Abstract
All cellular single-stranded (ss) DNA is rapidly bound and stabilized by single stranded DNA-binding proteins (SSBs). Replication protein A, the main eukaryotic SSB, is able to unwind double-stranded (ds) DNA by binding and stabilizing transiently forming bubbles of ssDNA. Here, we study the dynamics of human RPA (hRPA) activity on topologically constrained dsDNA with single-molecule magnetic tweezers. We find that the hRPA unwinding rate is exponentially dependent on torsion present in the DNA. The unwinding reaction is self-limiting, ultimately removing the driving torsional stress. The process can easily be reverted: release of tension or the application of a rewinding torque leads to protein dissociation and helix rewinding. Based on the force and salt dependence of the in vitro kinetics we anticipate that the unwinding reaction occurs frequently in vivo. We propose that the hRPA unwinding reaction serves to protect and stabilize the dsDNA when it is structurally destabilized by mechanical stresses.
INTRODUCTION
Single-stranded binding proteins (SSBs) are essential to all organisms. Their primary function is to rapidly bind ssDNA to stabilize and protect it. Replication protein A (RPA) is the main eukaryotic SSB (1,2). It is involved in a host of DNA-processing pathways of eukaryotic cellular metabolism including replication, recombination and, most, if not all, DNA repair pathways (3). The principal role of RPA in these processes stems from its ssDNA-binding properties. However, the ability of RPA to directly interact with a variety of proteins is also of crucial importance (4,5). Furthermore, RPA displays dsDNA binding and helix destabilizing activity. The affinity of RPA for double-stranded (ds) substrates increases when these substrates are damaged (6,7). Lao et al. (8) suggested that the dsDNA binding and helix destabilizing activities of RPA are in fact interrelated and that they can be explained by RPA binding to bubbles of ssDNA that are transiently formed in the dsDNA upon helix breathing (9). Once bound, RPA stabilizes these ssDNA bubbles and newly incoming RPA molecules can then further wedge into and progressively unwind the dsDNA. In contrast to the unwinding activity of helicases, the unwinding activity of RPA does not require ATP hydrolysis (10) and is likely dependent on mechanical energy stored in the DNA. Direct mechanistic insight, however, is lacking. In addition, it remains unclear how the reverse, rewinding reaction takes place. Acquiring these insights is crucial given the potential role of the RPA-unwinding activity in replication and DNA repair pathways. This paper reveals the kinetics and mechanism of the unwinding and rewinding reaction through single-molecule experiments.
Human RPA (hRPA) is a highly flexible protein complex composed of three tightly associated subunits with molecular weights of ∼70, 32 and 14 kDa (accordingly referred to as RPA70, RPA32 and RPA14). hRPA binds ssDNA with high affinity, low sequence specificity, and low cooperativity (cooperativity factor, ω = 10) (11,12). RPA contains four ssDNA-binding domains that bind DNA with decreasing affinities A–D. Binding of RPA to ssDNA proceeds in a multi-step process starting with an 8–10-nt binding mode (binding domains A–B) and culminating in a final 30-nt binding mode. This process of multi-step binding to ssDNA is accompanied by a profound conformational change from a globular to an extended conformation of the protein (4,13–15).
Using single-molecule magnetic tweezers (MTs), we studied the dsDNA-unwinding activity of hRPA. We find that the helix-unwinding activity of hRPA is very sensitive to and driven by torsional stress present in the DNA substrate. Previously it was suggested that the reaction only occurs at low salt concentrations (8,16,17). However, we find that hRPA-induced helix unwinding also occurs at relatively high, physiologically relevant salt concentrations (>100 mM NaCl) when a modest stretching force (∼0.6 pN) is applied, which generates an unwinding torque in a negatively supercoiled molecule. Dissociation of hRPA and the resulting rewinding of the duplex occur upon release of the unwinding torsional stress or upon application of rewinding torque. The strong torque sensitivity of the helix-unwinding reaction provides a means for tight mechanical regulation.
Many cellular processes have been described to generate torsional stress (18). Here, we argue that the helix unwinding activity of hRPA is important for protecting and stabilizing dsDNA that undergoes stress-induced structural transitions. We furthermore discuss the importance of hRPA-stimulated helix unwinding in DNA replication, recombination and repair.
MATERIALS AND METHODS
Expression and purification of human RPA
hRPA was expressed in Escherichia coli BL21(DE3)pLysS using the expression vector pET11d-hRPA and purified through Affigel-Blue (Biorad) and MonoQ anion-exchange chromatography (GE Healthcare) as described (19). This was followed by size-exclusion chromatography (Superdex 200, GE Healthcare) and fractions containing pure hRPA were concentrated using anion exchange chromatography. Aliquots containing 10% glycerol were frozen in liquid nitrogen and stored at −80°C until use.
DNA constructs
The ss- and dsDNA constructs were prepared as described previously(20).
MTs assay
A MTs setup was used in these experiments as described (21). Employing image processing, positional tracking of the bead with 5-nm position accuracy in all three dimensions (3D) was achieved (22). To exclude the effect of thermal drift, all positions were measured relative to a 3.2-μm polystyrene bead (Bang Laboratories, Carmel, IN, USA) that is fixed to the bottom of the flow cell. DNA constructs carrying a magnetic bead at one end were anchored to the bottom of a flow cell as described elsewhere (22). The size of the magnetic beads was chosen based upon the requirements to apply high forces in the experiments with ssDNA (bead size 2.8 μm) and to maximize the experimental resolution in the case of dsDNA (bead size 1.0 μm). Prior to protein experiments, the force-extension curve of single DNA molecules was measured and correct contour and persistence lengths were confirmed. In case of dsDNA, rotation curves at different forces were recorded additionally. In plots showing bead height, the red traces presented are the raw data acquired at 60 Hz. When shown, yellow lines correspond to data filtered at 0.5 Hz. All measurements were carried out at 22°C.
RPA/DNA reactions
The flow-cell final volume was ∼100 μl. All reactions were performed in Tris–HCl 30 mM (pH 7.5). Salt concentrations and hRPA concentrations were as described in the text. Interactions of hRPA with ssDNA and dsDNA molecules were monitored through measurement of the height of the bead.
Fitting procedures
On dsDNA, association and dissociation rates were extracted on the basis of fits of the linear portion of the extension traces. Occluded binding size was assumed to be 30 nt leading to the removal of three supercoils. Size of the supercoils for a given reaction were determined by fitting the slope of rotation curves acquired on bare DNA at the same force and [NaCl]. The step trace in Figure 2b was acquired by applying a step-fitting algorithm (23).
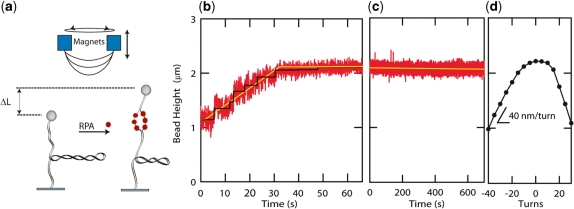
Figure 2.
hRPA dsDNA binding and helix unwinding. (a) Schematic of the experimental scheme. hRPA binds to ssDNA that is transiently exposed upon helix breathing. Further protein binding unwinds the helix. Unwinding leads to removal of negative supercoils that were previously applied to the molecule (n=−40). (b) Height of the magnetic bead versus time after introduction of hRPA. The increasing height signals hRPA binding. The binding curve reveals a reaction that progresses linearly in time (yellow line, guide to the eye). Step-like behavior in the binding traces indicates single enzyme binding events (black line, fit based on step finding algorithm). The reaction saturates when the length of the molecule reaches the length of the non-supercoiled molecule. (c) After removal of the unbound hRPA, hRPA remains stably bound to the ssDNA-bubble ([NaCl]=30 mM). (d) Rotation curve of molecule taken prior to the reaction. The slope of the rotation curve at experimental reaction conditions [NaCl]=30 mM; F=0.55 pN equals 40 nm/turn. A singly bound RPA molecule is expected to unwind the helix by ∼3 turns, thus leading to a length increase of 120 nm.
RESULTS
Dynamics of hRPA association on ssDNA
MT-based assays (24) enable studying the association and dissociation of hRPA on ssDNA and dsDNA. In a MT assay, an ssDNA or dsDNA molecule is at one end attached to the bottom glass slide of a flow cell and on the other end to a superparamagnetic bead. Positioning of external magnets allows the application of tunable stretching forces (0–60 pN). Video microscopy is used to accurately track bead positions in 3D, and thus to measure the end-to-end distance of the molecule (Figure 1a and ‘Materials and Methods’ section).
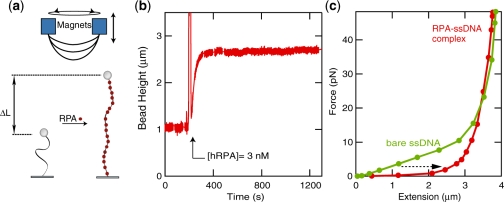
Figure 1.
hRPA binding to ssDNA. (a) In a magnetic tweezer setup, an ssDNA molecule is tethered in between a glass slide and a paramagnetic bead. (b) hRPA binding curve taken at a stretching force of 3 pN. The reaction is initiated upon introduction of hRPA in the flow cell. A fast increase in the end-to-end distance is measured. The exponential shape of the binding curve indicates a low binding cooperativity (ω = 10–30). (c) Force-extension curves for bare ssDNA (green) and an ssDNA–hRPA complex (red) reveal an increase in hRPA-induced flexural rigidity and the prevention of hairpin formation. [NaCl]=30 mM. [hRPA]=3 nM.
First, we examined hRPA-binding to a ssDNA molecule (7.3 kb). hRPA-binding reactions are initiated upon introduction of a buffer containing hRPA into the flow cell ([hRPA] = 3 nM, [NaCl] = 30 mM). Figure 1b displays an example of a time trace recorded at a stretching force of 3 pN. The end-to-end distance of the molecule quickly increases as hRPA protein binds to the ssDNA substrate, resulting in a stiffening of the molecule and an increase of its end-to-end distance. Fifty seconds after the initiation of the reaction, the ssDNA end-to-end distance reaches a plateau indicating termination of the reaction. The shape of the association curve allows extracting information on the cooperativity of the reaction (see ref. 25 and Supplementary Data section for details on the fitting procedures). Such an analysis reveals that hRPA binds ssDNA with low cooperativity (ω < 30), independent of [NaCl] in the range 30–600 mM, confirming earlier reports (12).
After completion of the reaction, force-extension curves were measured. The force-extension curve recorded for the ssDNA–hRPA complex is plotted in Figure 1c alongside the force-extension curve recorded for the bare ssDNA substrate. We observe a pronounced increase in the extension at low forces for the hRPA–ssDNA complex. This can be explained by the suppression of hairpin formation in combination with an increase in bending rigidity. From the extension data at high force, it is apparent that the hRPA–ssDNA complex has a slightly shorter contour length than ssDNA (3%).
Experimental configuration for probing the unwinding reaction
Next, we probed the dsDNA-unwinding activity of hRPA. A dsDNA molecule (8.0 kb) was tethered in the flow cell and rotationally constrained at both attachment points. Negative supercoils were induced in the construct prior to the experiment by rotating the external magnets (Figure 2a and ‘Materials and Methods’ section). Reactions were again initiated through the introduction of a hRPA-containing buffer.
As was described previously (8,9), helix unwinding induced by hRPA occurs upon binding and stabilizing of the ssDNA bubble that is transiently formed upon helix breathing. The formation and growth of such a hRPA-stabilized DNA bubble leads to a local change in DNA length and thereby a means is provided to follow the reaction in real time in the MT assay. More significantly however, the local unwinding of the duplex induced by hRPA binding also changes the helical twist. This change in property can be measured more easily in our instrument: since the linking number is constant, changes in twist give rise to a change in the number of supercoils in the DNA and accordingly to a pronounced change in length (40-nm change in end-to-end distance per plectonemic supercoil at an applied force of F=0.55 pN, see the rotation curve plotted in Figure 2d). Since hRPA has a 30 nt occluded binding size on ssDNA, binding to a ssDNA bubble in the duplex of the same size leads to unwinding of the helix by approximately three turns and consequently the removal of ∼3 supercoils, i.e. an increase in bead height of about 120 nm.
Progressive unwinding and reaction saturation
An example binding trace is displayed in Figure 2b (experimental conditions were 30 mM [NaCl], 25 nM [hRPA], 40 negative turns (n = –40), F = 0.55 pN). After nucleation of the binding reaction (after a lag time of about 5 s), the recorded bead height increased with a constant rate. This is a consequence of progressive duplex unwinding by hRPA proteins that bind and wedge the ssDNA bubble (8,9). Note that individual binding steps can be discerned as discrete steps in the time trace (black line), which likely can be associated with single proteins that bind the ssDNA bubble. In independent experiments under conditions where association and dissociation occur with similar rates, we have measured the occluded binding size for hRPA at the first binding step to be 31±1 nt, see below.
Interestingly, the reaction halted abruptly when the end-to-end distance of the molecule reached a length corresponding to the top of the rotation curve (Figure 2b), i.e. the length of the non-supercoiled molecule. As will be discussed in detail below, this is a consequence of the strong torque dependence of the reaction, being promoted by negative (unwinding) torque and suppressed by positive (rewinding) torque. Reactions performed on nicked dsDNA molecules did not yield any measurable change in length at the reaction conditions tested; [hRPA] ≤ 200 nM, [NaCl] ≥ 30 mM, force range 0.2–0.6 pN (data not shown) (9). This is a further indication that the reaction is sensitive to writhe and twist present in the molecule.
After completion of the reaction, we removed the unbound hRPA from the flow-cell in order to study the stability of the hRPA-stabilized dsDNA bubbles (Figure 2c). Under conditions of low salt ([NaCl] < 100 mM) and moderate applied force (F > 0.5 pN), we found that hRPA bubbles are highly stable and essentially no dissociation is measured (see Figure 2c, the measured end-to-end distance of the molecule does not change on a time scale of at least 10 min). At [NaCl] higher than 100 mM, dissociation does occur, in line with previous results (16), see also below.
Influence of salt on the kinetics of association
Stretching forces and twist can induce structural transitions in DNA. Unwinding and stretching of DNA, for example, can lead to local denaturation of the duplex DNA (26). This well-described property occurs at modest stretching forces (≥0.3 pN) and manifests itself in MT experiments by a dissimilar response of the duplex to positive and negative coiling, in turn associated with an asymmetry of the rotation curves (for an example of such rotation curve, see Figure 2d). The denaturated-DNA and B-DNA phases coexist, with the extent of denaturation being dependent on factors that influence helix stability. We investigated the effects of two such factors, the stretching force and the concentration of monovalent salt, to obtain more insight in the nature of the hRPA-unwinding reaction.
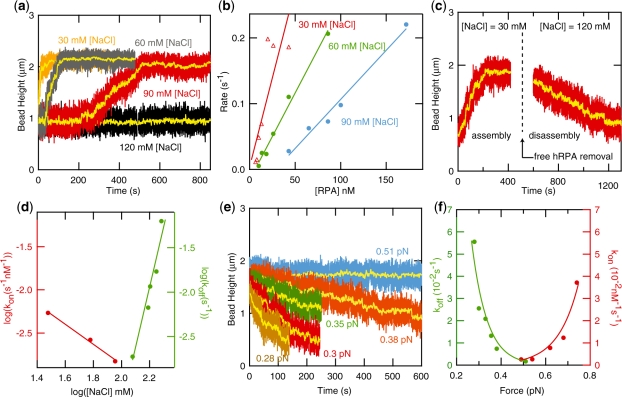
Figure 3a shows binding curves obtained at [hRPA]=40 nM and [NaCl]=30, 60, 90 mM and 120 mM, n=–40, F=0.5 pN. Increasing the salt concentration markedly alters the reaction kinetics. At high salt, the binding curves are characterized by a salt-dependent lag time, followed by a nucleation event that initializes the reaction. The slower nucleation of the reaction can be expected since higher [NaCl] leads to increased charge screening and thereby to enhanced helix stability and hence lower levels of DNA denaturation. After nucleation, the reaction progressed with a constant, salt-dependent reaction speed. At [NaCl]=120 mM, no reaction was observed. Figure 3b summarizes the hRPA binding rates obtained from experiments at various [hRPA] and [NaCl]. We confirmed that association is much faster than dissociation for the conditions used in these experiments (koff < 10–3 s–1, see dissociation data below) and the reactions can thus be regarded to be in the limit of strong, irreversible binding. The binding rates plotted in Figure 3b follow a clear linear trend as a function of [hRPA] with the intercepts of the linear fits crossing the x-axis close to zero, a further indication that dissociation is slow (27,28). From the slopes of the linear fits, we extracted the on-rates, kon. The values extracted for kon as a function of [NaCl] follow a linear trend on a log–log scale with 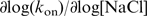 = –1.2 (Figure 3d). At low salt ([NaCl] < 0.1 M), the kinetics of hRPA binding to the transiently forming ssDNA bubble is diffusion limited. In a diffusion-limited reaction, an increased salt concentration affects the kinetics through a more efficient charge screening which facilitates the mutual approach of the overall negatively charged hRPA and ssDNA and thus leads to faster association (27,28). The effect of salt on the stability of the helix has the opposite effect, with kon decreasing with [NaCl] (29). The combination of these two effects leads to the weakly negative dependence of log(kon) on log([NaCl]) measured for the unwinding reaction.
= –1.2 (Figure 3d). At low salt ([NaCl] < 0.1 M), the kinetics of hRPA binding to the transiently forming ssDNA bubble is diffusion limited. In a diffusion-limited reaction, an increased salt concentration affects the kinetics through a more efficient charge screening which facilitates the mutual approach of the overall negatively charged hRPA and ssDNA and thus leads to faster association (27,28). The effect of salt on the stability of the helix has the opposite effect, with kon decreasing with [NaCl] (29). The combination of these two effects leads to the weakly negative dependence of log(kon) on log([NaCl]) measured for the unwinding reaction.
Figure 3.
Influence of helix (de-)stabilizing effects: salt and force. (a) Binding curves recorded at 0.5 pN and [RPA]=40 nM for [NaCl]=30, 60, 90 and 120 mM. The salt concentration has a marked effect on the binding kinetics. High-salt traces are characterized by a salt-dependent lag time and slower extension rates. (b) Extension rates at varying [hRPA] concentrations. The reaction rates are linearly dependent on [hRPA]. (c) Binding curve at low salt ([NaCl]=30 mM) followed by dissociation at high salt ([NaCl]=120 mM) after removal of hRPA. (d) Association and dissociation rates on a log–log scale. (e) Dissociation curves measured at varying stretching force. (f) Association and dissociation rates as function of force reveal a strong force dependence of the reaction kinetics.
Influence of salt on the kinetics of dissociation
We furthermore studied the salt dependence of protein dissociation from the ssDNA bubbles in dsDNA. To this end, after completion of the binding reaction at low salt and moderate applied force (0.4 pN), the unbound hRPA was removed from the flow cell and dissociation curves were recorded, see Figure 3c for an example trace. Dissociation rates are plotted versus [NaCl] on a log–log scale in Figure 3d. The value for the slope extracted for the dissociation rates amounts to 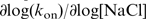 =7.0, much higher than the value found for the association rates. The dissociation rates measured for reactions on dsDNA can be compared with rates obtained for ssDNA-dissociation reactions with a surface plasmon resonance technique (30) (see Supplementary Figure S3). Dissociation of hRPA from ssDNA is much slower than dissociation from dsDNA (koff,ssDNA=2.2 10–5 s–1 versus koff,dsDNA=2 10–3 s–1 at [NaCl]=120 mM, F=0.5 pN). Furthermore, the kinetics of dissociation from ssDNA substrates display a weaker dependence on [NaCl] in comparison to dissociation from dsDNA:
=7.0, much higher than the value found for the association rates. The dissociation rates measured for reactions on dsDNA can be compared with rates obtained for ssDNA-dissociation reactions with a surface plasmon resonance technique (30) (see Supplementary Figure S3). Dissociation of hRPA from ssDNA is much slower than dissociation from dsDNA (koff,ssDNA=2.2 10–5 s–1 versus koff,dsDNA=2 10–3 s–1 at [NaCl]=120 mM, F=0.5 pN). Furthermore, the kinetics of dissociation from ssDNA substrates display a weaker dependence on [NaCl] in comparison to dissociation from dsDNA: 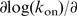 log[NaCl]=4.6 (see Supplementary Figure S3). These findings indicate that the dynamics of helix breathing contribute to the dissociation kinetics. Such behavior is indeed expected in case dissociation occurs as a multi-step process. In such a multi-step process, a partially bound protein remains after release of one or more binding domains. For partially dissociated hRPA proteins adjacent the dsDNA junction, helix breathing and re-association compete with completion of dissociation. Reaction conditions that lead to more frequent helix breathing are therefore expected to give rise to slower overall dissociation. A multi-step dissociation mechanism also explains the constant rate of dissociation observed in experiments (see Figure 3c), which indicates that dissociation predominantly occurs at the ends of the protein cluster. The experiments on the force dependence of the kinetics of dissociation that will be discussed in the next section also point at a multi-step dissociation process.
log[NaCl]=4.6 (see Supplementary Figure S3). These findings indicate that the dynamics of helix breathing contribute to the dissociation kinetics. Such behavior is indeed expected in case dissociation occurs as a multi-step process. In such a multi-step process, a partially bound protein remains after release of one or more binding domains. For partially dissociated hRPA proteins adjacent the dsDNA junction, helix breathing and re-association compete with completion of dissociation. Reaction conditions that lead to more frequent helix breathing are therefore expected to give rise to slower overall dissociation. A multi-step dissociation mechanism also explains the constant rate of dissociation observed in experiments (see Figure 3c), which indicates that dissociation predominantly occurs at the ends of the protein cluster. The experiments on the force dependence of the kinetics of dissociation that will be discussed in the next section also point at a multi-step dissociation process.
Strong force dependence of association and dissociation
We subsequently investigated the force dependence of the association and dissociation rates. Association data were taken at low salt ([NaCl]=30 mM), a condition for which dissociation can be disregarded. Dissociation curves were taken at various stretching forces and at [NaCl]=120 mM, after association at F=0.4 pN and [NaCl]=30 mM (Figure 3c). Figure 3e shows a collection of dissociation curves illustrating a strong dependence of the rate on force. In Figure 3f, rates of association and dissociation are plotted versus the applied force. The data reveal a strong force dependence of the kinetics of both association and dissociation. Fast association occurs at high force, whereas at lower forces dissociation is favored. The data was fit on the basis of the torque-based model that is discussed below.
Torsional regulation of the reaction kinetics
We examined whether reactions can be reinitiated after saturation by means of a mechanical application of unwinding turns. Figure 4a shows association curves obtained after application of an increasing number of unwinding turns (n=−40, −70 and −100). After each addition of new negative supercoils, a renewed association was observed confirming the possibility of reaction reinitiation.
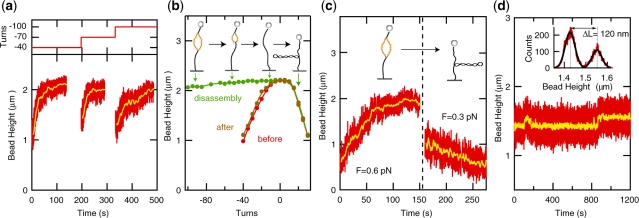
Figure 4.
Mechanochemical regulation of the reaction kinetics. (a) Binding curves obtained after application of an increasing number of unwinding turns (n=–40, −70 and −100). Reactions are reinitiated and binding of hRPA to an extended stretch of DNA occurs after each increase in the number of negative turns. (b) Dissociation induced by application of positive (rewinding) coiling (from n=−100 to n=+30). The length of the molecule is recorded in five turn intervals. The resulting rotation curve is displayed in Figure 4b (green). The long plateau indicates that protein dissociation is induced by positive coiling. The rotation curve taken after dissociation (brown) closely matches the rotation curve measured for the bare DNA molecule prior to the experiment (red) (F=0.55 pN, [NaCl]=30 mM). (c) Shift of reaction equilibrium by a modest change in force. At F=0.6 pN, hRPA association is observed ([hRPA]=100 nM). At 0.3 pN, rapid dissociation occurs for the exact same buffer condition ([NaCl]=120 mM). (d) hRPA-induced dsDNA unwinding under conditions where kon ≈ koff, [NaCl]=150 mM, F=0.8 pN, [hRPA]=120 nM, n=−30. The end-to-end-distance of the molecule fluctuates between two stable levels indicative of an individual molecule hopping on and off the dsDNA substrate. Change in length, ΔL=120 nm (see histogram of filtered extension data in inset), corresponds to the removal of 3.1±0.1 supercoils during the binding event, indicative of an hRPA occluded binding size of 31±1 nt.
The high sensitivity of the dissociation kinetics to writhe and twist furthermore raises the question whether hRPA can be dislodged from the DNA by mechanical rewinding of the helix. To investigate this possibility, we first induced extended binding of hRPA (up to n = –100 as above). Next, we flushed the unbound hRPA out of the flow cell and turned the magnet in the opposite direction from n=−100 to n=+30, thereby applying a rewinding twist. We recorded the end-to-end distance of the molecule with five turn intervals. The resulting rotation curve is displayed in Figure 4b (green symbols). A long plateau is measured, indicating that mechanical rewinding does not lead to positive supercoiling since supercoiling would result in a reduction of the measured end-to-end distance. Apparently, hRPA molecules are instead dislodged from the helix and the DNA molecule is rewound. Indeed, the rotation curve that was measured subsequently (brown) closely matches the rotation curve that was recorded before the start of the experiment (red), confirming that the bound hRPA proteins were all removed from the helix, leaving the DNA molecule in its initial bare state. Control over the superhelical density of the molecule therefore provides a direct handle for shifting the reaction equilibrium back and forth from a state where association is favored to a state where dissociation is invoked.
The data obtained on the force dependence of the reaction rates predict that a drastic shift in the reaction equilibrium can also be induced through a simple change in the magnitude of applied force. On the basis of the data in Figure 3f, it is expected that a decrease in applied force from 0.6 to 0.3 pN increases the equilibrium constant, koff/kon, of the reaction by a factor of 400. We experimentally assessed this notion (see Figure 4c): at the physiological [NaCl] of 120 mM, hRPA association and dsDNA unwinding were induced at an applied force of 0.6 pN. Subsequently, the force was lowered to 0.3 pN without changing the reaction buffer. A fast dissociation of the protein–DNA complex is observed in accordance with a drastic shift of the reaction equilibrium.
On the basis of the [NaCl] and force dependencies of the dissociation and association rates, it is possible to tune the reaction toward a regime where koff ≈ kon. Figure 4d shows an MT trace recorded under such conditions, [NaCl]=150 mM, F=0.8 pN, [hRPA]=120 nM, n=−30. The recorded end-to-end distance fluctuates between two stable levels, indicative of individual hRPA proteins hopping on and off the dsDNA substrate. A histogram of the extension data (inset of Figure 4d) allows extracting the change in dsDNA length, ΔL, associated with binding of a single hRPA molecule. ΔL=120 nm, corresponding to the removal of 3.1±0.1 supercoils under the reaction conditions used, and hence indicative of an occluded binding size of 31±1 nt, in line with the reported size of the largest binding mode for hRPA (30 nt).
DISCUSSION
Mechanistic model for helix unwinding by hRPA
The above observations are captured in a model for the helix unwinding reaction presented in Figure 5a. hRPA is represented with its four DNA-binding domains labeled A–D. Binding is assumed to occur in a multi-step pathway starting with binding of DNA-binding domains A–B and proceeding with a 5′-to-3′ polarity (31). In line with previous studies (9), the model assumes that reaction nucleation and subsequent hRPA-binding occur upon local helix breathing, which transiently exposes a ssDNA substrate to which hRPA can bind. The linear kinetics of subsequent unwinding measured in the experiments indicates that further denaturation and protein binding occur primarily at the junction between the dsDNA and hRPA-stabilized bubbles, where a DNA-fork structure is formed and where the helix stability is reduced. This gives rise to contiguous binding of newly incoming proteins, consistent with the cooperative nature of the reaction observed in our experiments.
Figure 5.
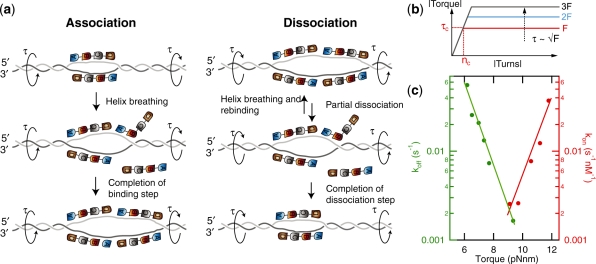
Proposed model for hRPA-driven helix unwinding. (a) hRPA binds to ssDNA that is exposed upon helix breathing. Helix breathing is strongly promoted by unwinding torque. Much like association, dissociation (right panel) utilizes a multi-step pathway. After partial dissociation, helix breathing and subsequent re-association competes with complete dissociation. (b) Relation between torque and force in a supercoiled molecule. Torque reaches maximum and constant value (with τc approximately proportional to 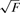 ) for |n| > nc, with nc the critical coiling number required for supercoil generation (27). (c) Reaction rates from Figure 3f replotted as a function of torque. Linear dependencies on this log-linear plot indicate an exponential dependence of the rates on torque.
) for |n| > nc, with nc the critical coiling number required for supercoil generation (27). (c) Reaction rates from Figure 3f replotted as a function of torque. Linear dependencies on this log-linear plot indicate an exponential dependence of the rates on torque.
For the reaction conditions used in the association experiments, koff << kon, and the association reaction can be represented as
| (1a) |
| (1b) |
where ka is the intrinsic protein binding on-rate and ko and kc represent the rates of opening and closing of the helix over a minimally required number of basepairs. The minimal distance of helix breathing required for protein binding likely corresponds to the footprint required for the first binding step in the multi-step binding pathway (see below). The apparent association rate, kon, for the above reaction can be expressed as
| (2) |
Herein, α = ko/(ko + kc) can be interpreted as the net fraction of time over which a ssDNA-binding template for hRPA is available.
The stability of the dsDNA directly adjacent to the fork structures is expected to be highly sensitive to torsional stress present in the molecule. Our model therefore assumes that the applied force primarily acts by modulating the torsional stress. A number of models have been proposed to describe the torque, τ, that is generated by stretching force applied to coiled DNA (26,32). A few features are general to these models: at low supercoiling density, σ, τ is approximately independent of force and linearly dependent on σ. The formation of plectonemes requires the torque to exceed a force-dependent critical value, τc. In the postbuckling regime, where supercoiled and extended DNA domains coexist, τ is independent of σ, i.e. τ=τc. The dependence of torque on the number of turns and force is visualized in Figure 5b.
For a torque-driven reaction, α and consequently the apparent on-rate, are expected to depend on τ according to (33)
| (3) |
where δθ is the rotational angle, the relevant reaction coordinate in our model. The constant reaction rate observed in the association traces indicates that the rate of extension is independent of writhe, i.e. of the number of supercoils. This finding can now be understood from the fact that the torque is independent of writhe (τ = τc). The saturation of the reaction is furthermore explained by the reduction of the driving torque once all supercoils have been consumed.
The strong dependence of the dissociation rates on force indicates that the dynamics of helix breathing also contribute to the kinetics of dissociation. An involvement of helix breathing in the dissociation kinetics can be understood when dissociation occurs as a multi-step process. In such process, after release of one or several binding domains, a partially bound protein remains. At the junction with the dsDNA, completion of dissociation competes with helix breathing and rebinding of the released units (see Figure 5a). Note that the study of the salt dependence of dissociation kinetics indeed also suggests a step-by-step dissociation process. If the multi-step dissociation model holds, the off-rate, koff is expected to follow a dependence on torque similar to the association rate (with unwinding torque leading to faster breathing and faster rebinding of dissociated subunits and thus slower overall dissociation):
| (4) |
In order to validate the proposed model, the data of Figure 3e on the force dependence of association and dissociation rates were plotted as a function of torque in Figure 5c. Here, the torque exerted on the supercoiled DNA molecule was calculated on the basis of the model by Marko (32):
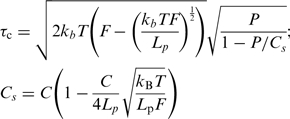 |
(5) |
where, Lp is the persistence length of dsDNA (Lp=50 nm) and Cs and P are the twist stiffnesses of the molecule in the extended and plectonemic state respectively, P=24 nm, C=95 nm (32). Recent direct measurements of the torque generated in a supercoiled molecule have validated this model (34). Linear trends are evident from the plot in Figure 5c, demonstrating that the data are consistent with a torque-based model. Fitting of the linear slopes in Figure 5c results in δθ=0.65±0.1 turns for association and δθ=0.72±0.05 turns for dissociation. Considering the helical repeat of a relaxed DNA molecule (10.5 bp/turn) these values correspond to breathing lengths of ∼7.0±1 bp and 7.5±0.5 bp, respectively. For the association reaction, this value is physically very plausible, i.e. in accordance with the size of the smallest binding mode reported for hRPA (8 bp).
Biological function of torsional regulation of the unwinding reaction
The high binding affinity of RPA for ssDNA substrates and the abundance of RPA in the nucleus (1 × 104–2 × 105 copies per cell) (35) imply that any ssDNA that is formed in the cell is quickly bound and protected by RPA. ssDNA and substrates with single-stranded character, such as damaged or denatured dsDNA, are frequent intermediates of cellular metabolism, hence the critical involvement of RPA in a great variety of DNA-processing pathways.
Here, we studied the dsDNA binding and helix-unwinding activity of hRPA. This activity is a manifestation of its high-affinity ssDNA binding capability: hRPA is able to rapidly bind and stabilize bubbles of ssDNA in the dsDNA that are transiently exposed upon helix breathing. We conclude that the helix unwinding activity is torsionally regulated, with unwinding torque promoting the reaction and rewinding torque suppressing the reaction. As previously described, the reaction kinetics are strongly dependent on salt, with conditions of higher salt inhibiting the reaction. However, we find that these inhibitory effects can easily be overcome by the generation of unwinding torque, with reactions readily taking place at [NaCl] > 100 mM under application of modest stretching forces (F ∼ 0.6 pN). The unwinding activity is self-limiting as helix unwinding goes hand in hand with removal of supercoils, ultimately removing the driving torsional stress. The reaction can easily be reverted: release of tension or the application of a rewinding torsional stress leads to protein dissociation and helix rewinding. Our data on the kinetics of dissociation from dsDNA indicate that dissociation utilizes a multi-step pathway, much like the association process.
A plausible cellular function of the hRPA-induced helix unwinding activity is akin to its main role on ssDNA: the stabilization and protection of torsionally destabilized dsDNA. Many cellular processes can generate sufficient torsional stress to destabilize the helix and invoke helix–coil transitions (18). The process of transcription, for example, is known to generate torsional stress, leading to supercoiled DNA in topological domains. Transcribing RNA polymerases are even able to generate sufficient torsion to transiently perturb DNA structure even on DNA templates that are rotationally unconstrained (36,37). Another example is found in ATP-dependent chromatin remodelers that generate negative superhelical torsion and can give rise to structural transitions in DNA (38).
Our findings have implications for many of the DNA-processing pathways in which RPA-induced helix unwinding was previously implicated to be important: replication-origin firing, damage recognition and incision in excision repair (10,39), and finally, promotion of joint molecule formation in homologous recombination (HR). Our study calls for a careful consideration of the role of torsional regulation in these processes. Interestingly, for example, the human Rad54 translocase protein was previously proposed to stimulate joint molecule formation in HR by supercoiling DNA domains (40).
The torsional regulation of the reverse, rewinding reaction is equally important. Annealing of RPA-bound ssDNA is required in the second-end capture step in HR (41). Also in HR, the re-annealing of homologous strands after strand invasion needs to be prevented, a function to which the post-strand invasion activity of RPA has been attributed (42). Finally, the exertion of torsional stress may constitute a possible mechanism for the dissociation of RPA-stabilized ssDNA bubbles by annealing helicases (43).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The ‘Stichting voor Fundamenteel Onderzoek der Materie (FOM)’, which is financially supported by the ‘Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)’; NWO chemical sciences VIDI (to J.L.); a NWO chemical sciences TOP grant; The Netherlands genomic initiative/NWO; and the DNA-in-action consortium.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Susanne Hage for technical assistance, Marc Wold for his kind gift of the hRPA expression plasmid, and Claire Wyman for discussions.
REFERENCES
- 1.Wold M. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Iftode C, Daniely Y, Borowiec J. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 3.Zou Y, Liu Y, Wu X, Shell S. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fanning E, Klimovich V, Nager A. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles C, Edwards A, Chazin W. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 6.Lao Y, Gomes XV, Ren Y, Taylor J-S, Wold MS. Replication Protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry. 2000;39:850–859. doi: 10.1021/bi991704s. [DOI] [PubMed] [Google Scholar]
- 7.Patrick SM, Turchi JJ. Replication protein A (RPA) binding to duplex cisplatin-damaged DNA is mediated through the generation of single-stranded DNA. J. Biol. Chem. 1999;274:14972–14978. doi: 10.1074/jbc.274.21.14972. [DOI] [PubMed] [Google Scholar]
- 8.Lao Y, Lee CG, Wold MS. Replication Protein A interactions with DNA. 2. characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry. 1999;38:3974–3984. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 9.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J. Mol. Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 10.Georgaki A, Strack B, Podust V, Hubscher U. DNA unwinding activity of replication protein-A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- 11.Iftode C, Borowiec JA. 5′ ->3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39:11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 12.Kim C, Wold M. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 13.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai L, Roginskaya M, Qu Y, Yang Z, Xu Y, Zou Y. A structural characterization of human RPA sequential binding to single-stranded DNA using ssDNA as a molecular ruler. Biochemistry. 2007;46:8226. doi: 10.1021/bi7004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochkareva E, Belegu V, Korolev S, Bochkarev A. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 2001;20:612. doi: 10.1093/emboj/20.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treuner K, Eckerich C, Knippers R. Chromatin association of replication protein A. J. Biol. Chem. 1998;273:31744–31750. doi: 10.1074/jbc.273.48.31744. [DOI] [PubMed] [Google Scholar]
- 17.Georgaki A, Hubscher U. DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res. 1993;21:3659. doi: 10.1093/nar/21.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front. Biosci. 2007;12:4409–4423. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 19.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein-A- Expression, complex-formation and functional-characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 20.van Loenhout M, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: Formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 24.Strick T, Allemand J, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 25.van der Heijden T, Dekker C. Monte Carlo Simulations of Protein Assembly, Disassembly, and Linear Motion on DNA. Biophys. J. 2008;95:4560–4569. doi: 10.1529/biophysj.108.135061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strick T, Dessinges M, Charvin G, Dekker N, Allemand J, Bensimon D, Croquette V. Stretching of macromolecules and proteins. Re. Progr. Phys. 2003;66:1–45. [Google Scholar]
- 27.Lohman T, Kowalczykowski S. Kinetics and mechanism of the association of the bacteriophage T4 gene 32 (helix destabilizing) protein with single-stranded nucleic acids. Evidence for protein translocation. J. Mol. Biol. 1981;152:67. doi: 10.1016/0022-2836(81)90096-6. [DOI] [PubMed] [Google Scholar]
- 28.Kowalczykowski SC, Lonberg N, Newport JW, Vonhippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. 1. Characterization of the binding interactions. J. Mol. Biol. 1981;145:75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 29.Yakovchuk P, Protozanova E, Frank-Kamenetskii M. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubert F, Zettl H, Hafner W, Krauss G, Krausch G. Comparative thermodynamic analysis of DNA-protein interactions using surface plasmon resonance and fluorescence correlation spectroscopy. Biochemistry. 2003;42:10288–10294. doi: 10.1021/bi034033d. [DOI] [PubMed] [Google Scholar]
- 31.Iftode C, Borowiec J. 5′→3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39:11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 32.Marko J. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys. Rev. E. 2007;76:21926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 33.Koster D, Croquette V, Dekker C, Shuman S, Dekker N. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 34.Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys. Rev. Lett. 2008;100:4. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seroussi E, Lavi S. Replication protein A is the major single-stranded-DNA binding-protein detected in mammalian-cell extracts by gel retardation assays and UV- cross-linking of long and short single-stranded-DNA molecules. J. Biol. Chem. 1993;268:7147–7154. [PubMed] [Google Scholar]
- 36.Nelson P. Transport of torsional stress in DNA. Proc. Natl Acad. Sci. USA. 1999;96:14342. doi: 10.1073/pnas.96.25.14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouzine F, Liu JH, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 38.Havas K, Flaus A, Phelan M, Kingston R, Wade P, Lilley D, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 39.He Z, Henricksen L, Wold M, Ingles C. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 40.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54–DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA. 2001;98:8454. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Haber J. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.