Abstract
The role of parent-of-origin effects (POE) in the etiology of complex diseases such as type 2 diabetes (T2DM) and obesity is currently of intense interest, but still largely unclear. POE are transmittable genetic effects whereby the expression of the phenotype in the offspring depends upon whether the transmission originated from the mother or father. In mammals, POE can be caused by genetic imprinting, intrauterine effects, or maternally inherited mitochondrial genes. In this paper, we describe the different mechanisms underlying POE, characterize known examples of POE in rare forms of diabetes, and review the evidence from linkage and association studies for POE in T2DM and obesity. Finally, we summarize some of the new and established statistical and experimental approaches commonly used to detect POE. Through this paper, we hope emphasizes the potentially significant importance of POE in the etiology of T2DM and obesity.
Keywords: Parent-of-origin effects, imprinting, mitochondrial genome, intrauterine environment, epigenetics, type 2 diabetes, obesity
I. Introduction
Parent-of-origin effects (POE) refer to a class of genetic effects that are transmitted from parents to offspring whereby the expression of the phenotype in the offspring depends upon whether the transmission originated from the mother or father. POE are frequently equated with the concept of imprinting, in which an allele of a specific gene is silenced (via epigenetic mechanisms such as methylation) when inherited from one parent and expressed when inherited from the other. However, other parent-of-origin effects have also been described. In general, there are three types of parent-specific transmittable effects: 1) those arising from epigenetic regulation of gene expression (such as imprinting); 2) those arising from the effects of the maternal intrauterine environment on the developing fetus; and 3) those arising from genetic variation in the maternally inherited mitochondrial genome. The defining characteristics of all three types of POE are that: 1) parental transmission of alleles must involve some mechanism other than classical Mendelian segregation of nuclear genes, and 2) offspring expression of the phenotype is at least partly dependent upon whether the transmission occurred from the mother or father.
POE are of particular relevance to the study of diabetes and obesity because some of the mechanisms involved are often associated with growth and development. In fact, most known imprinted genes influence pathways that involve growth or placental development [1, 2]. There are several known examples of POE in largely monogenic forms of diabetes as outlined below. However, researchers have only begun to assess the existence of parent-of-origin effects in more common forms of diabetes.
The purpose of this review is to describe the major mechanisms underlying POE and provide known examples of each, to identify and characterize known examples of POE in rare forms of diabetes, to describe how POE may influence susceptibility to type 2 diabetes (T2D) and obesity, and to summarize some of the new and established statistical and experimental approaches commonly used to detect POE and their role in future studies.
II. Mechanisms of POE
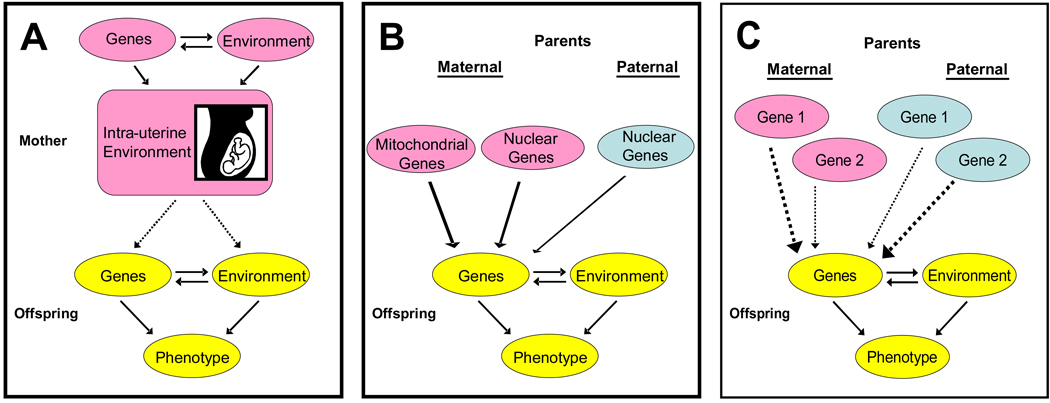
There are three mechanisms which may underlie parent-specific transmittable effects: 1) epigenetic regulation of gene expression (such as imprinting), 2) influence of the maternal intrauterine environment on fetal development, and 3) expression of genetic variation in the maternally-inherited mitochondrial genome. The defining characteristics of these classes of POE are that: 1) they require that parental transmission involve some mechanism other than classical Mendelian segregation of nuclear genes, and 2) offspring expression of the phenotype is at least partly dependent upon whether the transmission occurred from the mother or father. These mechanisms are illustrated pictorially in Figure 1.
Figure 1.
Mechanisms of Parent-of-Origin Effects include (A) the influence of the maternal intrauterine environment on gene expression patterns, (B) exclusive maternal inheritance of functional genes in the mitochondrial genome, and (C) differential expression of gene copies depending on from which parent, mother or father, they are inherited (paternally-dependent expression shown). Pathways corresponding to parent-of-origin effects indicated by dashed arrows.
Imprinting
Genomic imprinting is one mechanism of POE in which gene expression is dependent on the parent of origin of the inherited allele [3]. With genomic imprinting, the molecular modifications of DNA occur in germline cells and occur in different patterns depending upon the sex of the parent. These molecular modifications are termed “epigenetic” because they involve changes to DNA structure other than those involving changes to the DNA sequence. Like sequence changes, these modifications can be stably transmitted through several generations of cells or organisms. Unlike sequence changes, these epigenetic modifications can also be reset, or undone, under appropriate conditions such as during primordial germ cell development [4]. Known molecular epigenetic mechanisms involved in imprinting include changes in histone deacetylation and modification of cytosine methylation [3]. Histone modifications are post-translational modifications of the core histone proteins that constitute the nucleosome. Some evidence suggests, although not conclusively, that these modifications are used to program genes for activation during specific steps in cellular differentiation [5]. DNA methylation occurs when a hydrogen atom of the cytosine base is replaced by a methyl group [5]. In humans, this phenomenon is restricted mainly to cytosines in CpG clusters. The role of methylation in imprinting, which involves parent-specific methylation of CpG-rich domains during gametogenesis, is not completely understood, but evidence based on mouse knock-out models implicates control by a group of proteins including methyltransferases, methyl-binding proteins, and histone-modifying proteins [3]. Methylation causes the structural change of chromatin and can produce gene silencing when it occurs in a promoter region [6]. As a result, the methylated allele is not transcribed. For example, if the maternal allele is methylated, then only the paternal allele is transcribed.
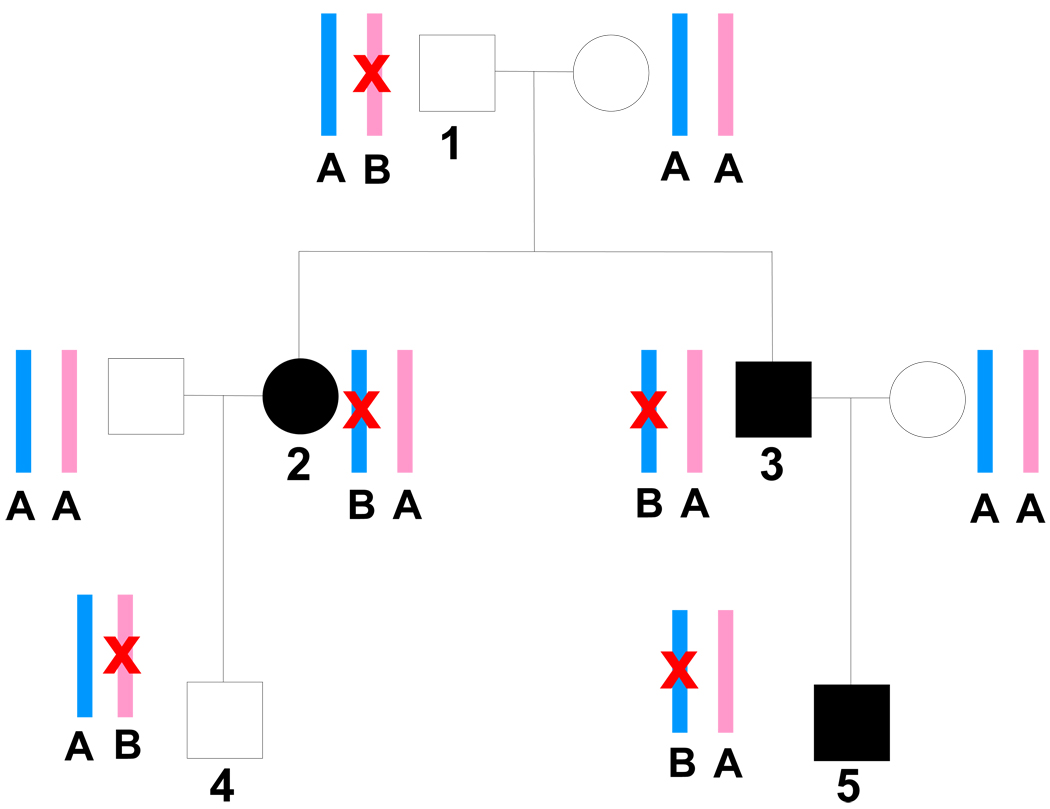
Based on our current understanding, we know that imprinting is reversible during gametogenesis, is ‘reset’ at each successive generation, and is not uniform across different tissue types and stages of development [4]. We also know that imprinted genes tend to be found in evolutionarily-conserved imprinted domains [4]. Although it is estimated that only ~1% of the human genome is imprinted, this phenomenon may be particularly important in the development of type 2 diabetes (T2DM) and obesity [7–12]. An example of how a maternally imprinted allele that is transmitted within a family might influence susceptibility to T2DM is depicted in Figure 2.
Figure 2.
Pedigree depicting a maternally-imprinted, paternally-expressed type 2 diabetes gene. Affected individuals are represented by solid squares and circles. Maternally-inherited alleles are shown in pink, paternally-inherited alleles in blue. The A allele represents a normal functioning copy of the gene and the B allele represents a mutant copy of the gene (also indicated by red “X”). Offspring are affected with type 2 diabetes when the mutant allele B is inherited from the father because the normal maternal copy is also nonfunctional due to silencing by imprinting. If mutant allele B is inherited from the mother, the offspring will be unaffected because the paternal (expressed) copy is normal. Note that for simplification, the mutation shown here is assumed to be fully penetrant and the sole possible cause of the disease, which are not typical assumptions for type 2 diabetes susceptibility genes. Note that individuals 1–5 all have a copy of the mutant allele B. Individuals 2, 3, and 5 received allele B from their father and are therefore affected. In contrast, individuals 1 and 4 received allele B from their mother and are unaffected.
There is much speculation from an evolutionary point of view as to why imprinting exists. The main hypothesis is the ‘gametic conflict’ or ‘parental conflict’ theory which states that, while the father’s sole interest is in the development of offspring that share his genes, the mother’s interests are both in providing equal resources to all offspring and in maintaining her own reproductive viability. Thus it would be predicted that evolutionary pressure would favor silencing by the father of growth-restricted genes and silencing by the mother of growth-promoting genes [13–15]. This theory is supported by the fact that imprinting has been observed in fetal and placental tissues in eutherian (placental) mammals as well as in metatherian (marsupial) mammals, in which maternal resources are continually utilized by offspring during their early development, but not in monotreme (egg-laying) mammals or birds [15].
Maternal Effects on the Intrauterine Environment
At birth, the phenotype of placental mammals has already been heavily influenced by environmental factors, specifically, the maternal intrauterine environment. Therefore, the mother’s genes, regardless of which ones are passed to the offspring, can have maternal-specific influences on offspring phenotype. This is especially true for genes influencing maternal nutrition and metabolism. Sorting out maternal influences on the fetus due to genetic vs. non-genetic causes may be challenging since maternal environment and behavior can also affect fetal growth and development. Parental genetic effects on the fetus can become even more complicated to sort out when one takes into account how differences in genotype inherited from both the mother and father influence the developing fetus’s response to the intrauterine environment.
Mitochondrial Effects
Mitochondrial maternal effects refer to the phenotypic effects of sequence variation in the mitochondrial genome on offspring. Mitochondria are cellular organelles containing their own circular genome. Oocytes contain approximately 1000 times more mitochondrial DNA than sperm, which along with the destruction of the sperm midpiece and its mitochondria shortly after fertilization apparently due to the actions of the protein ubiquitin [16–18], severely limits the number of paternal mitochondria contributing to animal progeny [19]. With only a few very notable exceptions, including one in mussels [20] and a single example of paternal inheritance of a mitochondrial disease in humans [21], only a maternal contribution to offspring mitochondrial DNA has been observed in animals.
III. Examples of POE Relevant to Diabetes
Each of the three mechanisms of POE has shown unequivocal involvement in the development of diabetes in a minority of diabetes cases: imprinting in transient neonatal diabetes, maternal in utero effects in MODY2 and mitochondrial inheritance in maternally inherited diabetes and deafness. Examples of each are provided below and summarized in Table 1.
Table 1.
Selected disorders involving parent-of-origin effects
| Condition | POE Mechanism |
Specific POE Effect | Phenotype | Selected References |
|---|---|---|---|---|
| Transient neonatal diabetes mellitus |
Paternally imprinted chromosome 6q24 |
Paternal duplication or paternal UPD of chromosome 6q24, methylation defect of maternal ZAC/HYMAI |
Diabetes mellitus occurring before one week of age and resolving by three months |
Temple and Shield (2002) [22] Mackay et al (2005) [27] |
| Beckwith- Wiedemann syndrome |
Imprinted chromosome 11p15.5 |
Complex variety of mechanisms including maternal germline CDKN1C mutations, mosaic paternal UPD 11p15.5, loss of maternal imprinting of IGF2, loss of maternal expression of H19, deletion of paternal LIT1 |
Numerous features including overgrowth, macroglossia, abdominal wall defects, embryonic tumors |
Engel et al (2000) [67] Niemitz et al (2004) [68] |
| Prader-Willi syndrome |
Imprinted chromosome 15q11-13 |
Paternal deletion 15q11- 13, maternal UPD 15q11- 13, imprinting defect 15q11-13 |
Neonatal hypotonia/failure to thrive, hyperphagia, obesity, hypogonadisim, short stature, small hands/feet, mild mental retardation |
Nicholls et al (1998) [69] |
| Angelman syndrome | Imprinted chromosome 15q11-13 |
Maternal deletion 15q11- 13, paternal UPD 15q11- 13 (rare), imprinting defect 15q11-13 |
Ataxia, tremor, seizures, sleep disorder, severe mental retardation, happy/laughing affect |
Nicholls et al (1998) [69] |
| Albright hereditary osteodystrophy |
Paternally imprinted chromosome 20q13 |
GNAS1 mutation; maternally transmitted AHO cases additionally have multiple hormone resistance due to tissue- specific paternal GNAS1 imprinting |
Short stature, obesity, developmental delay, skeletal abnormalties ± multiple hormone resistance |
Weinstein et al (2001) [70] |
| Russell-Silver syndrome (approximately 10% of cases) |
Maternally imprinted chromosome 7 |
Maternal UPD 7 | Short stature, asymmetry, triangular face |
Hannula et al (2002) [71] |
| Maternally inherited diabetes and deafness |
Maternal inheritance via mitochondrial genome |
Mitochondrial 3243 A➔ G mutation |
Early onset non- insulin requiring diabetes without obesity, neurosensory hearing loss |
Guillausseau et al (2001) [38] |
| Maturity Onset Diabetes of the Young 2 (MODY2) and birthweight |
Maternal intrauterine effects |
Maternal glucokinase (GCK) mutation associated with gestational diabetes mellitus and high offspring birth weight, fetal GCK mutation associated with low birth weight, maternal/fetal concordance for GCK mutation associated with normal birth weight |
MODY2: Modestly elevated fasting blood glucose, susceptibility to gestational diabetes and diabetes with minimal complications |
Hattersley et al (1998) [34] |
Imprinting: Transient Neonatal Diabetes Mellitus (TNDM)
Several disorders having clear associations with imprinted genes involve obesity- or growth-related phenotypes, including Prader-Willi syndrome, Angelman syndrome Beckwith-Wiedeman syndrome, Albright hereditary osteodystrophy, Russell-Silver syndrome, and transient neonatal diabetes mellitus (TNDM). We describe below as an example, the mechanisms underlying TNDM.
TNDM is a rare condition (affecting ~ 1/400,000 live births) that occurs in growth retarded infants in the first few weeks of life. The condition generally resolves within one year, although there is an elevated risk of insulin-requiring diabetes recurring later in life [22]. Involvement of imprinting in TNDM was first suspected in a 1995 case study when, in a search for the parental and chromosomal origin of a supernumerary marker chromosome in a child with TNDM, intrauterine growth retardation, and a large tongue, the child was found to have chromosome 6 microsatellite data consistent with two paternal copies (paternal uniparental disomy (UPD)) of chromosome 6 and no maternal copies other than the region contained in the marker chromosome [8]. Testing two unrelated patients with TNDM for UPD yielded one who also had UPD for chromosome 6, and these data combined with a patient reported in the literature with paternal UPD, methylmalonic acidemia (a rare recessive condition caused by mutations in a gene on chromosome 6) [23], strongly suggested an imprinting mechanism for TNDM. Subsequently an association was found between TNDM and paternal, but not maternal, duplications of chromosome 6q24 [24] including all familial cases. The critical overlapping duplicated region was found to comprise 440 kb [25] and contain two imprinted genes, ZAC and HYMAI [26]. Most recently it has been estimated based on 55 cases that 85% of individuals with TNDM have one of the following abnormalities of chromosome 6q24: paternal UPD6, paternal duplication of 6q24, or an isolated methylation defect of 6q24 [27].
Maternal Intrauterine Effects: Maturity Onset Diabetes of the Young 2 (MODY2)
The relationship of birth weight with future risk of T2DM is complex, with both low and high birth weight babies at increased risk for developing T2DM in later years [28]. The cause of increased disease risk in low birth weight babies is controversial but thought by some to be mediated by compromised organ development arising from the metabolic and nutritional milieu of the intrauterine environment [29] and others to be caused by neonatal overfeeding [28]. As a result of extensive studies of birth weight/insulin resistance relationships, particularly in populations in India, Yajnik has proposed a more complex “life course model of evolution of insulin resistance” [30] which states in part that undernourished fetuses might preferentially accumulate fat tissue (vs. lean), particularly central adiposity, as a ready source of energy and that this accumulation combined with accelerated post-natal growth leads to increased insulin resistance. In this way, both high and low birth weight babies would have increased adiposity (relative to lean body mass in the latter case) and would both be at risk for insulin resistance. In any case, one mechanism for diabetes susceptibility in high birth weight babies, particularly offspring of diabetic mothers, is thought to be related to effects of early hypersecretion of insulin on weight gain and future insulin sensitivity [28, 31]. When a woman has diabetes during pregnancy, a hyperglycemic intrauterine environment is created, leading to increased fetal insulin production, growth and high birth weight. An example described below involving a relatively rare single gene form of diabetes underscores the further complicating role the fetal genotype plays in the relationship among intrauterine environment, genetics, birth weight and diabetes.
Maturity onset diabetes of the young (MODY) is an autosomal dominant, genetically heterogeneous form of diabetes. Eight MODY genes have been described to date [32–34], with mutations in two of these genes, glucokinase (GCK; MODY2) and hepatic nuclear factor 1-alpha (HNF-1α; MODY 3) accounting for the majority of cases. Glucokinase, encoded by the MODY2 gene, is an enzyme catalyzing the phosphorylation of glucose in pancreatic beta cells, signaling the cells to release insulin; thus defects in GCK cause a delayed insulin response to glucose. MODY2 is generally considered a disorder of glucose sensing. MODY2 generally causes mild hyperglycemia and is associated with a low rate of complications. Approximately 50% of GCK mutation carriers are estimated to develop gestational diabetes (GDM). Women with GDM are at increased risk of giving birth to babies with macrosomia, or increased birth weight. However, it has been shown in a study of 58 offspring of couples with one parent carrying a GCK mutation that a fetal GCK mutation is associated with decreased birth weight [35]. In this study, babies who did not have GCK mutations but were born to women with GCK mutations were, on average, of high birth weight (86th percentile adjusted for sex, birth order and gestational age) as would be expected if the mothers had GDM. In contrast, babies with GCK mutations not inherited from their mothers were small (24th percentile). Finally, in cases in which both mother and fetus had a GCK mutation, birth weight was in the average range (53rd percentile). The mechanism appears to be as follows: Women with GCK mutations experience a hyperglycemic in-utero environment, leading the fetus to increase insulin production to utilize the excess glucose, leading to a high birth weight. On the other hand, a fetus with a GCK mutation is unable to produce adequate insulin in response to normal amounts of blood glucose, leading to low glucose utilization and a low birth weight. It is believed that this is a direct result of the poor glucose sensing in the pancreas of the fetus, which in turn causes low insulin secretion and poor development. In mutation-concordant mother-infant pairs, it appears that the maternal hyperglycemia compensates for the poor glucose sensing in the fetus. A case report in which a woman was treated for GDM with diet and insulin gave birth to a baby in the lowest 1st percentile for weight showed that the baby was found to have a GCK mutation inherited from his mother; his younger brother who did not carry the mutation was of normal birth weight [36].
Mitochondrial Inheritance: Maternally Inherited Diabetes and Deafness (MIDD)
Confirmation of mitochondrial genome involvement in maternally inherited diabetes was first reported by two groups in 1992. Ballinger et al. noted maternal transmission of diabetes and/or deafness in a three generation pedigree and identified a maternally transmitted 10 kb deletion of the mitochondrial genome [37]. At about the same time, van den Ouweland et al. studied an extensive three generation pedigree with maternal transmission of diabetes and/or deafness and through a formal segregation analysis found a 250 to 1 odds favoring transmission consistent with mitochondrial vs. autosomal dominant inheritance [38]. A point mutation in the LEU tRNA codon (3243 A➔G), previously associated with a phenotypically different disorder known as MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes, was found to be the cause of diabetes and deafness in this pedigree [38]. It is now estimated that approximately 0.5 to 2.8% of individuals with diabetes mellitus possess this point mutation [39].
IV. POE in Complex Forms of T2D and Obesity
The documented examples of POE effects shown to be associated with monogenic forms of diabetes and obesity (see descriptions above) lead to the speculation that POE effects might also contribute to the more commonly occurring complex forms of diabetes and obesity. In fact, it is possibly the added complexity of the POE effects that makes identification of the genes involved difficult. In this section we review the evidence supporting the role of POE in T2DM and obesity along with the statistical approaches for detection of POE in families.
Possible Over-representation of Maternal Transmission of Diabetes in Population Studies
It is well known that type 2 diabetes risk increases with the number of type 2 diabetic parents. This was first demonstrated using prospective data in the Pima Indians [40]. Moreover, while offspring of diabetic parents clearly had an increased incidence of T2DM compared to offspring of nondiabetic parents, there was little difference in diabetes incidence in this study between offspring of mothers and father’s with diabetes. In contrast, a subsequent number of studies from a variety of populations have reported that individuals with diabetes are more likely to report that their mother had diabetes than their father [41–49]. However, cautions about this type of study design were expressed by Mitchell et al., who speculated that reliance on self reporting of parental diabetes status might be subject to a number of reporting biases since subjects may be more likely to know health status of their mothers than of their fathers [42]. Indeed, individuals in a study of Mexican Americans were more likely to report a maternal history of diabetes [42], although in a second study from this same target population in which parents were actually tested for diabetes, no evidence for a specific maternal or paternal effect was observed [42]. Similarly, in the Framingham Heart Study, maternal and paternal diabetes conferred equivalent risk for type 2 diabetes being present in the offspring, although offspring with maternal diabetes were slightly more likely to have abnormal glucose tolerance compared with those with paternal diabetes [50].
Evidence from Linkage Studies Incorporating POE
Several studies have employed linkage analysis methods to map the locations of obesity or diabetes-related genes having parent-of-origin effects. Some of these linkage methods were designed for outcome variables that are discrete in nature (e.g., obesity or T2D), while others were designed for continuously distributed outcome variables (e.g., body mass index, glucose or insulin levels). Names of some of the commonly used linkage analysis programs and characteristics of each are summarized in Table 2. The different linkage modeling approaches each depend on distinguishing between alleles that are transmitted from the mother and those that are transmitted from the father. Under the traditional parametric linkage approach, POE can be incorporated by specification of separate male and female recombination fractions [51]. Alternatively, two heterozygote penetrance parameters (instead of one) can be specified to indicate paternal and maternal origin of the disease allele [52]. Then, a POE imprinting test can be performed by comparing the four-penetrance model (with penetrances given as P(+/+), P(m/+), P(+/m), and P(m/m), where m=variant allele), with a standard three-penetrance model (with penetrances given as P(+/+), P(m/+ or +/m), and P(m/m)). In contrast to the standard three penetrance model which assumes that the heterozygotes have equal penetrances , the four penetrance model allows paternal transmissions P(m/+) to be treated differently from maternal transmissions P(+/m) [52].
Table 2.
Linkage methods that incorporate parent-of-origin effects.
| Software | Trait type | Algorithm | Parametric / Nonparametric |
Pedigree size | N Loci | Reference |
|---|---|---|---|---|---|---|
| ACT package - MULTIC |
Quantitative | Variance- components |
Nonparametric | >40 individuals | >20 | Shete et al. (2002) [59]; Shete S. et al. (2003) [72] |
| Allegro | Discrete | Lander-Green | Nonparametric | <32 individuals | >20 | Gudbjartsson DF. et al. (2000) [73]; Karason et al. (2003) [74] |
| Genehunter- Imprinting |
Discrete | Lander-Green | Semi-parametric | 18–20 individuals | >20 | Strauch K. et al. (2000) [51] |
| Genehunter- Modscore |
Discrete | Lander-Green | Semi-parametric | 18–20 individuals | >20 | Dietter J et. al. (2007) [75] |
| Linkage-Imprint | Discrete | Elston- Stewart |
Parametric | >40 individuals | 4–5 | Shete S. and Zhou X. et al. (2005) [76] |
| SAS | Quantitative | Variance- components |
Nonparametric | 18–20 individuals | >20 | Hanson et al. (2001) [58] |
| n/a | Quantitative | Variance- components |
Nonparametric | Affected sibpairs | >20 | Wu et al. (2005) [77] |
Abbreviations : loci = genetic marker; pedigree = family; parametric = assumes an underlying genetic model; non-parametric = does not make assumptions about the genetic model or mode of inheritance
Nonparametric (model-free) approaches are based on conventional allele-sharing methods. In these approaches, a correlation is made across multiple pairs of relatives between phenotypic similarity and genetic similarity, estimated as the probability that both relatives in the pair share two alleles identical by descent. The statistical test is then based either on regressing the probability of allele-sharing with some metric of the difference in phenotype between the relative pair or alternatively, by assessing whether the total trait variation may be explained partially by allele-sharing probability at the locus in question. In the parent of origin extension to these nonparametric approaches, the parental origin of each allele is also considered.
Results of published linkage studies investigating POE effects on diabetes and obesity are summarized in Table 3. To date, the most striking linkages (based on the LOD-score statistic) for POE have been reported by Dong et al., who carried out an initial linkage analysis of body mass index and other obesity-related traits in a set of 1,297 individuals from 260 families and then performed additional linkage analyses in two smaller replication samples [53]. For obesity analyzed as a discrete trait, they detected strong evidence for linkage across all three samples to a locus transmitted from the mother. The peak LOD score was 4.52, occurring at chromosome 10p12, implying that a maternally-transmitted DNA variant in this region influenced risk of obesity. These analyses were based on testing the hypothesis that obese siblings were more likely to share the maternal (or paternal) allele than would be expected by chance. When BMI was analyzed as a quantitative trait, additional evidence for a maternal effect was detected in the region 12q24 (multipoint LOD = 4.01 and 3.69 for BMI and waist circumference, respectively), and evidence for a paternal effect was detected in the region 13q32 (multipoint LOD = 3.72 for BMI). However, these linkages were detected in the European Caucasian sample only. The quantitative trait analyses utilized a regression-based approach that assessed the correlation in parent-specific allele-sharing between two siblings and their phenotypic similarity.
Table 3.
Studies of parent-of-origin effects of type 2 diabetes, obesity, and related traits in humans.
| Maternal | Paternal | ||||||
|---|---|---|---|---|---|---|---|
| Study | Trait | Sample | Dataset | Region | LOD | Region | LOD |
|
Lindsay R.S. (2001) [55] |
T2DM | Pima Indians | 332 nuclear families + 112 extended pedigrees |
5p13.3 | 1.5 | 1q21-24 | 2.6 |
| 6q16.1 | 3 | 5p15 | 1.7 | ||||
| 14q32.12 | 1.6 | ||||||
| Lindsay R.S. (2001) [55] | BMI | Pima Indians | 453 affected sib-pairs | 5p13.2 | 1.7 | 10p15.3 | 1.7 |
| 11q23.3 | 1.6 | ||||||
| Reynisdottir I. (2003) [54] | Icelandic | 227 families | 5q34-q35.2 | 3.5 | |||
| Gorlova, O.Y. (2003) [56] | BMI | >99% Caucasian | 893 sib-pairs | 4q31.1-q32 | 1.9 | 3p23-p24 | 1.8 |
| 10p14-q11 | 1.9 | ||||||
| 12p12-pter | 1.8 | ||||||
| 4q31-qter | |||||||
| 8p11.2 | 2 | ||||||
|
Dong, C. (2005) [52] |
Obesity (BMI ≥ 27) BMI |
African American + Caucasian |
260 families |
10p12 |
4.5 |
||
| 12q24 | 4 | 13q32 | 3.7 | ||||
| Waist circumference | 12q24 | 3.7 | |||||
|
Guo YF. (2006) [53] |
BMI / Percent body fat |
Caucasian | 883 families | 3q24 | 2 | 3p14 | 2.2 |
| 19q13 | 1.8 | ||||||
| 2q31 | 3.3 | ||||||
Efforts to detect parent-specific linkages to obesity were also undertaken by Guo et al. in their linkage analysis carried out in a population of 4,000 individuals [54]. In their initial analysis with no modeling of POE effects, they detected suggestive evidence for linkage at 2q31 (LOD = 2.23) and 16q22 (LOD = 1.87) for BMI and 2q37 (LOD = 2.23) for BMI and percent fat mass. When modeling POE effects, more compelling evidence for linkage was then detected at 2q37 for maternal linkage with both BMI and percent fat mass (LOD=3.34), and suggestive evidence for parent-specific effects were detected for BMI at three additional genomic regions, including 3p14 (LOD = 2.20, paternal), 3q24 (LOD = 1.97, maternal), and 19q13 (LOD = 1.81, maternal).
Few linkage studies of T2D incorporating POE have performed. One of the few studies to have reported such effects was carried out in Icelanders by Reynisdottir et al. [55]. In an initial nonparametric multipoint linkage analysis, linkage to 5q34-q35.2 (LOD = 2.90, P=1.29 × 10−4) was observed for T2D in the full Icelandic sample. Further analyses revealed evidence for linkage to be confined exclusively to non-obese (BMI < 30 kg/m2) diabetic subjects (LOD = 3.64, P=2.12 × (10−5) in the non-obese diabetics only. Yet a further analysis conditioning on maternal transmission to non-obese diabetics resulted in a LOD score of 3.48 (P=3.12 × 10−5) in the same region, whereas conditioning on paternal transmission led to a substantial drop in the LOD score. A handful of other studies have been reported [56, 57], but with the exception of those described above, none have demonstrated compelling evidence for linkage nor have they provided much evidence for replication for parent of origin-specific effects on chromosomes 10p, 12q, 13q, or 2q. Thus, taken together, these data do not lend support to any specific replicated regions of imprinting. However, this could be due to heterogeneity of sample populations owing to different ethnicities, trait definitions, or genetic effects.
It is debatable whether conducting POE-specific linkage analyses should be restricted to regions where initial linkages (not modeled for POE) have been observed. On the one hand, it has been shown that ignoring POE in the analysis model, when in fact POE effects are present can result in a loss of power to detect linkage [58]. However, the opposite is also true; that is, incorrectly modeling POE effects when there are none can also lead to reduced power [59, 60]. Thus, as a guideline it may be advisable to incorporate POE in linkage analyses to specific regions where linkages have already been identified or for disorders where a priori hypotheses of imprinting exists (ie., for developmental disorders).
Evidence of POE from Association Studies
The availability of smaller nuclear families (i.e., parent-offspring trios) can allow assessment of whether the transmission of susceptibility alleles is disproportionate and biased towards one parent. We have outlined the general characteristics of family-based association tests of POE in Table 4. Overall, the majority of these tests are similar in that they can be used to assess risk specific to the child’s genotype, maternal genotype or maternal-child genotype interaction. In some cases, the Expectation-Maximization (EM) algorithm is used to handle ambiguity of parental origin of the variant allele inherited by children in families where all individuals are heterozygous, or where parental genotypes are unavailable. Sinsheimer (2003) further developed a maternal-fetal incompatibility test which determines whether a combination of maternal-fetal genotypes adversely affects the development of the fetus [61].
Table 4.
Characteristics of family-based association tests of parent-of-origin effects.
| Model Characteristics | CPG1 | CEPG2 | Combined LRT3 |
Log-linear models4 |
MFG5 | PO-LRT6 | QPL7 |
|---|---|---|---|---|---|---|---|
| Child genotype risk effect |
X | X | X | X | X | X | |
| Maternal genotype risk effect |
X | X | X | X | X | X | |
| Maternal-child genotype risk effect |
X | X | X | X | X | X | |
| POE effect | X | X | X | X | X | X | |
| Quantitative trait | X | ||||||
| Dichotomous trait | X | X | X | X | X | X | |
| Handles multi-allelic markers, multiple markers |
X | X | |||||
| Excludes ambiguous (1,1,1) triads |
X | X | |||||
| Handles missing parental data |
X | X | X | X | |||
| Uses unaffected siblings | X | X | |||||
| References | Cordell et al . 2004 [78] |
Cordell et al. 2004 [79] |
Rampersaud et al. 2007 [80] |
Weinberg et al. 1998 [81] |
Shinsheimer et al. 2003 [60] |
Weinberg et al 1998 [82] |
Kistner et al. 2006 [83] |
CPG-Conditional on parental genotypes;
CEPG -Conditional on exchangeable parental genotypes;
Combined likelihood ratio test;
Log-linear model;
MFG- Maternal-fetal incompatibility test;
PO-LRT - Parent of origin likelihood ratio test;
Quantitative polytomous logistic model.
To date, few candidate gene studies of T2D or obesity incorporating POE have been performed in humans. The best example is by Huxtable et al. (2000) who found excess paternal allelic transmission to 91 offspring with T2D for class III alleles of the variable number tandem repeat minisatellite 5’ of the insulin gene (INS-VNTR) [10]. In another study by Klupa et al., age at onset of T2D (MODY) was related to differential transmission of the HNF-1α mutation [62]. Earlier age at onset of T2D (~15 yrs old) was observed in families where the mutation was maternally inherited, whereas in families where the mutation was paternally inherited, the age at onset was older (~25 yrs old). Saxena et al (2006) examined whether common (>1%) mitochondrial DNA variants were associated with metabolic traits in a sample of 3,304 diabetics and 3,304 matched nondiabetic individuals, but did not find any evidence to suggest that these mtDNA variants were associated with T2D.
Some investigation has been made of the effects of common maternal/fetal diabetes susceptibility variants on birthweight via maternal intrauterine effects as an extension of the observation of effects of maternal and fetal MODY2/GCK mutations on birth weight apparently through intrauterine effects on glucose availability/utilization [35]. A common GCK polymorphism has been associated with increased birth weight in the offspring of mothers who carry it [63]; the TCF7L2 rs7903146 variant has as well [64].
Molecular Techniques to Assess Imprinting and Mitochondrial Activity
Research on the role of POE in diabetes and obesity included molecular studies of animal and human models of diabetes. Molecular technologies applicable to the investigation of imprinting focus on two main mechanisms: DNA methylation and histone and chromatin modification. Briefly, methods for investigating methylation involve approaches such as hybridization and precipitation techniques to mark and identify regions of interest and quantify the levels of methylation. Methods for investigating histone/chromatin modification involve inhibiting or enhancing deacetylation of histones bound to genomic DNA, and examining what effects these changes have on the functionality of genes. A comprehensive review of histone modification techniques has been provided elsewhere [65].
A general strategy for investigating DNA methylation on a gene-specific or global level has also been outlined by Ho and Tang [65]. Gene-specific techniques include methylation CpG island amplification-representational difference analysis (MCA-RDA), differential methylation hybridization (DMH), methylation-specificoligonucleotide microarrays (MSO), and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS)/high-performance liquid chromatography (HPLC). Global profiling methods include methylation sensitive restriction fingerprinting (MSRF), restriction landmark genomic scanning (RLGS), and chromatin immunoprecipitation (ChIP)-on chips promoter arrays. Details of the methodology underlying each technique are available elsewhere [65].
A novel method of experimentally screening for novel imprinted genes in humans on a genome-wide basis was recently presented in 2007 [66]. Using high-density oligonucleotide arrays, the authors searched for differential allelic expression (DAE) in 7,109 common coding SNPs (cSNPs) in lymphoblastoid cell lines (LCL) from 67 ethnically diverse unrelated individuals. While this approach is currently the most sophisticated in its ability to screen multiple genes, it is still limited. First, detecting tissue-specific imprinting is problematic since imprinted genes showing monoallelic expression in certain tissues might show bi-allelic expression in LCLs [67]. This problem can be overcome by obtaining mRNA from a many different types of tissues. Secondly, individuals heterozygous for coding SNPs are most informative for detecting DAE. Hence additional characterization of such SNPs will be necessary to exhaustively scan for genomic imprinting. Improvements in these technologies are being made at a rapid pace, allowing us in years to come to more thoroughly investigate imprinting as a mechanism for disease susceptibility.
V. Summary
Parent of origin effects (POE) refer to differential expression of a trait that is dependent on the sex of the parent from which transmission takes place. In mammals, POE can be caused by imprinting, which is an epigenetic mechanism, as well as intrauterine effects or location of causative or susceptibility genes on the maternally inherited mitochondrial genome. The role of POE in modifying risk of common forms of type 2 diabetes or obesity is unclear; however studies aimed at elucidating a possible connection seem warranted given several lines of evidence. Current statistical and experimental techniques are available for such studies, although each is limited. However, future improvements of these methods will allow for focused and comprehensive examination of this phenomenon.
ACKNOWLEDGEMENTS
This work was supported in part by NIH research grants R01-DK54261, R01-DK073490, R01-AR43351, and R01-HL088119. Dr. Rampersaud was funded by a postdoctoral NIH/NHLBI sponsored NRSA training grant (T32HL072751). Dr. Naj was funded by NIH/NHLBI Cardiovascular Diseases Training Grant (T32HL007024).
REFERENCES
- 1.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29:275–276. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oakey RJ, Beechey CV. Imprinted genes: identification by chromosome rearrangements and post-genomic strategies. Trends Genet. 2002;18:359–366. doi: 10.1016/s0168-9525(02)02708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Hitchins MP, Moore GE. Genomic imprinting in fetal growth and development. Expert Rev Mol Med. 2002;4:1–19. doi: 10.1017/S146239940200457X. [DOI] [PubMed] [Google Scholar]
- 5.Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- 6.Crouse HV. The Controlling Element in Sex Chromosome Behavior in Sciara. Genetics. 1960;45:1429–1443. doi: 10.1093/genetics/45.10.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C. Parental genomic imprinting of the human IGF2 gene. Nat Genet. 1993;4:98–101. doi: 10.1038/ng0593-98. [DOI] [PubMed] [Google Scholar]
- 8.Temple IK, James RS, Crolla JA, Sitch FL, Jacobs PA, Howell WM, Betts P, Baum JD, Shield JP. An imprinted gene(s) for diabetes? Nat Genet. 1995;9:110–112. doi: 10.1038/ng0295-110. [DOI] [PubMed] [Google Scholar]
- 9.Gardner RL, Squire S, Zaina S, Hills S, Graham CF. Insulin-like growth factor-2 regulation of conceptus composition: effects of the trophectoderm and inner cell mass genotypes in the mouse. Biol Reprod. 1999;60:190–195. doi: 10.1095/biolreprod60.1.190. [DOI] [PubMed] [Google Scholar]
- 10.Huxtable SJ, Saker PJ, Haddad L, Walker M, Frayling TM, Levy JC, Hitman GA, O'Rahilly S, Hattersley AT, McCarthy MI. Analysis of parent-offspring trios provides evidence for linkage and association between the insulin gene and type 2 diabetes mediated exclusively through paternally transmitted class III variable number tandem repeat alleles. Diabetes. 2000;49:126–130. doi: 10.2337/diabetes.49.1.126. [DOI] [PubMed] [Google Scholar]
- 11.Polychronakos C, Kukuvitis A. Parental genomic imprinting in endocrinopathies. Eur J Endocrinol. 2002;147:561–569. doi: 10.1530/eje.0.1470561. [DOI] [PubMed] [Google Scholar]
- 12.Wren JD, Garner HR. Data-mining analysis suggests an epigenetic pathogenesis for type 2 diabetes. J Biomed Biotechnol. 2005;2005:104–112. doi: 10.1155/JBB.2005.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haig D, Westoby M. An earlier formulation of the genetic conflict hypothesis of genomic imprinting. Nat Genet. 2006;38:271. doi: 10.1038/ng0306-271. [DOI] [PubMed] [Google Scholar]
- 14.Abu-Amero S, Monk D, Apostolidou S, Stanier P, Moore G. Imprinted genes and their role in human fetal growth. Cytogenet Genome Res. 2006;113:262–270. doi: 10.1159/000090841. [DOI] [PubMed] [Google Scholar]
- 15.Murphy SK, Jirtle RL. Imprinting evolution and the price of silence. Bioessays. 2003;25:577–588. doi: 10.1002/bies.10277. [DOI] [PubMed] [Google Scholar]
- 16.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63:582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- 17.Hiraoka J, Hirao Y. Fate of sperm tail components after incorporation into the hamster egg. Gamete Res. 1988;19:369–380. doi: 10.1002/mrd.1120190408. [DOI] [PubMed] [Google Scholar]
- 18.Szollosi D. The fate of sperm middle-piece mitochondria in the rat egg. J Exp Zool. 1965;159:367–377. doi: 10.1002/jez.1401590309. [DOI] [PubMed] [Google Scholar]
- 19.Birky CW., Jr The inheritance of genes in mitochondria and chloroplastslaws: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 20.Zouros E. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet Syst. 2000;75:313–318. doi: 10.1266/ggs.75.313. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 22.Temple IK, Shield JP. Transient neonatal diabetes, a disorder of imprinting. J Med Genet. 2002;39:872–875. doi: 10.1136/jmg.39.12.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramowicz MJ, Andrien M, Dupont E, Dorchy H, Parma J, Duprez L, Ledley FD, Courtens W, Vamos E. Isodisomy of chromosome 6 in a newborn with methylmalonic acidemia and agenesis of pancreatic beta cells causing diabetes mellitus. J Clin Invest. 1994;94:418–421. doi: 10.1172/JCI117339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cave H, Polak M, Drunat S, Denamur E, Czernichow P. Refinement of the 6q chromosomal region implicated in transient neonatal diabetes. Diabetes. 2000;49:108–113. doi: 10.2337/diabetes.49.1.108. [DOI] [PubMed] [Google Scholar]
- 25.Gardner RJ, Mackay DJ, Mungall AJ, Polychronakos C, Siebert R, Shield JP, Temple IK, Robinson DO. An imprinted locus associated with transient neonatal diabetes mellitus. Hum Mol Genet. 2000;9:589–596. doi: 10.1093/hmg/9.4.589. [DOI] [PubMed] [Google Scholar]
- 26.Arima T, Drewell RA, Oshimura M, Wake N, Surani MA. A novel imprinted gene, HYMAI, is located within an imprinted domain on human chromosome 6 containing ZAC. Genomics. 2000;67:248–255. doi: 10.1006/geno.2000.6266. [DOI] [PubMed] [Google Scholar]
- 27.Mackay DJ, Temple IK, Shield JP, Robinson DO. Bisulphite sequencing of the transient neonatal diabetes mellitus DMR facilitates a novel diagnostic test but reveals no methylation anomalies in patients of unknown aetiology. Hum Genet. 2005;116:255–261. doi: 10.1007/s00439-004-1236-1. [DOI] [PubMed] [Google Scholar]
- 28.Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am J Epidemiol. 2007;165:849–857. doi: 10.1093/aje/kwk071. [DOI] [PubMed] [Google Scholar]
- 29.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–67. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- 30.Yajnik CS. Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr. 2004;134:205–210. doi: 10.1093/jn/134.1.205. [DOI] [PubMed] [Google Scholar]
- 31.Plagemann A. 'Fetal programming' and 'functional teratogenesis': on epigenetic mechanisms and prevention of perinatally acquired lasting health risks. J Perinat Med. 2004;32:297–305. doi: 10.1515/JPM.2004.055. [DOI] [PubMed] [Google Scholar]
- 32.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 33.Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, Dina C, Hamid YH, Joly E, Vaillant E, Benmezroua Y, Durand E, Bakaher N, Delannoy V, Vaxillaire M, Cook T, Dallinga-Thie GM, Jansen H, Charles MA, Clement K, Galan P, Hercberg S, Helbecque N, Charpentier G, Prentki M, Hansen T, Pedersen O, Urrutia R, Melloul D, Froguel P. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raeder H, Bjorkhaug L, Johansson S, Mangseth K, Sagen JV, Hunting A, Folling I, Johansen O, Bjorgaas M, Paus PN, Sovik O, Molven A, Njolstad PR. A hepatocyte nuclear factor-4 alpha gene (HNF4A) P2 promoter haplotype linked with late-onset diabetes: studies of HNF4A variants in the Norwegian MODY registry. Diabetes. 2006;55:1899–1903. doi: 10.2337/db05-1677. [DOI] [PubMed] [Google Scholar]
- 35.Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet. 1998;19:268–270. doi: 10.1038/953. [DOI] [PubMed] [Google Scholar]
- 36.Spyer G, Hattersley AT, Sykes JE, Sturley RH, MacLeod KM. Influence of maternal and fetal glucokinase mutations in gestational diabetes. Am J Obstet Gynecol. 2001;185:240–241. doi: 10.1067/mob.2001.113127. [DOI] [PubMed] [Google Scholar]
- 37.Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, Koontz DA, Wallace DC. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 38.van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 39.Guillausseau PJ, Massin P, Dubois-LaForgue D, Timsit J, Virally M, Gin H, Bertin E, Blickle JF, Bouhanick B, Cahen J, Caillat-Zucman S, Charpentier G, Chedin P, Derrien C, Ducluzeau PH, Grimaldi A, Guerci B, Kaloustian E, Murat A, Olivier F, Paques M, Paquis-Flucklinger V, Porokhov B, Samuel-Lajeunesse J, Vialettes B. Maternally inherited diabetes and deafness: a multicenter study. Ann Intern Med. 2001;134:721–728. doi: 10.7326/0003-4819-134-9_part_1-200105010-00008. [DOI] [PubMed] [Google Scholar]
- 40.Knowler WC, Pettitt DJ, Savage PJ, Bennett PH. Diabetes incidence in Pima indians: contributions of obesity and parental diabetes. Am J Epidemiol. 1981;113:144–156. doi: 10.1093/oxfordjournals.aje.a113079. [DOI] [PubMed] [Google Scholar]
- 41.Alcolado JC, Alcolado R. Importance of maternal history of non-insulin dependent diabetic patients. Bmj. 1991;302:1178–1180. doi: 10.1136/bmj.302.6786.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell BD, Valdez R, Hazuda HP, Haffner SM, Monterrosa A, Stern MP. Differences in the prevalence of diabetes and impaired glucose tolerance according to maternal or paternal history of diabetes. Diabetes Care. 1993;16:1262–1267. doi: 10.2337/diacare.16.9.1262. [DOI] [PubMed] [Google Scholar]
- 43.Karter AJ, Rowell SE, Ackerson LM, Mitchell BD, Ferrara A, Selby JV, Newman B. Excess maternal transmission of type 2 diabetes. The Northern California Kaiser Permanente Diabetes Registry. Diabetes Care. 1999;22:938–943. doi: 10.2337/diacare.22.6.938. [DOI] [PubMed] [Google Scholar]
- 44.DeSilva S, Hana M, Sutija VG, Raziuddin K. Effect of amino acids on glucose tolerance and hyperkalemia in very low birth weight infants. J Perinat Med. 2002;30:128–131. doi: 10.1515/JPM.2002.015. [DOI] [PubMed] [Google Scholar]
- 45.Bjornholt JV, Erikssen G, Liestol K, Jervell J, Thaulow E, Erikssen J. Type 2 diabetes and maternal family history: an impact beyond slow glucose removal rate and fasting hyperglycemia in low-risk individuals? Results from 22.5 years of follow-up of healthy nondiabetic men. Diabetes Care. 2000;23:1255–1259. doi: 10.2337/diacare.23.9.1255. [DOI] [PubMed] [Google Scholar]
- 46.Fischbacher CM, Bhopal R, Unwin N, Walker M, White M, Alberti KG. Maternal transmission of type 2 diabetes varies by ethnic group: cross-sectional survey of Europeans and South Asians. Diabetes Care. 2001;24:1685–1686. doi: 10.2337/diacare.24.9.1685-a. [DOI] [PubMed] [Google Scholar]
- 47.Momiyama Y, Suzuki Y, Ohsuzu F, Atsumi Y, Matsuoka K, Kimura M. Maternally transmitted susceptibility to non-insulin-dependent diabetes mellitus and left ventricular hypertrophy. J Am Coll Cardiol. 1999;33:1372–1378. doi: 10.1016/s0735-1097(98)00689-5. [DOI] [PubMed] [Google Scholar]
- 48.Young CA, Kumar S, Young MJ, Boulton AJ. Excess maternal history of diabetes in Caucasian and Afro-origin non-insulin-dependent diabetic patients suggests dominant maternal factors in disease transmission. Diabetes Res Clin Pract. 1995;28:47–49. doi: 10.1016/0168-8227(94)01058-8. [DOI] [PubMed] [Google Scholar]
- 49.Klein BE, Klein R, Moss SE, Cruickshanks KJ. Parental history of diabetes in a population-based study. Diabetes Care. 1996;19:827–830. doi: 10.2337/diacare.19.8.827. [DOI] [PubMed] [Google Scholar]
- 50.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49:2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 51.Smalley SL. Sex-specific recombination frequencies: a consequence of imprinting? Am J Hum Genet. 1993;52:210–212. [PMC free article] [PubMed] [Google Scholar]
- 52.Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66:1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong C, Li WD, Geller F, Lei L, Li D, Gorlova OY, Hebebrand J, Amos CI, Nicholls RD, Price RA. Possible genomic imprinting of three human obesity-related genetic loci. Am J Hum Genet. 2005;76:427–437. doi: 10.1086/428438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo YF, Shen H, Liu YJ, Wang W, Xiong DH, Xiao P, Liu YZ, Zhao LJ, Recker RR, Deng HW. Assessment of genetic linkage and parent-of-origin effects on obesity. J Clin Endocrinol Metab. 2006;91:4001–4005. doi: 10.1210/jc.2006-0549. [DOI] [PubMed] [Google Scholar]
- 55.Reynisdottir I, Thorleifsson G, Benediktsson R, Sigurdsson G, Emilsson V, Einarsdottir AS, Hjorleifsdottir EE, Orlygsdottir GT, Bjornsdottir GT, Saemundsdottir J, Halldorsson S, Hrafnkelsdottir S, Sigurjonsdottir SB, Steinsdottir S, Martin M, Kochan JP, Rhees BK, Grant SF, Frigge ML, Kong A, Gudnason V, Stefansson K, Gulcher JR. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am J Hum Genet. 2003;73:323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindsay RS, Kobes S, Knowler WC, Bennett PH, Hanson RL. Genome-wide linkage analysis assessing parent-of-origin effects in the inheritance of type 2 diabetes and BMI in Pima Indians. Diabetes. 2001;50:2850–2857. doi: 10.2337/diabetes.50.12.2850. [DOI] [PubMed] [Google Scholar]
- 57.Gorlova OY, Amos CI, Wang NW, Shete S, Turner ST, Boerwinkle E. Genetic linkage and imprinting effects on body mass index in children and young adults. Eur J Hum Genet. 2003;11:425–432. doi: 10.1038/sj.ejhg.5200979. [DOI] [PubMed] [Google Scholar]
- 58.Strauch K, Fimmers R, Windemuth C, Hahn A, Wienker TF, Baur MP. Linkage analysis with adequate modeling of a parent-of-origin effect. Genet Epidemiol. 1999;17 Suppl 1:S331–S336. doi: 10.1002/gepi.1370170756. [DOI] [PubMed] [Google Scholar]
- 59.Hanson RL, Kobes S, Lindsay RS, Knowler WC. Assessment of parent-of-origin effects in linkage analysis of quantitative traits. Am J Hum Genet. 2001;68:951–962. doi: 10.1086/319508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shete S, Amos CI. Testing for genetic linkage in families by a variance-components approach in the presence of genomic imprinting. Am J Hum Genet. 2002;70:751–757. doi: 10.1086/338931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinsheimer JS, Palmer CG, Woodward JA. Detecting genotype combinations that increase risk for disease: maternal-fetal genotype incompatibility test. Genet Epidemiol. 2003;24:1–13. doi: 10.1002/gepi.10211. [DOI] [PubMed] [Google Scholar]
- 62.Klupa T, Warram JH, Antonellis A, Pezzolesi M, Nam M, Malecki MT, Doria A, Rich SS, Krolewski AS. Determinants of the development of diabetes (maturity-onset diabetes of the young-3) in carriers of HNF-1alpha mutations: evidence for parent-of-origin effect. Diabetes Care. 2002;25:2292–2301. doi: 10.2337/diacare.25.12.2292. [DOI] [PubMed] [Google Scholar]
- 63.Weedon MN, Clark VJ, Qian Y, Ben-Shlomo Y, Timpson N, Ebrahim S, Lawlor DA, Pembrey ME, Ring S, Wilkin TJ, Voss LD, Jeffery AN, Metcalf B, Ferrucci L, Corsi AM, Murray A, Melzer D, Knight B, Shields B, Smith GD, Hattersley AT, Di Rienzo A, Frayling TM. A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am J Hum Genet. 2006;79:991–1001. doi: 10.1086/509517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freathy RM, Weedon MN, Bennett A, Hypponen E, Relton CL, Knight B, Shields B, Parnell KS, Groves CJ, Ring SM, Pembrey ME, Ben-Shlomo Y, Strachan DP, Power C, Jarvelin MR, McCarthy MI, Davey Smith G, Hattersley AT, Frayling TM. Type 2 diabetes TCF7L2 risk genotypes alter birth weight: a study of 24,053 individuals. Am J Hum Genet. 2007;80:1150–1161. doi: 10.1086/518517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho SM, Tang WY. Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: An emphasis on fetal-based adult diseases. Reprod Toxicol. 2007;23:267–282. doi: 10.1016/j.reprotox.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pollard KS, Serre D, Wang X, Tao H, Grundberg E, Hudson TJ, Clark AG, Frazer K. A genome-wide approach to identifying novel-imprinted genes. Hum Genet. 2008;122:625–634. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- 67.Zhou H, Jungbluth H, Sewry CA, Feng L, Bertini E, Bushby K, Straub V, Roper H, Rose MR, Brockington M, Kinali M, Manzur A, Robb S, Appleton R, Messina S, D'Amico A, Quinlivan R, Swash M, Muller CR, Brown S, Treves S, Muntoni F. Molecular mechanisms and phenotypic variation in RYR1-related congenital myopathies. Brain. 2007;130:2024–2036. doi: 10.1093/brain/awm096. [DOI] [PubMed] [Google Scholar]
- 68.Engel JR, Smallwood A, Harper A, Higgins MJ, Oshimura M, Reik W, Schofield PN, Maher ER. Epigenotype-phenotype correlations in Beckwith-Wiedemann syndrome. J Med Genet. 2000;37:921–926. doi: 10.1136/jmg.37.12.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niemitz EL, DeBaun MR, Fallon J, Murakami K, Kugoh H, Oshimura M, Feinberg AP. Microdeletion of LIT1 in familial Beckwith-Wiedemann syndrome. Am J Hum Genet. 2004;75:844–849. doi: 10.1086/425343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 71.Weinstein LS. The role of tissue-specific imprinting as a source of phenotypic heterogeneity in human disease. Biol Psychiatry. 2001;50:927–931. doi: 10.1016/s0006-3223(01)01295-1. [DOI] [PubMed] [Google Scholar]
- 72.Hannula K, Lipsanen-Nyman M, Kristo P, Kaitila I, Simola KO, Lenko HL, Tapanainen P, Holmberg C, Kere J. Genetic screening for maternal uniparental disomy of chromosome 7 in prenatal and postnatal growth retardation of unknown cause. Pediatrics. 2002;109:441–448. doi: 10.1542/peds.109.3.441. [DOI] [PubMed] [Google Scholar]
- 73.Shete S, Zhou X, Amos CI. Genomic imprinting and linkage test for quantitative-trait Loci in extended pedigrees. Am J Hum Genet. 2003;73:933–938. doi: 10.1086/378592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 75.Karason A, Gudjonsson JE, Upmanyu R, Antonsdottir AA, Hauksson VB, Runasdottir EH, Jonsson HH, Gudbjartsson DF, Frigge ML, Kong A, Stefansson K, Valdimarsson H, Gulcher JR. A susceptibility gene for psoriatic arthritis maps to chromosome 16q: evidence for imprinting. Am J Hum Genet. 2003;72:125–131. doi: 10.1086/345646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dietter J, Mattheisen M, Furst R, Ruschendorf F, Wienker TF, Strauch K. Linkage analysis using sex-specific recombination fractions with GENEHUNTER-MODSCORE. Bioinformatics. 2007;23:64–70. doi: 10.1093/bioinformatics/btl539. [DOI] [PubMed] [Google Scholar]
- 77.Shete S, Zhou X. Parametric approach to genomic imprinting analysis with applications to Angelman's syndrome. Hum Hered. 2005;59:26–33. doi: 10.1159/000084734. [DOI] [PubMed] [Google Scholar]
- 78.Wu CC, Shete S, Amos CI. Linkage analysis of affected sib pairs allowing for parent-of-origin effects. Ann Hum Genet. 2005;69:113–126. doi: 10.1046/j.1529-8817.2004.00139.x. [DOI] [PubMed] [Google Scholar]
- 79.Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- 80.Cordell HJ. Properties of case/pseudocontrol analysis for genetic association studies: Effects of recombination, ascertainment, and multiple affected offspring. Genet Epidemiol. 2004;26:186–205. doi: 10.1002/gepi.10306. [DOI] [PubMed] [Google Scholar]
- 81.Rampersaud E, Morris RW, Weinberg CR, Speer MC, Martin ER. Power calculations for likelihood ratio tests for offspring genotype risks, maternal effects, and parent-of-origin (POO) effects in the presence of missing parental genotypes when unaffected siblings are available. Genet Epidemiol. 2007;31:18–30. doi: 10.1002/gepi.20189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kistner EO, Infante-Rivard C, Weinberg CR. A method for using incomplete triads to test maternally mediated genetic effects and parent-of-origin effects in relation to a quantitative trait. Am J Epidemiol. 2006;163:255–261. doi: 10.1093/aje/kwj030. [DOI] [PubMed] [Google Scholar]