Abstract
A-74528 is a recently discovered natural product of Streptomyces sp. SANK 61196 that inhibits 2′,5′-oligoadenylate phosphodiesterase (2′-PDE), a key regulatory enzyme of the interferon pathway. Inhibition of 2′-PDE by A-74528 reduces viral replication, and therefore shows promise as a new type of antiviral drug. The complete A-74528 gene cluster, comprising of 29 open reading frames, was cloned and sequenced, and shown to possess a type II polyketide synthase (PKS) at its core. Its identity was confirmed by analysis of a mutant generated by targeted disruption of a PKS gene, and by functional expression in a heterologous Streptomyces host. Remarkably, it showed exceptional end-to-end sequence identity to the gene cluster responsible for biosynthesis of fredericamycin A, a structurally unrelated antitumor antibiotic with a distinct mode of action. Whereas the fredericamycin producing strain, Streptomyces griseus, produced undetectable quantities of A-74528, the A-74528 gene cluster was capable of producing both antibiotics. The biosynthetic roles of three genes, including one that represents the only qualitative difference between the two gene clusters, were investigated by targeted gene disruption. The implications for the evolution of antibiotics with different biological activities from the same gene cluster are discussed.
INTRODUCTION
The design of effective antiviral drugs represents a difficult but important frontier for contemporary science. The ability of viruses to incorporate their own genome into that of the host and thereby orchestrate control over host cell machinery presents a challenge to design therapeutic agents that specifically disrupt virus replication with minimal deleterious effect to the host cell. In order to treat patients with multi-drug resistant viral strains, drugs with new mechanisms of activity and the requisite specificity must be developed.
A-74528 is a metabolite of Streptomyces sp. SANK 61196, discovered by Ogita and coworkers during the screening of microbial extracts for antiviral compounds that activate the interferon (INF) system via 2′,5′-oligoadenylate phosphodiesterase (2′-PDE) inhibition.1,2 Viral infection of cells induces the production of INFs, which in turn up-regulates 2′,5′-oligoadenylates (2–5A).3 In the antiviral pathway, 2–5A promotes RNase L activity, leading to the degradation of RNA, and ultimately apoptosis. In an alternative pathway, 2–5A is degraded into AMP and ATP by 2′-PDE,2,4 suppressing INF-induced cell death. The inhibition of 2′-PDE prolongs the half-life of 2–5A, which in turn activates the antiviral pathway.5 Thus, the ability of A-74528 to modulate 2′-PDE represents a unique therapeutic approach to regulate the physiological response to viral infection.3 In addition, 2′-PDE inhibitors may show promise in targeting other diseases in which a significant decrease in INF biosynthesis is believed to contribute to pathogenesis, including AIDS and cancer.6,7
The structure of A-74528 (Figure 1) was elucidated by a rigorous NMR investigation,1 as any attempt to crystallize the compound for X-ray crystallographic studies failed due to the instability of the compound. Whereas A-74528 represents an unprecedented chemotype, its oxidized polyphenolic structure suggests that the compound is synthesized via a type II aromatic polyketide synthase (PKS). Such bacterial PKSs are responsible for the biosynthesis of a large family of structurally diverse natural products, many of which have emerged as clinically useful drugs.8–12 Recent progress in utilizing molecular tools and recombinant methods to understand PKS-mediated biosynthesis at the genetic and biochemical level has set the stage for engineering novel aromatic polyketide derivatives. The elucidation of the PKS enzymes responsible for the synthesis of A-74528 will present opportunities for the biosynthetic engineering of novel compounds that may be relevant either in the clinic as new therapeutic agents, or in the laboratory as tools to investigate the mechanism of action of 2′-PDE inhibitors.
Figure 1.
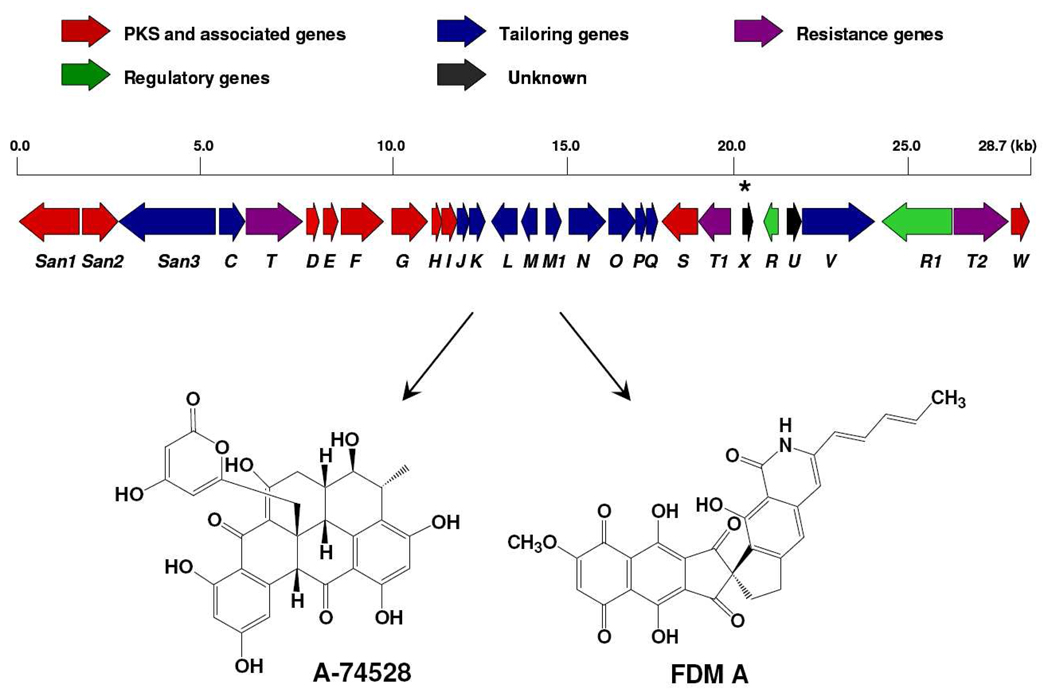
Map of the 28.7 Kb region of Streptomyces sp. SANK 61196 hosting the san biosynthetic gene cluster and chemical structures of the metabolites A-74528 and FDM A. The ORF sanX represents the only qualitative difference between the fdm and san gene clusters and is denoted with an asterisk.
Our interest in characterizing the gene cluster responsible for the production or A-74528 extends beyond its bioactivity to its structural novelty. For example, the C30 polyphenol is likely formed from a pentadecaketide, the longest chain length identified thus far in the processing of a polyketide through a type II PKS complex. Fredericamycin (FDM), an anticancer agent featuring two sets of peri-hydroxy tricyclic aromatic groups connected through a chiral spiro carbon center, is the only other C30 type II poyketide known. Thus, characterization of the A-74528 chain length factor (CLF) will likely assist in the rational design of novel polyketides with longer carbon skeletons. Moreover, the fused core of six contiguous rings with a highly oxygenated periphery suggests that the biosynthetic pathway may involve unique oxidation and cyclization patterns. Last but not least, the six stereocenters in A-74528 make this compound highly unusual among natural products of type II PKSs. Therefore, understanding the factors that control ring formation in A-74528 biosynthesis will provide new and exciting targets for the diversification of engineered natural products.
MATERIALS AND METHODS
General
E. coli XL1-Blue competent cells were used for routine subcloning and plasmid preparations. SuperCos I (Stratagene) and pHU20413 were used for cosmid library preparations. Streptomyces lividans K4-11414 was the host for heterologous expression. E. coli cells were grown in LB medium with appropriate antibiotics, where necessary. Streptomyces sp. SANK 61196 was obtained from Daiichi Sankyo. Streptomyces griseus ATCC 49344 was from ATCC (American Type Culture Collection, Rockville, MD). Common biochemicals were from standard commercial sources unless otherwise noted. Streptomyces sp. SANK 61196 was cultured on semi-solid R5 medium to prepare spore stocks, and was inoculated in YEME medium for genomic DNA isolation. R5, YEME, 2X YT, and MS medium were prepared according to standard protocols.15
Probe preparation
Degenerate primers KS-FP and KS-RP16 were used to amplify putative type-II KS-CLF-containing genes from Streptomyces sp. SANK 61196. The produced PCR products were cloned into pCRBlunt (Invitrogen) and sequenced to verify the presence of KS-CLF homologs. Sequencing of the captured PCR products showed three KS-CLF sequences in SANK 61196 genome. One sequence showed 99% sequence identity to the act KS-CLF sequence (octaketide), the second had 84% and 91% identity to the fdm KS-CLF (pentadecaketide) and the third was closely related to the whiE spore pigment PKS. Primers 5′-CAGGCGATGGACATATAC-3′and 5′-TCCATGGTCGGGCACTCG-3′were designed based on the sequence obtained from the fdm homolog to prepare a PCR product as a probe for cosmid isolation. The PCR fragment of 760-bp was gel purified and labeled with digoxigenin (DIG) DNA labeled Kit (Roche Diagnostics Corp., Indianapolis, IN) for genomic library screening.
Cosmid library construction
Cosmid libraries of Streptomyces sp. SANK 61196 were prepared using two different cosmids – SuperCos I (Stratagene) and the shuttle vector pHU204.17 The SuperCos I library was constructed according to the manufacturer’s protocol using genomic DNA partially digested with MboI of Streptomyces sp. SANK 61196 and dephosphorylated with calf intestine alkaline phosphatase. The pHU204 library was prepared using partially digested genomic DNA with MboI without dephosphorylation. Both libraries were prepared using fragments in the 30–40 kb range. Ligation reactions were packaged into lambda phage for E. coli infection. The packaging reaction was tested to determine the size of each cosmid library and then amplified for cosmid isolation.
Isolation and sequencing of pKZ1 and pKZ2
Nine positive colonies were isolated from the SuperCos library by colony blot hybridization using the KS probe. Their identity was tested by PCR amplification. Six of these clones produced the expected 760-bp PCR product. They were subsequently digested with BamHI; their digestion patterns revealed three distinct cosmid clones, designated pKZ1, pKZ2 and pKZ3. Cosmids pKZ1 and pKZ2 were fully sequenced by a combination of primer walking, shotgun sequencing and subcloning approaches. Contig assembly was performed using DNASTAR-SeqMan II and Redasoft Visual Cloning 3.2 software’s, and individual ORFs were identified and assigned with the assistance of BLAST analyses. The entire nucleotide sequence of the A-74528 gene cluster has been deposited in GenBank database under accession number GU937384.
Isolation of pKZ11
Anticipating that functional expression of the A-74528 gene cluster would lead to a pigmented phenotype, we screened the pHU204 cosmid library for pigment production. Protoplasts of Streptomyces lividans K4-114 were prepared according to standard procedures,15 and transformed with the DNA cosmid library. Transformants were selected on R2YE thiostrepton (50 µg/ml) plates. One colony, producing a black pigment similar to wild-type Streptomyces sp. SANK 61196, was purified by repeated restreaking; its plasmid was designated pKZ11.
Construction of a KS mutant of Streptomyces sp. SANK 61196
To genetically inactivate the putative A-74528 gene cluster, an internal fragment from the KS gene was amplified using two primers, KS-Fwd (5′-GGTCTGGAAGATGATCGTCAAGG-3′) and KS-Rev (5′-AGTACAGATGCGGCTTCATGTAG-3′), and cloned into the conjugation vector pJQ200SK in the ApaI/SacI restriction sites, yielding pJQ200SK+KS. This plasmid was introduced into E. coli ET12567 (pUZ8002), and conjugally transferred to Streptomyces sp. SANK 61196 as follows. A single colony of the E. coli transformant was grown overnight in liquid LB media supplemented with 25 µg/mL gentamicin at 37 °C. Meanwhile, fresh spores from Streptomyces sp. SANK 61196 were harvested from R5 agar plates (5 × 30-mL plates), resuspended in 2 mL of TES buffer, and germinated by heating at 50 °C for 10 min. To the heat shocked suspension, 2 mL of 2X YT were added. The donor and recipient cultures were mixed in a falcon tube, spread on MS agar plates without antibiotic, and grown overnight at 30 °C. The plates were overlaid with gentamicin and nalidixic acid (100 µg and 200 µg, respectively), and incubated at 30 °C for 4–7 days. Genomic DNA was prepared from drug resistant Streptomyces colonies, and verified by PCR analysis. One of these colonies was designated Streptomyces sp. SANK 61196ΔKS.
Preparation of in-frame deletion mutants in selected ORFs on pKZ11
To interrogate the biosynthetic roles of genes of interest, three in-frame deletion mutants of pKZ11 were constructed using the PCR targeting system and FLP-recombination (Table S1).18 Specifically, san2, sanM and sanX were disrupted, yielding plasmids pKZ12, pKZ13, pKZ14, respectively (Figure S1). To inactivate each gene, long primers were used to amplify cassettes harboring the deletion of interest, apramycin resistance and OriT loci. To prepare pKZ12, oligos San2-20Fwd/San2-19Rev (Table S1) were designed to amplify the Aprar-cassette flanked with homologous regions (39 nucleotides) of san2. An apramycin resistant disrupted mutant was generated by the addition of L-arabinose, which induced homologous recombination of the amplified fragment with the corresponding regions in pKZ11. Colonies resistant to apramycin were selected for PCR screening using primers San2-OutFwd/San2-OutRev (Table S1), designed outside of the homologous region. The disrupted mutants were then introduced in DH5α containing the FLP-recombinase to resolve the Apra/OriT cassette and produce the in-frame deletion mutants. Colonies sensitive to apramycin and resistant to carbenicillin were selected for another PCR screening with primers San2-OutFwd/San2-OutRev. Mutants pKZ13 and pKZ14 were constructed in a similar manner, using primers SanM-20Fwd/SanM-19Rev and SanX-20Fwd/SanX1-19Rev respectively to amplify the Apra/OriT cassette and primers SanM-OutF/SanM-OutR and SanX-OutF/SanX-OutR for PCR screening (Table S1). The mutant plasmids pKZ12, pKZ13, and pKZ14 were each introduced into Streptomyces lividans K4-114 by transformation.
Analysis of A-74528 production by Streptomyces spp
Strains of interest were grown in 1 L R5 media at 30 °C. After 4 days, the supernatant was collected from each culture by centrifugation. Ammonium sulfate was added to a final concentration of 0.5 g/mL, and the solution was stirred at room temperature for 1 h. After further incubation of this solution at 4 °C for 12 h, the precipitate was collected by centrifugation and extracted with MeOH (3 × 4 mL). The MeOH solution was then analyzed by LC-MS for the presence of A-74528. LC-MS analysis was performed on a Micromass Q-ToF hybrid quadrupole-time of flight instrument (Zorbax C18 column 150 × 0.5 mm, 7 µL/min flow rate, 5 µL injection). An isocratic mobile phase composed of 60% solvent A (0.1% formic in water) and 40% solvent B (0.1% formic acid in acetonitrile) was used. The LC-MS data of each extract was compared to an authentic A-74528 standard, which eluted at 9.7 minutes (MH+ = 561.12). Compound identity was confirmed in each case on a Micromass Quattro Premier triple quadrupole LC-MS instrument (Zorbax CB-C18 column, 2.1 × 30 mm 3.5 µm column, 250 µL/min flow rate, 20-fold dilution in 50% methanol, 10 µL injection). An authentic standard of A-74528 eluted at 5.7 min (isocratic elution, 70% solvent A, 30% solvent B), and displayed three characteristic fragments of m/z = 435, 417 and 375.
Analysis of fredericamycin production by Streptomyces spp
Strains of interest were each grown in 1 L R5 media at 30 °C. After 4 days of incubation, the mycelia were harvested from each culture by centrifugation. The mycelia were extracted with acetone with 1% 2 N HCl (3 × 10 mL). Crude extracts were analyzed by HPLC as described previously.16 In brief, the extract was injected on a C18 column equilibrated with 50% solvent A (1% acetic acid in water) and 50% solvent B (1% acetic acid in acetonitrile) and developed as follows: 0–5 min, 50% A/50% B; 5–10 min, a linear gradient from 50% A/50% B to 5% A/95%B; 15–25 min, 5% A/95% B at a flow rate of 0.8 mL/min and UV detection at 375 nm. HPLC and ESI-MS spectra of each crude extract were compared to an authentic FDM A standard isolated from S. griseus16 to determine if fredericamycin was produced by the strain.
RESULTS
Cloning and sequencing of the A-74528 gene cluster
Previously published primers KS-FP and KS-RP16 were used for PCR amplification on genomic DNA from Streptomyces sp. SANK 61196. A PCR product of 760 bp was purified, cloned and sequenced showing an 84% and 91% identity to the fdm KS-CLF (which synthesizes a pentadecaketide product). This DNA fragment was used as a probe to screen two cosmid libraries, constructed in SuperCos I and pHU204, of genomic DNA from Streptomyces sp. SANK 61196. Three different cosmid clones (pKZ1, pKZ2, and pKZ3) were identified as positive hits from the SuperCos I library by colony blot hybridization, and were confirmed by PCR and Southern blot analyses. One colony from the pHU204 library in S. lividans K4-114 displayed a dark pigmentation phenotype similar to the wild-type Streptomyces sp. SANK 61196 strain. The cosmid (pKZ11) from this colony was isolated, introduced via transformation into E. coli, and analyzed for the presence of the KS probe by PCR (data not shown).
Since both FDM and A-74528 are likely to be derived from C30 polyketide backbones, it was anticipated that their PKS gene sequences would be homologous. Indeed, sequence analysis of the gene cluster encoded within cosmids pKZ1, pKZ2 and pKZ3 revealed minimal PKS genes sanF (KS), sanG (CLF) and sanH (ACP), which share high sequence similarity to their fdm homologs.16 What was entirely unexpected was that the putative san PKS gene cluster had extraordinarily high end-to-end sequence similarity to the fdm gene cluster (Table 1), including all but one biosynthetic gene as well as resistance and regulatory genes. Even the organization of these genes within their respective clusters is nearly identical (Figure 1). The ORF of unknown function, sanX located between sanT1 and sanR, represents the only qualitative difference between the two gene clusters.
Table 1.
Description of open reading frames (ORFs) and proposed functions of gene products identified in the A-74528 biosynthetic gene cluster
| Gene | Sizeaa | Protein homolog (Gene bank accession numbers, similarity identity (%) |
Proposed function |
|---|---|---|---|
| san1 | 564 | orf1 (AAQ08990) 93/89 | Acyl-CoA decarboxylase |
| san2 | 342 | orf2 (AAQ08910) 74/68 | Acyl-ACP thioesterase24 |
| san3 | 932 | orf3 (AAQ08911 )74/66 | Quinone-forming monoxygenase26 |
| sanC | 246 | fdmC (AAQ08912 )89/83 | 3-Ketoacyl ACP-reductase |
| sanT | 511 | fdmT (AAQ08913) 88/81 | Peptide transporter |
| sanD | 106 | fdmD (AAQ08914) 94/88 | Polyketide cyclase |
| sanE | 141 | fdmE (AAQ08915) 87/82 | Polyketide cyclase |
| sanF | 421 | fdmF (AAQ08916) 96/92 | Ketosynthase (KS) |
| sanG | 423 | fdmG (AAQ08917) 90/84 | Chain Length Factor (CLF) |
| sanH | 85 | fdmH (AAQ08918) 89/79 | Acyl Carrier Protein (ACP) |
| sanI | 151 | fdmI (AAQ08919) 90/82 | Polyketide cyclase/aromatase |
| sanJ | 114 | fdmJ (AAQ08920) 82/75 | Monooxygenase |
| sanK | 144 | fdmK (AAQ08921) 87/80 | Monooxygenase |
| sanL | 246 | fdmL (AAQ08922) 86/79 | Monooxygenase |
| sanM | 148 | fdmM (AAQ08923) 89/84 | Hydroxylase25 |
| sanM1 | 155 | fdmM1 (AAQ08924) 87/82 | Hydoxylase25 |
| sanN | 350 | fdmN (AAQ08925) 85/78 | O-methyltransferase |
| sanO | 249 | fdmO (AAQ08926) 93/88 | Ketoreductase |
| sanP | 106 | fdmP (AAQ08927) 88/78 | Monooxygenase |
| sanQ | 105 | fdmQ (AAQ08928) 82/74 | Monooxygenase |
| sanS | 335 | fdmS (AAQ08929) 91/85 | 3-Ketoacyl ACP synthase |
| sanT1 | 338 | fdmT1(AAQ08930) 86/78 | Transporter |
| sanX | 101 | Unknown | |
| sanR | 143 | fdmR (AAQ08931) 89/80 | Regulator |
| sanU | 146 | fdmU (AAQ08932) 75/61 | Unknown |
| sanV | 634 | fdmV (AAQ08933) 83/77 | Asparagine synthetase |
| sanR1 | 610 | fdmR1 (AAQ08934) 82/73 | Regulator |
| sanT2 | 510 | fdmT2 (AAQ08935) 84/79 | Transporter |
| sanW | 159 | fdmW (AAQ089136) 74/68 | Phosphopantetheinyl transferase23 |
Verifying the identity of the cloned A-74528 gene cluster
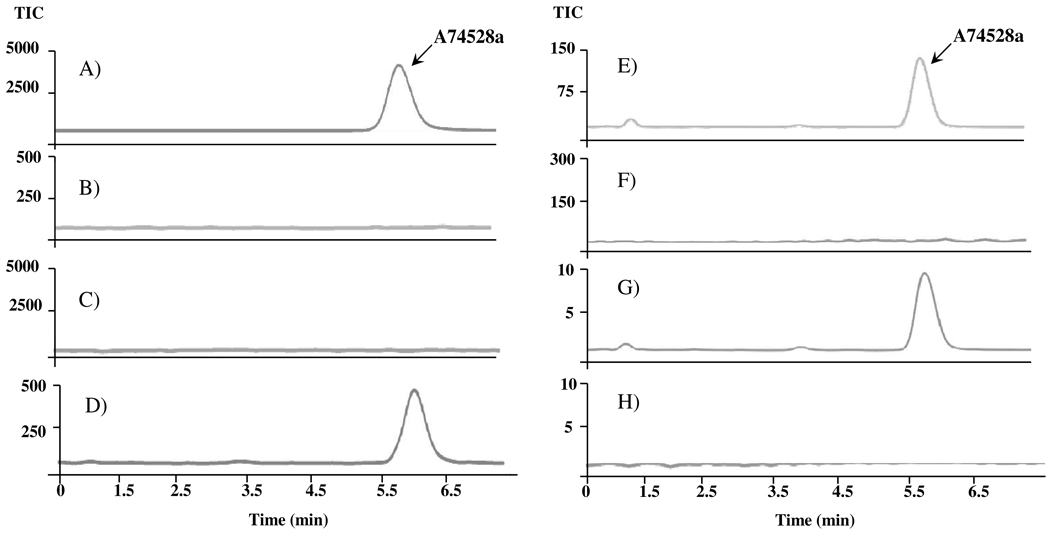
Two distinct strategies were pursued in order to verify the identity of the putative gene cluster described above. First, the gene encoding the KS subunit of the minimal san PKS was disrupted. A 450-bp internal fragment from the KS gene was amplified by PCR from cosmid pKZ1, and cloned into a gene disruption vector pJQ200SK. Mating of E. coli ET12567/pUZ8002 carrying this plasmid with Streptomyces sp. SANK 61196 yielded gentamicin resistant colonies. Out of 12 resistant colonies, one was found by PCR analysis to have resulted from homologous recombination. Specifically, a 465-bp DNA band was amplified using primers T3 and KS1, and a 460-bp DNA band was obtained using primers T7 and KS2. No PCR product was produced with genomic DNA from the wild-type strain or other candidate clones. The correct mutant clone, designated Streptomyces sp. SANK 61196ΔKS, had a pale yellow phenotype, suggesting that the disrupted PKS is also responsible for synthesizing the dark-colored pigment in wild-type Streptomyces sp. SANK 61196. Cultures of this mutant were extracted as described in the Materials and Methods section, and analyzed by LC-MS for the presence of A-74528. As shown in Figure 2, Streptomyces sp. SANK 61196ΔKS has lost its ability to synthesize A-74528.
Figure 2.
LC-MS-MS analysis of the metabolite A-74528. Triple quadrupole LC-MS multiple reaction monitoring detection of A-74528 in extracts from (A) wild-type Streptomyces. sp SANK 61196, (B) wild-type Streptomyces griseus, (C) Streptomyces lividans K4-114/pKZ11, (D) mutant Streptomyces sp. SANK 61196ΔKS, (E) Streptomyces lividans K4-114/pKZ11, (F) Streptomyces lividans K4-114/pKZ12, (G) Streptomyces lividans K4-114/pKZ13, and (H) Streptomyces lividans K4-114/pKZ14. The peaks shown correspond to a metabolite with a parent ion of 561 (m/z) and a fragment ion of 417 (presumably corresponding to the loss of the lactone moiety of A-74528)
In an alternative strategy for verifying the function of the san gene cluster, cosmid pKZ11 was introduced into Streptomyces lividans K4-114 by protoplast transformation.15 The resulting transformant was analyzed by quadrupole-time of flight mass spectrometry, and revealed the presence of a new compound with the predicted exact mass (M + Na = 583.12157 calc’d; 583.12157 obs) and retention time (9.7 min) as the A-74528 standard. The identity of A-74528 derived from pKZ11 was further confirmed by triple quadrupole LC-MS (Figure 2). In good agreement with MS/MS analysis of the A-74528 authentic standard, the product of the recombinant strain showed −126 and −144 fragmentation patterns, which likely result from the loss of the lactone and dehydration plus loss of lactone, respectively. Taken together, these results unambiguously establish that the identified gene cluster encodes for A-74528 biosynthesis.
Differential analysis of san and fdm gene clusters
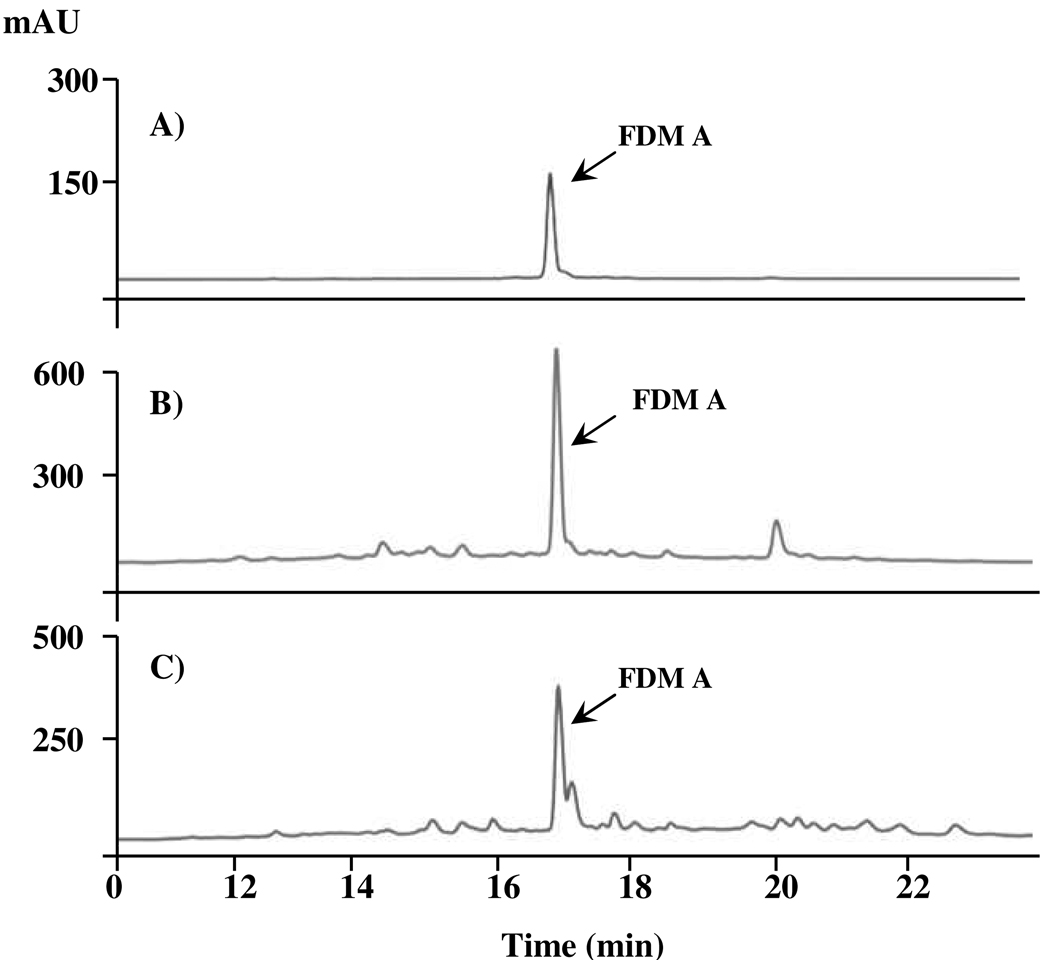
Given that A-74528 and FDM A are produced from remarkably similar gene clusters, we investigated the ability of S. griseus, Streptomyces sp. SANK 61196, and S. lividans K4-114/pKZ11 to synthesize both products. Mycelia were harvested, extracted, and analyzed by HPLC for the presence of FDM A, whereas the supernatant was treated with ammonium sulfate and analyzed by LC-MS for the presence of A-74528. As shown in Figure 3, FDM A was extracted from all three microbial sources, whereas A-74528 was only detected in culture media of Streptomyces sp. SANK 61196 and S. lividans K4-114/pKZ11 (Figure 2).
Figure 3.
Reverse phase HPLC analysis of the metabolite FDM A in Streptomyces griseus ATCC 49344 and Streptomyces lividans K4-114/pKZ11. (A) Authentic FDM A standard, (B) wild-type S. griseus, (C) S. lividans K4-114/pKZ11.
Construction and analysis of in-frame deletions in selected genes of the A-74528 cluster
As a first step towards understanding the role of individual san genes in A-74528 biosynthesis, we generated three in-frame deletion mutants in cosmid pKZ11. Mutant cosmid pKZ12 carries an in-frame deletion in the san2 gene, which encodes a putative acyl-ACP thioesterase. Oligonucleotides San2-20Fwd and San2-19Rev (Table-S1) were used to amplify the Aprar cassette, which includes 39 bp from each end of the san2 gene flanking the Aprar-gene. Upon induction with L-arabinose, the transfected cassette underwent homologous recombination with the san gene cluster present on pKZ11 to yield an apramycin-resistant mutant plasmid. To remove the Aprar-gene and generate an in-frame deletion mutant in san2, FLP mediated recombination was induced at 42°C in a cell line carrying this intermediate plasmid. Colonies sensitive to apramycin and resistant to carbenicillin were screened by PCR to verify loss of the Aprar-cassette. Similar in-frame deletions were also constructed in the sanM hydroxylase gene and the sanX gene of unknown function using primers SanM-20Fwd/SanM-19Rev and SanX-20Fwd/SanX1-19Rev (Table S1), respectively. The corresponding plasmids were designated pKZ13 and pKZ14, respectively. Protoplasts of S. lividans K-4114 were transformed with mutant plasmids pKZ12, pKZ13 and pKZ14. S. lividans K4-114/pKZ13 was intensely pigmented, whereas K4-114/pKZ12 and K4-114/pKZ14 were only faintly pigmented. Individual transformants were grown and extracted as described in the Materials and Methods section, and analyzed by LCMS for A-74528 and FDM A production. Whereas the sanM deletion mutant (pKZ13) showed a peak with a retention time of 5.4 minutes corresponding to A-74528, deletion mutants in san2 (pKZ12) and sanX (pKZ14) resulted in a loss of A-74528 production to below the detection limit (Figure 2). In contrast, low but detectable quantities of FDM A were observed in cultures of K4-114/pKZ12, K4-114/pKZ13, and K4-114/pKZ14 (data not shown). Taken together, these results suggest that, whereas SanX and the San2 acyl-ACP thioesterase play a role in the biosynthesis of both A-74528 and FDM A, the SanM hydroxylase exclusively participates in of FDM A biosynthesis. 23
DISCUSION
In this study we have shown that the recently discovered 2′-PDE inhibitor A-74528 from Streptomyces sp. SANK 61196 is synthesized by a pathway with a type II PKS at its core. Its high sequence similarity to the PKS responsible for the biosynthesis of fredericamycin A (FDM A) supports the hypothesis that A-74528 backbone is also a pentadecaketide. Furthermore, the observed homology between fdmS and sanS, both of which encode a ketosynthase responsible for chain initiation, suggests that both polyketide backbones are derived from a C6 primer unit that is further elaborated via condensation of 12 malonyl-CoA derived extender units.
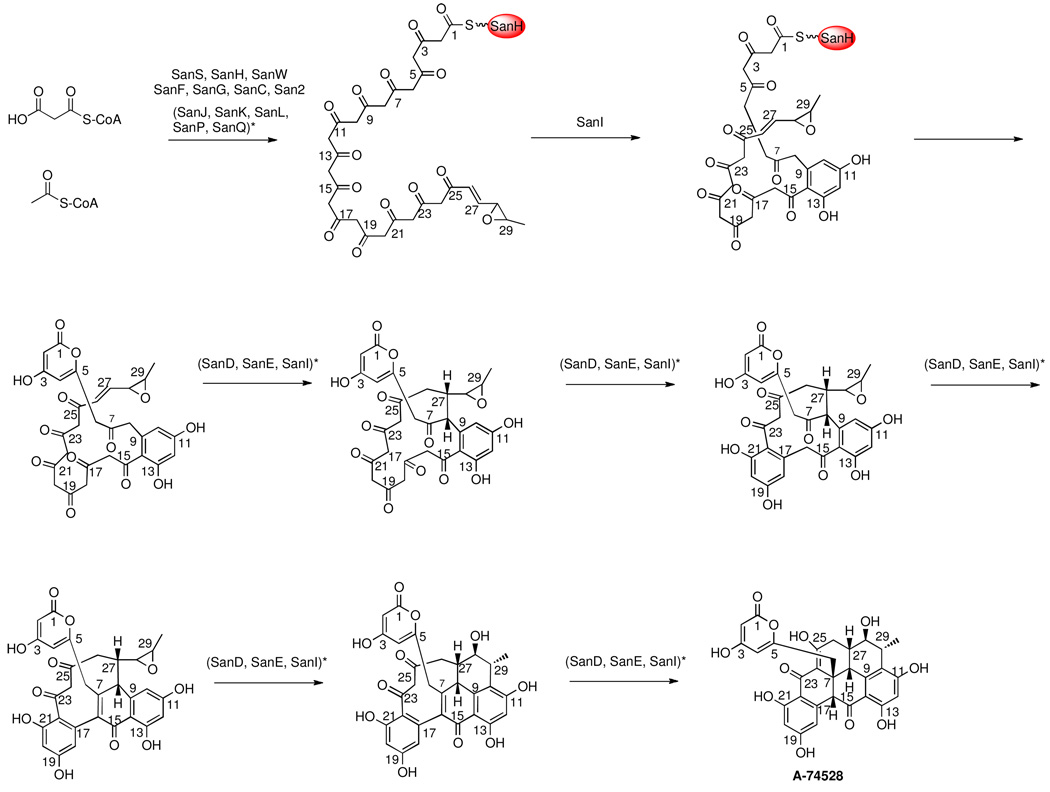
Our proposed biosynthetic pathway for the production of A-74528 is outlined in Figure 4. Specifically, the nascent C30 polyketide chain is likely formed through the concerted enzymatic activity of a bimodular PKS comprised of SanS, SanH, SanC, SanF and SanG. Such bimodular PKSs have been implicated in the biosynthetic pathways of several type II PKS products, including FDM A.12,16,19–22 Chain initiation is catalyzed by SanS, SanH and SanC. Apo-SanH is activated to holo-SanH by the phosphopantethienyl transferase SanW.23 From our data and earlier precedent24, San2 is responsible for purging any inappropriate primers that might transfer onto holo-SanH, thereby ensuring accurate chain initiation. The assignment of SanC as the ketoreductase responsible for reduction of the C-27 carbonyl is based on the enzyme’s high sequence similarity to FdmC, which has been biochemically shown to be involved in hexadienyl-ACP formation (A. Das, P.H. Szu J. T. Fitzgerald & C. Khosla, accompanying manuscript). A malonyl-CoA:ACP transacylase (MAT) from the endogenous fatty acid synthase also participates in chain initiation. The source of the dehydratase activity in the initiation PKS module is uncertain, but this enzyme is presumably also borrowed from the endogenous fatty acid synthase. The resulting C6 intermediate is extended by the SanF-SanG KS-CLF, SanH and the MAT into a full-length polyketide backbone, which must then undergo several tailoring reactions.
Figure 4.
Proposed mechanism of A-74528 biosynthesis. For transformations in which multiple enzymes are bracketed and denoted with *, the precise enzyme(s) involved in the polyketide tailoring step is unclear. (For example, SanJ, SanK, SanL, SanP, and SanQ are all possible candidates for the monooxygenase responsible for the C28–29 epoxide formation.) For all other steps, enzyme function is assigned based on previously characterized functions of paralogs in other type II PKS gene clusters. For details, see Discussion section.
Our model for A-74528 biosynthesis implicates critical differences in the early stages of the FDM A and A-74528 pathways. Specifically, we propose that the C28-C29 bond is oxidized to an epoxide during the earliest stages of A-74528 biosynthesis (i.e. during the formation or elongation of the C6 primer unit), affording the enzyme-bound C30 carbon chain shown in Figure 4. The installation of an epoxide early on during A-74528 would presumably serve two purposes. First, it could direct the regio- and stereo-chemistry of the unique cyclization cascade leading to A-74528, as shown in Figure 4. Second, it introduces a branch-point between A-74528 and FDM A biosynthesis at the earliest stages of the two overlapping metabolic pathways. Specifically, whereas formation of a hexadienyl unit enables access to FDM A, formation of the C6 mono-unsaturated epoxide intermediate yields A-74528. Several monooxygenase candidates exist within the san gene cluster (sanJ, sanK, sanL, sanP, and sanQ) that could generate this putative C28–29 epoxide. While other oxygenase enzymes are also present within the san gene cluster (san3, sanM, and sanM1), sequence homology suggests that these enzymes likely utilize aromatic substrates.25,26 Our data also rules out a role for SanM in A-74528 production. Once formed, the C30 polyketide chain bearing the monounsaturated epoxide appendage could undergo six consecutive ring-closing reactions through spontaneous and enzyme-controlled ring closures and Michael additions to generate aromatic ring systems. Based on earlier precedent27, the cyclase/aromatase SanI is likely responsible for the first ring closure (C9–C14), whereas SanD, SanE and/or SanI may be responsible for the remaining enzyme-catalyzed cyclization steps involved in creating the highly fused A-74528 skeleton. Finally, the transporters (SanT, SanT1, and SanT2) presumably shuttle A-74528 out of the cell. Our observation that A-74528 resides in the culture supernatant validates the role of transporters in the export of A-74528.
Importantly, our results indicate that SanX must also be involved in A-74528 biosynthesis, since deletion of this gene also decreases the A-74528 titer to below the detection limit of the LC-MS. However, as this 101 amino acid protein lacks any homologs in the sequence database, its function is unclear, and further biochemical investigations are warranted to clarify the chemistry of this prototypical protein. The presence of sanX provides a mark of evolution and may originate from horizontal gene transfer. We speculate that the fdm gene cluster was transferred to Streptomyces sp. SANK 61196, or vice-versa, and that further recombination and rearrangements resulted in the insertion or deletion of the sanX gene, leading to loss of synteny between the two gene clusters.
Perhaps most intriguingly, our studies represent the first example to our knowledge of a naturally occurring gene cluster that is capable of making two structurally and biologically distinct antibiotics, and suggests that the generally accepted “one cluster one antibiotic” paradigm may be overly restrictive. Earlier studies on polyketide pathways have highlighted the capacity of these multi-enzyme assemblies to synthesize structurally diverse products, although those products had no known antibiotic activity.28–30 If the controlled, multi-purpose deployment of PKS gene clusters for antibiotic biosynthesis is indeed a general phenomenon, then the medicinal relevance of the large (and growing) collection of gene clusters available through sequence databases may be considerably higher than previously imagined.
Supplementary Material
Acknowledgement
This research was supported by NIH grant R01 CA077248 to C.K. We thank Daiichi Sankyo for the generous gift of Streptomyces sp. SANK 61196 strain. We are grateful to Dr. Pavel Aronov of the Stanford University Mass Spectrometry Facility for support in obtaining LC-MS data, Dr. Tiangang Liu for practical guidance, and Jay Fitzgerald for helpful discussions. K. Z-R. is the recipient of a CONACYT postdoctoral fellowship, Mexico City. L. K. C. was supported by a postdoctoral fellowship (F32 CA137944) from the National Cancer Institute.
Footnotes
Abbreviations: INF, interferon; 2–5A, 2′,5′-oligoadenylates; 2′-PDE, 2′,5′-oligoadenylate phosphodiesterase; PKS, polyketide synthase; KS, ketoacyl synthase; CLF, chain length factor; ACP, acyl carrier protein; FDM, fredericamycin; LC-MS, liquid chromatography-mass spectrometry; MAT, malonyl-CoA:ACP transacylase
Supporting Information Available: Table S1 and Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Fujita Y, Kasuya A, Matsushita Y, Suga M, Kizuka M, Iijima Y, Ogita T. Bioorg Med Chem Lett. 2005;15:4317–4321. doi: 10.1016/j.bmcl.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 2.Kubota K, Nakahara K, Ohtsuka T, Yoshida S, Kawaguchi J, Fujita Y, Ozeki Y, Hara A, Yoshimura C, Furukawa H, Haruyama H, Ichikawa K, Yamashita M, Matsuoka T, Iijima Y. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 3.Player MR, Torrence PF. Pharmacol Ther. 1998;78:55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt A, Zilberstein A, Shulman L, Federman P, Berissi H, Revel M. FEBS Lett. 1978;95:257–264. doi: 10.1016/0014-5793(78)81006-0. [DOI] [PubMed] [Google Scholar]
- 5.Johnston MI, Hearl WG. J Biol Chem. 1987;262:8377–8382. [PubMed] [Google Scholar]
- 6.Silverman RH. Biochemistry. 2003;42:1805–1812. doi: 10.1021/bi027147i. [DOI] [PubMed] [Google Scholar]
- 7.Chadha KC, Ambrus JL, Jr, Dembinski W, Ambrus JL., Sr Exp Biol Med (Maywood) 2004;229:285–290. doi: 10.1177/153537020422900402. [DOI] [PubMed] [Google Scholar]
- 8.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 9.Khosla C, Gokhale RS, Jacobsen JR, Cane DE. Annu Rev Biochem. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- 10.Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 11.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 12.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 13.Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. J Biol Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 14.Ziermann R, Betlach MC. Biotechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA, editors. Practical Streptomyces Genetics. Norwich: Crowes; 2000. [Google Scholar]
- 16.Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, Kwon H, Hutchinson CR, Shen B. J. Am. Chem. Soc. 2005;127:16442–16452. doi: 10.1021/ja054376u. [DOI] [PubMed] [Google Scholar]
- 17.Marti T, Hu Z, Pohl NL, Shah AN, Khosla C. J. Biol. Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 18.Gust B, Kieser T, Chater K. PCR targeting systems in Streptomyces coelicolor A3(2), Norwich Resarch. [Google Scholar]
- 19.Moore BS, Hertweck C. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 20.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. Chem. Biol. 2004;11:959–969. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z, Metsa-Ketela M, Hertweck C. J Biotechnol. 2009;140:107–113. doi: 10.1016/j.jbiotec.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 22.Oja T, Palmu K, Lehmussola H, Lepparanta O, Hannikainen K, Niemi J, Mantsala P, Metsa-Ketela M. Chem Biol. 2008;15:1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Wendt-Pienkowski E, Shen B. J. Biol. Chem. 2006;281:29660–29668. doi: 10.1074/jbc.M604895200. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Koppisch AT, Khosla C. Biochemistry. 2004;43:9546–9555. doi: 10.1021/bi049157k. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Wendt-Pienkoski E, Rajski SR, Shen B. J Biol Chem. 2009;284:24735–24743. doi: 10.1074/jbc.M109.014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funa N, Funabashi M, Yoshimura E, Horinouchi S. J Biol Chem. 2005;280:14514–14523. doi: 10.1074/jbc.M500190200. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel R, Khosla SE, Hopwood DA, Khosla C. J. Am. Chem. Soc. 1994;116:10855–10859. [Google Scholar]
- 28.Fu H, McDaniel R, Hopwood DA, Khosla C. Biochemistry. 1994;33:9321–9326. doi: 10.1021/bi00197a036. [DOI] [PubMed] [Google Scholar]
- 29.Yu TW, Shen Y, McDaniel R, Floss HG, Khosla C, Hopwood DA, Moore BS. J. Am. Chem. Soc. 1998;120:7749–7759. [Google Scholar]
- 30.Shen Y, Yoon P, Yu TW, Floss HG, Hopwood D, Moore BS. Proc Natl Acad Sci U S A. 1999;96:3622–3627. doi: 10.1073/pnas.96.7.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.