang H, Woo J, Cho Y. High frequency of drug resistance mutations in human immunodeficiency virus type-1 infected Korean patients treated with HAART. AIDS Res Hum Retroviruses. 2007;23:1223–1229. doi: 10.1089/aid.2007.0008. [DOI] [PubMed] [Google Scholar]
- 13.Ferradini L, Jeannin A, Pinoges L, Izopet J, Odhiambo D, Mankhambo L, et al. Scaling up of highly active antiretroviral therapy in a rural district of Malawi: an effectiveness assessment. Lancet. 2006;367:1335–1342. doi: 10.1016/S0140-6736(06)68580-2. [DOI] [PubMed] [Google Scholar]
- 14.Brenner BG, Oliveire M, Doualla-Bell F, Moisi DD, Ntemgwa M, Frankel F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20:F9–F13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 15.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard H, Kuritzkes D, Pillay D, et al. Update on the drug resistance mutations in HIV-1: Spring 2008. Top HIV Med. 2008;16:62–68. doi: 10.1007/s11750-007-0034-z. [DOI] [PubMed] [Google Scholar]
- 16.Bacheler Lee T, Anton Elizabeth D, Kudish P, Baker D, Bunville J, Krakowski K, et al. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob Agents Chemother. 2000;44:2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Gamarnik A, Limoli K, Petropoulous CJ, Whitcomb J. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J Virol. 2003;77:1512–1523. doi: 10.1128/JVI.77.2.1512-1523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Lerma JG, MacInnes H, Bennett D, Reid P, Nidtha S, Weinstock H, et al. A novel genetic pathway of human immunodeficiency virus type 1 resistance to stavudine mediated by the K65R mutation. J Virol. 2003;77:5685–5693. doi: 10.1128/JVI.77.10.5685-5693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamya MR, Mayanja-Kissa H, Kambugu A, Bakeera-Kitaka S, Semitala F, Mwebaze-Songa P, et al. Predictors of long-term viral failure among Ugandan children and adults treated with antiretroviral therapy. J Acquir Immune Defic Syndr. 2007;46:187–193. doi: 10.1097/QAI.0b013e31814278c0. [DOI] [PubMed] [Google Scholar]
- 20.Parikh UM, Barnas DC, Faruki H, Mellors JW. Antagonism between the HIV-1 reverse-transcriptase mutation K65R and thymidine-analogue mutations at the genomic level. J Infect Dis. 2006;194:651–660. doi: 10.1086/505711. [DOI] [PubMed] [Google Scholar]
- 21.Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21:1405–1414. doi: 10.1097/QAD.0b013e3281ac229b. [DOI] [PubMed] [Google Scholar]
- 22.Parikh UM, Bacheler L, Koontz D, Mellors JW. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J Virol. 2006;80:4971–4977. doi: 10.1128/JVI.80.10.4971-4977.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coutsinos D, Invernizzi CF, Xu H, Moisi D, Oliveira M, Brenner BG, Wainberg MA. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J Virol. 2009;83:2029–2033. doi: 10.1128/JVI.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sungkanuparph S, Manosuthi W, Kiertiburanakul S, Saekang N, Pairoj W, Chantratita W. Prevalence and risk factors for developing K65R mutations among HIV-1 infected patients who fail an initial regimen of fixed-dose combination of stavudine, lamivudine, and nevirapine. J Clin Virol. 2008;41:310–313. doi: 10.1016/j.jcv.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Marcelin A, Jarrousse A, Derache A, Ba M, Dakouo ML, Doumbia A, et al. HIV drug resistance after the use of generic fixed-dose combination stavudine/lamivudine/nevirapine as standard first-line regimen. AIDS. 2007;21:2341–2342. doi: 10.1097/QAD.0b013e328235a527. [DOI] [PubMed] [Google Scholar]
- 26.Isaakidis P, Raguenaud ME, Phe T, Khim S, Kuoch S, Khem S, et al. Evaluation of a systematic substitution of zidovudine for stavudine-based HAART in a program setting in rural Cambodia. J Acquir Immune Defic Syndr. 2008;49:48–54. doi: 10.1097/QAI.0b013e31817bec19. [DOI] [PubMed] [Google Scholar]
- 27.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arribas JR, Pulido F, Delgado R, Lorenzo A, Miralles P, Arranz A, et al. Lopinavir/ritonavir as single-drug therapy for maintenance of HIV-1 viral suppression: 48 week results of a randomized, controlled, open-label, proof-of-concept pilot clinical trial (OK study) J Acquir Immune Defic Syndr. 2005;40:280–287. doi: 10.1097/01.qai.0000180077.59159.f4. [DOI] [PubMed] [Google Scholar]
- 29.Phillips A, Pillay D, Miners A, Bennett D, Gilks C, Lundgren J. Outcomes from monitoring of patients on antiretroviral therapy in resource-limited settings with viral load, CD4 cell count, or clinical observation alone: a computer simulation model. Lancet. 2008;371:1443–1451. doi: 10.1016/S0140-6736(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 30.Choe SS, Stawiski E, Parkin NT. Interpretation of drug susceptibility and replicative capacity results from subtype C HIV-1 protease/RT is not influenced by the subtype of the resistance test vector [abstract #107]. XV International HIV Drug Resistance Workshop; 12–16 June 2007; Bridgetown, Barbados. 2007. [Google Scholar]


