Abstract
The engineering of new enzymes that efficiently and specifically modify DNA sequences is necessary for the development of enhanced gene therapies and genetic studies. To address this need, we developed a robust strategy for evolving site-specific recombinases with novel substrate specificities. In this system, recombinase variants are selected for activity on new substrates based on enzyme-mediated reassembly of the gene encoding β-lactamase that confers ampicillin resistance to Escherichia coli. This stringent evolution method was used to alter the specificities of catalytic domains in the context of a modular zinc finger-recombinase fusion protein. Gene reassembly was detectable over several orders of magnitude, which allowed for tunable selectivity and exceptional sensitivity. Engineered recombinases were evolved to react with sequences from the human genome with only three rounds of selection. Many of the evolved residues, selected from a randomly-mutated library, were conserved among other members of this family of recombinases. This enhanced evolution system will translate recombinase engineering and genome editing into a practical and expedient endeavor for academic, industrial and clinical applications.
INTRODUCTION
Site-specific recombinases (SSRs) have been widely employed to catalyze recombination reactions that result in the targeted addition or removal of genetic material within cellular genomes or plasmid DNA (1,2). For example, the Cre/loxP and Flp/FRT systems have been extensively used for basic genetic studies in a variety of organisms (3). More recently, these enzymes have been developed commercially into simplified strategies for laboratory molecular cloning and cell line engineering, such as the Gateway® and Flp-InTM technologies developed by Invitrogen (4,5). These naturally occurring enzymes have evolved to react with specific DNA recognition sequences present in their host organisms (1). Therefore, their use is limited to applications in which this target site has been artificially introduced into the plasmid and/or genomic DNA. The development of new DNA-modifying enzymes that can react with any desired target sequence is critical if the promise of the Genomic Revolution is to be realized. To address this need, directed evolution strategies have recently been devised to alter the substrate specificities of SSRs (6,7). These approaches aim to reprogram a SSR to react with a particular target site of interest. If successful, this work will enable the engineering of enzymes that can cut and paste DNA at any desired location in a cellular genome.
Current methods for SSR evolution are based on substrate-linked protein evolution (SLiPE). This method involves introducing a library of SSR variants into an expression plasmid that also acts as the substrate for a recombination reaction catalyzed by the expressed enzyme (7). The plasmid is introduced into a cellular host, followed by SSR expression and subsequent SSR-mediated plasmid recombination. The recombined plasmids then serve as markers for active SSR variants and are recovered based on selective PCR amplification or modulation of a fluorescent or colorimetric reporter gene (7–9). This pool of active SSR variants is then amplified and reinserted into the substrate plasmid for additional rounds of selection and evolution. By altering the sequences of the recombination target sites on the plasmid, SSR variants that are reactive with those new sequences can be selected. We and others have used this method to alter the substrate specificity profiles of several SSRs (7–13).
More recently, our work with engineered zinc finger-recombinase fusion proteins (RecZFs) has inspired an interest in developing a more stringent SLiPE system (10,14). RecZFs are modular SSRs: The catalytic domain derived from a serine recombinase (1) is fused to a synthetic zinc finger DNA-binding protein (10,14,15). The polydactyl zinc finger protein is derived from our extensive library of synthetic zinc finger motifs (16) and targets the enzymes to specific DNA sequences flanking the recombination site (Figure 1A) (10). The catalytic domain is responsible for coordinating the recombination reaction and also functions in a sequence-dependent manner on the 20 bp core of the recombination site (Figure 1A) (10). We have used these enzymes to excise a transgene from the human genome (10) and to integrate a plasmid into a specific genomic target with exceptional efficiency and specificity (14). Importantly, these studies used recombination target sites containing the 20-bp spacer sequence from the native target site of the catalytic domain. The enzymes did not catalyze recombination when the spacer sequence was altered (10,14). However, our goal is to use these enzymes to add or remove DNA at any location in the human genome via targeting with the synthetic zinc finger proteins. In order to engineer enzymes capable of this task, the sequence specificity of the catalytic domain must be broadened through directed evolution.
Figure 1.
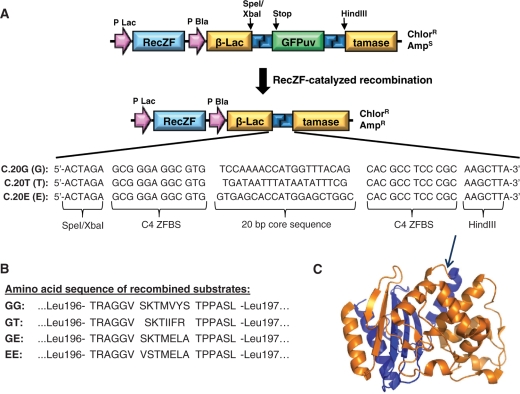
Recombinase-dependent split gene reassembly system. (A) Structure of evolution vector before and after recombinase-mediated excision of the GFPuv fragment, with recombination target sites shown in blue. SpeI and HindIII restriction sites were inserted into the β-lactamase gene between Leu196 and Leu197 for convenient insertion of a GFPuv transgene flanked by recombination sites. The structure of a RecZF target site consists of a 20 bp core sequence flanked by zinc finger-binding sites (ZFBS). The C4 ZFBS was used in all plasmids and has been described previously (14). The sequences of various 20-bp core sequences used in this study are shown, including sequences derived from the natural Gin invertase and Tn3 resolvase target sites (20G and 20T, respectively), and a 20-bp sequence derived from the promoter of ErbB2 on human chromosome 17 (20E). (B) The predicted sequences of the 19-amino acid peptide that is grafted into the reassembled β-lactamase protein after excision of GFPuv are shown for the four evolution vectors used in this study. (C) Structure of TEM-1 β-lactamase. The N-terminal fragment is shown in orange, and the C-terminal fragment is shown in blue. The arrow indicates the insertion site of the peptide encoded by the recombination site, between Leu 196 and Leu 197 in the loop connecting helices 9 and 10.
The conventional PCR-based recovery of active SSRs typically relies on selective amplification of a recombined plasmid that is resistant to restriction digest and produces a uniquely sized PCR product (7). However, we have found that unreacted substrate plasmid is capable of interfering with productive PCR amplification of rare (<10–3) recombination events. Additionally, this approach is susceptible to PCR-borne mutations and artifacts such as parasites that have confounded many of our selections. Alternatively, colorimetric and fluorescent assays (8,9) are relatively low-throughput and labor-intensive making recovery of rare active SSRs from large (>106) libraries impractical.
The goal of this work is to develop a SLiPE system that is quantitative, high-throughput, capable of recovering rare recombinants from large libraries of SSRs, and independent of PCR-based amplification methods. To achieve this goal, a SLiPE system was designed with selection based on recombinase-mediated reconstitution of TEM-1 β-lactamase that confers ampicillin resistance to bacterial hosts. A selection system based on antibiotic resistance is highly desirable, as it is theoretically possible to recover a single recombination event that occurs in a large bacterial population by culturing in the presence of antibiotic. This system is shown to be highly selective and stringent in efficiently selecting active SSRs on novel substrates. Using this system, SSRs are evolved to react with sequences derived from the human genome with only one round of mutagenesis and three rounds of selection. Additionally, comparison of the evolved variants with other members of this family of SSRs shows that this evolution system may be used to identify conserved residues critical to enzyme function and catalysis.
MATERIALS AND METHODS
Plasmids
The split gene reassembly vector was derived from pBC SK(–) (Stratagene), a derivative of the pBluescript II phagemid in which the ampicillin resistance gene has been replaced with the chloramphenicol resistance gene. The gene encoding TEM-1 β-lactamase, including the pBla promoter, was amplified in two fragments from pcDNA3.1 (Invitrogen), which were subsequently fused by overlap PCR such that SpeI and HindIII restriction sites were inserted between the codons for Leu196 and Leu197. The 5′ fragment was amplified with primers 5′-XbaI pBla and 3′-AmpR mid-Spe-Hind. The 3′ fragment was amplified with primers 5′-AmpR mid-Spe-Hind and 3′-KpnI AmpR. The products from these two PCR reactions were subsequently used as the template in a PCR reaction with 5′-XbaI pBla and 3′-KpnI AmpR. An extra adenine nucleotide was added 3′ of the HindIII site so that the reading frame of the gene would be intact following insertion of a 44 bp recombination site between SpeI and HindIII (Figure 1A). This final PCR product was ligated into the XbaI and KpnI sites of pBC SK(–). Additionally, the SS stuffer (17) was ligated into the SacI and XbaI sites of this new vector to create pBCS-Bla.
Recombination target sites were generated as previously described (10). Briefly, the gene encoding GFPuv, a brighter variant of GFP, was amplified by PCR with primers containing recombination sites at both 5′ and 3′ ends. Recombination sites consisted of 12 bp C4 zinc finger target sites flanking a 20 bp spacer that is the core sequence of the target site (Figure 1A) (14). For example, the ‘GE’ evolution vector was generated by amplifying GFPuv with 5′-XbaI C4-20G-GFP and 3′-HindIII C4-20E-GFP. This PCR product was then digested with XbaI and HindIII and ligated into the SpeI and HindIII sites of pBCS-Bla to create pBCS-Bla GE.
Primer sequences are provided in the Supplementary Data.
Recombination assays
The genes for GinC4, GinC3 and GinC2 were amplified by PCR from existing mammalian expression vectors (14) with primers ResGin-cat fo1 prim1 and 3′ZF SS-AXEX prim2 in order to add a 5′ SacI site and Shine-Dalgarno sequence and a 3′ XbaI site. This PCR product was then ligated into the SacI and XbaI sites of a digested and CIP-treated pBCS-Bla with the indicated pair of recombination sites (GG, GT, GE or EE). Ligations were ethanol precipitated and transformed by electroporation into TOP10F′ Escherichia coli (Invitrogen) as described in published protocols (17). After 1 h of recovery in SOC medium, cultures were plated immediately onto LB agar with the indicated antibiotics or diluted with 100 ml of Super Broth, cultured at 37°C and 225 rpm, and plated at subsequent time points at appropriate dilutions for counting individual colonies. Transformation efficiencies were typically ∼5 × 106 colonies per microgram of vector DNA. Recombination activity was calculated as the number of colonies growing on LB agar plates with chloramphenicol (30 μg/ml) and carbenicillin (100 μg/ml), an ampicillin analog, divided by the number of colonies growing on plates with only chloramphenicol (CarbR/ChlorR). Colony number was determined by automated counting using a GelDoc XR imaging system with Quantity One 1-D analysis software (Bio-Rad).
Directed evolution
Libraries of RecZF variants were generated as previously described (10,18). The hyperactive Gin catalytic domain was amplified by error-prone PCR with dNTP analogs dPTP and 8-oxo-dGTP under conditions that generated approximately three amino acid substitutions per gene. The library of Gin variants was then fused to an error-free C4 zinc finger domain (14) by overlap PCR. The PCR product was digested with SacI and XbaI and ligated into the indicated split gene reassembly vector. Ligations were transformed as described above. Library size was determined by transformation efficiency, which was typically ∼5 × 106 CFU. After 16 h of culture with chloramphenicol, plasmid DNA was purified by miniprep (Invitrogen) of a 5 ml sample of the 100 ml culture. One microgram of this plasmid DNA was then retransformed into E. coli, as described above, and cultured in chloramphenicol and carbenicillin. Dilutions of this transformation were plated onto LB agar with the indicated antibiotics to determine recombination activity of the library population. Following 16 h of culture in chloramphenicol and carbenicillin, the plasmid DNA was purified from the 100 ml culture by maxiprep (Invitrogen). Thirty micrograms of this plasmid DNA was then digested with SacI and XbaI to recover the active RecZFs from the recombined carbenicillin-resistant plasmids by agarose gel electrophoresis and DNA purification (Qiagen). This recovered RecZF population was then re-ligated into the evolution vector for the next round of selection.
Data analysis and statistics
Data are presented from representative experiments as the mean of triplicate samples ± standard error of the mean (mean ± SEM). Replicates represent independent ligations and transformations. Statistical analyses included one-way ANOVA (Figure 2) or two-way ANOVA (Figure 5) accounting for both recombination substrate (vector) and RecZF variant (enzyme). Analyses were performed with Microsoft Office Excel 2007 with alpha = 0.05. Sequence alignments were performed with software packages including Vector NTI (Invitrogen) and ClustalX with the Jalview alignment editor.
Figure 2.
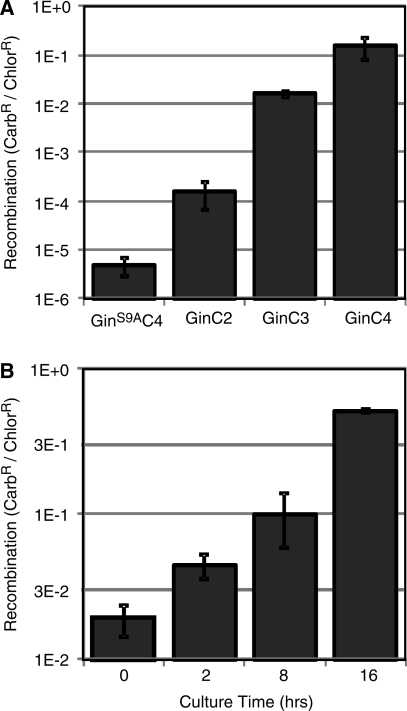
Gene reassembly of an evolution vector containing two C.20G sites (GG) by RecZFs containing a Gin catalytic domain was measured as the fraction of carbenicillin-resistant E. coli colonies (carbenicillin is an ampicillin analogue). (A) Recombination activity was dependent on the number of zinc finger domains on the RecZF, with four fingers showing more activity than two or three (P < 9E–7). Carbenicillin resistance in the presence of the catalytically inactive GinS9AC4 mutant represents the background level of recombination in this system. (B) Recombination activity by the RecZF with four fingers, GinC4, was also dependent on time in culture (P < 7E–5).
Figure 5.
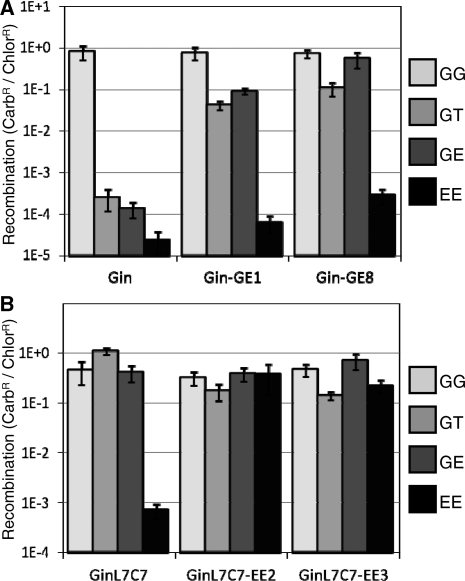
Recombination activity of the parent and evolved RecZF variants on diverse substrate sequences. The non-mutated starting clone and two unique variants from round 3 output libraries of (A) Gin selected on GE (variants GE1 & GE8) and (B) GinL7C7 selected on EE (variants EE2 & EE3) were tested for activity on vectors containing recombination sites of combinations of natural and unnatural core sequences. P-values with respect to vector & enzyme are (A) P < 4E–15, P < 2E-10 and (B) P < 6E–6, P < 3E–3.
RESULTS
In order to create a system in which recombination activity was linked to antibiotic resistance, we first tested a variety of strategies in which the gene encoding β-lactamase was separated from its promoter. However, attempts to design a plasmid with an intact β-lactamase coding sequence were unsuccessful in producing ampicillin-sensitive E. coli, presumably due to leaky expression levels of the efficient enzyme. Therefore, we pursued a split gene reassembly strategy, in which the coding sequence for β-lactamase was interrupted by a stuffer fragment encoding a GFPuv transgene flanked by recombination target sites for the zinc finger-recombinase fusion proteins (Figure 1A). This stuffer was designed to interrupt the expression of β-lactamase, rendering the E. coli host sensitive to ampicillin. When the appropriate RecZF is expressed from this plasmid, however, the enzyme will catalyze recombination at its target sites, resulting in excision of the GFPuv stuffer. This restores the β-lactamase coding sequence, and ampicillin resistance of the host bacterium. Importantly, a recombination target site (57 bp, including flanking restriction digest sites) will remain within the reassembled β-lactamase gene, resulting in a 19-amino acid peptide grafted within the β-lactamase protein (Figure 1B). The location of the insertion site of the stuffer fragment in the β-lactamase gene is critical as the grafted peptide cannot disrupt expression or activity of the β-lactamase enzyme. Therefore, we inserted the stuffer fragment between Leu 196 and Leu 197 in the loop connecting helices 9 and 10 (Figure 1C). This site has been used previously for reconstructing the β-lactamase enzyme without significantly disrupting function (19–21).
The split gene reassembly system was first characterized with the previously described GinC2, GinC3 and GinC4 RecZFs and the C.20G recombination site, consisting of the 20 bp spacer sequence from the native target site of the Gin invertase flanked by C4 zinc finger binding sites (Figure 1A) (14). These RecZFs are comprised of the catalytic domain of a hyperactive Gin invertase from bacteriophage Mu fused to 2, 3 or 4 zinc finger domains that recognize 6, 9 or 12 bp of DNA, respectively. DNA encoding each RecZF was ligated into an evolution vector containing C.20G target sites on both sides of GFPuv (GG), transformed into E. coli, and cultured for 16 h with chloramphenicol. To determine the extent of recombination, plasmid was purified from these cultures, retransformed into E. coli, and plated onto agar containing only chloramphenicol or agar containing chloramphenicol and carbenicillin. The level of RecZF-mediated recombination was measured as the fraction of carbenicillin-resistant colonies. The level of recombination increased with the number of zinc finger motifs due to enhanced DNA-binding affinity and specificity (Figure 2A). Maximal activity with the GinC4 enzyme corresponded to 15.1 ± 7.1% recombined plasmids, representing greater than four orders of magnitude increase over background recombination observed with the catalytically inactive GinS9AC4 mutant. This large dynamic range of quantifiable enzyme activity indicated that this system is well suited for directed evolution of SSR activity.
The extent of recombination was also monitored over time (Figure 2B). Ligations of GinC4 into the C.20G vector were transformed into E. coli. At the indicated time points, samples from the cultures were diluted and plated onto agar with antibiotics. Cultures that were plated immediately following transformation had 1.90 ± 0.47% carbenicillin-resistant colonies, representing the cells in which recombination occurred more rapidly than antibiotic-induced cell death. After 16 h of culture, the fraction of carbenicillin-resistant cells was 50.9 ± 1.8%. Importantly, these resistant cells contain a mixture of recombined and unrecombined plasmids, demonstrated by purifying the plasmid DNA, retransforming bacteria to redistribute the plasmids into individual cells and plating directly onto carbenicillin. These retransformed samples showed only 15.1% recombination (Figure 2A).
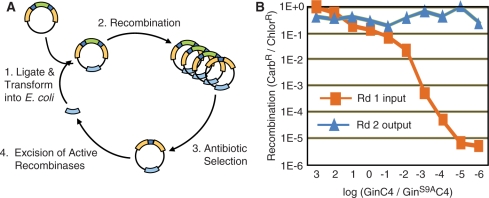
In order to test the sensitivity of this selection method in recovering active variants from a pool of RecZFs, C.20G vector containing the GinC4 enzyme was diluted into C.20G vector containing the inactive GinS9AC4 mutant. These mixtures then underwent two rounds of selection in the split gene reassembly system (Figure 3). Each round consisted of (i) transforming the mixture into E. coli, (ii) culturing for 16 hrs with chloramphenicol to allow recombination to occur, (iii) purifying and retransforming the plasmid for overnight culture in chloramphenicol and carbenicillin to select for recombined plasmids, and (iv) purification of selected plasmid DNA (Figure 3A). RecZF fragments were digested from these samples and inserted into a fresh evolution vector to begin the next round of selection. For each round, dilutions of the transformations were plated onto agar with the indicated antibiotics to determine the extent of recombined plasmids in the mixture. Prior to carbenicillin selection in Round 1, serial dilutions of GinC4 into GinS9AC4 from 103:1 to 1:106 showed a decrease in carbenicillin-resistant colonies of over five orders of magnitude (Figure 3B). However, after two rounds of selection all of the original dilutions reached the same level of activity as pure GinC4, demonstrating the successful enrichment of one GinC4-containing plasmid per 106 GinS9AC4-containing plasmids. Parallel controls of solely GinS9AC4 showed no increase in activity over two rounds of evolution. This high level of selectivity demonstrated the efficiency of the split gene reassembly system.
Figure 3.
(A) Cyclical selection strategy for the directed evolution of SSRs by split gene reassembly. (B) The selectivity and detection limit of the evolution system were determined by diluting GinC4-containing plasmid into catalytically inactive GinS9AC4 plasmid and performing two rounds of selection. The split gene reassembly system was capable of recovering one active GinC4 enzyme per 106 inactive GinS9AC4 enzymes.
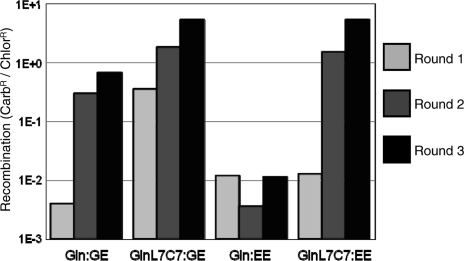
The split gene reassembly system was tested for its ability to select evolved RecZF catalytic domains that react with sequences present in the human genome. We have previously targeted the ErbB2 locus on chromosome 17 with artificial zinc finger proteins (22,23). A 20 bp sequence from the ErbB2 promoter (20E) was chosen due to its overlap with the previously targeted sites and presence of an AT core dinucleotide sequence that is compatible with recombination of the 20G and 20T sequences used in our previous work (Figure 1A) (10). Evolution vectors were created containing either 20G sites (GG), 20E sites (EE) or a combination of sites (GT, GE) flanking the GFPuv transgene. Ligation of GinC4 into these vectors led to high levels of activity on the GG substrate and background levels of recombination on any substrate containing an unnatural core sequence, as demonstrated previously (Figure 5A) (10,14). The GinL7C7 catalytic domain is a Gin variant that had previously been evolved to react with substrates containing one natural (20G) and one unnatural (non-20G) core sequence (10). When fused to the C4 zinc finger domain, this enzyme reacted efficiently with the three vectors containing a 20G sequence, but did not react with the EE substrate (Figure 5B). For directed evolution of enzymes that react with the 20E sequence, both catalytic domains were amplified by error-prone PCR such that approximately three amino acid substitutions were incorporated per domain. The libraries of catalytic domains were attached to the non-mutated C4 zinc finger protein and underwent three rounds of evolution with the GE or EE substrates (Figure 4). The activity of each library, with the exception of Gin on the EE substrate, increased several orders of magnitude over the three rounds. The inability of Gin to directly evolve for reactivity with two unnatural substrates (EE) indicates that an intermediate evolution on only one unnatural sequence may be necessary to obtain a domain that can react on two unnatural core sequences. This is supported by the success of GinL7C7, which was previously engineered to react with the GT substrate (10), to evolve for reactivity with EE in this study. The majority of variants of GinL7C7 selected on the GE substrate did not show any amino acid substitutions after three rounds, as expected. Individual variants from the round 3 outputs of Gin selected on the GE substrate and GinL7C7 selected on the EE substrate were independently characterized (sequences of variants available in Supplementary Data). Gin variants evolved on GE showed enhanced activity profiles, including relaxed specificity on substrates containing one natural core sequence, but background levels of recombination on the EE substrate (Figure 5A). In contrast, GinL7C7 clones demonstrated high levels of activity on all substrates, suggesting that sequence specificity of these domains had been lost (Figure 5B). Selected mutations were distributed throughout the domain and were similar to dominant mutations observed in previously evolved RecZFs (Figure 6, Supplementary Figures S1 and S2) (10). The evolved catalytic domains showed between 100- and 1000-fold enhanced activities on alternative target sequences.
Figure 4.
Directed evolution of recombinase specificity by split gene reassembly. Libraries of Gin and GinL7C7 variants underwent three rounds of selection with evolution vectors containing 20G and 20E (GE) or two 20E (EE) recombination sites. Activity of the library on the corresponding substrate increased with each round, except for Gin on the EE plasmid (Gin:EE).
Figure 6.
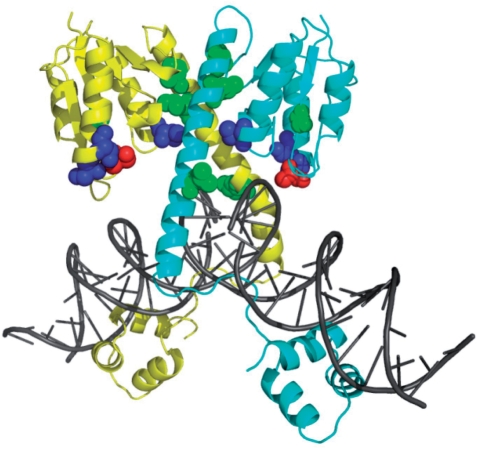
Residues evolved to conserved mutations from Gin:GE (blue; N11, N14, M70) and GinL7C7:EE (green; V6, F104, M108, I120) evolutions mapped onto the structure of a DNA-bound γδ resolvase dimer (yellow and cyan) (32). The catalytic serine (S9) is highlighted in red and DNA depicted in gray.
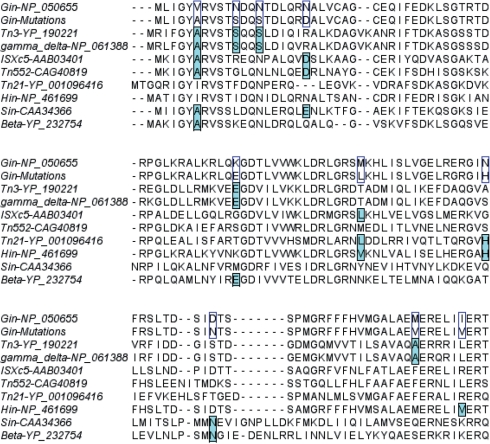
The utility of the split gene reassembly system is also supported by the nature of the mutations selected in the evolved variants. Although a relatively small number of evolved clones were sequenced, we noticed that many of the selected mutations were conserved among other members of the serine recombinase family (Figure 7). For example, three of the eight unique GinL7C7 variants selected to recombine the EE substrate contained a V6A mutation (Supplementary Figure S2). Notably, alanine is conserved at this position in almost all of the other highly homologous serine recombinases (Figure 7). Similarly, N11S and N14S were included in 13 of the 30 sequences recovered from the evolutions of Gin on the GE substrate, comprising two of the nine unique sequences (Supplementary Figure S1). Serine is also present at both of these positions in the highly homologous Tn3 and gamma-delta serine recombinases (Figure 7). Finally, four of the nine unique variants and 17 of the 30 total sequences recovered from the evolutions of Gin on the GE substrate contained a mutation of M70 to either valine or leucine (Supplementary Figure S1). Valine or leucine is also present at this position in three of the eight other family members included in this analysis (Figure 7). Other conserved mutations are summarized in Supplementary Table S2. The ability to recover highly conserved mutations from the randomly mutated gene sequences used in this study underscores the effectiveness of this evolution method.
Figure 7.
Selected mutations are conserved among the family of serine recombinases. The name of the recombinase and associated accession number are shown on the left. The top row shows the wild-type Gin sequence and the second row shows selected mutations in this study (boxed) that share homology to other family members at that position (shaded).
DISCUSSION
The Genomic Revolution has provided scientists with the information necessary to understand the relationships of DNA structure and sequence to molecular, cellular and organismal processes. This includes the cause-and-effect relationship that genome sequence shares with genetic diseases and the mechanisms of tissue regeneration. However, we currently do not have the tools necessary for capitalizing on this powerful information. In particular, methods that allow for the precise, efficient and economical restructuring of genomes and other DNA sequences would enable a new realm of possibilities in genetic medicine and research. SSR engineering represents a promising approach to this challenge, but current methods are laborious and often unsuccessful. For example, a recent breakthrough in SSR evolution demonstrated the evolution of a Cre recombinase variant capable of excising HIV proviral DNA from the human genome (11). However, this proof-of-principle study required 126 rounds of evolution, representing several years of continuous work. Clearly, this is not an effective means of routine protein engineering for commercial biotechnology or medicine. Nevertheless, this work has demonstrated the tremendous utility of successful SSR engineering and the need for methods that will enhance this approach.
In this study, we have established and validated a novel strategy for selecting unique DNA-modifying enzymes. Using one round of mutagenesis and three rounds of selection, we isolated RecZFs reactive with non-native sequences. This is in contrast to our previous results with conventional SLiPE, which required at least twice as many rounds of selection, as well as multiple rounds of DNA shuffling that were not necessary in the current study (10). These results indicate that the split gene reassembly system is several orders of magnitude more selective in the recovery of active SSRs than conventional SLiPE methods. Additionally, the quantitative nature of the split gene reassembly system allows for straightforward measurement of library enrichment and comparison of activity among SSR variants.
The evolutions performed in this study have generated catalytic domains that can act on exclusively unnatural substrates. This is in contrast to our previous work which developed catalytic domains capable of reaction with one natural and one unnatural substrate sequence (10). Consequently, the specificity of the new enzymes is determined solely by the zinc finger-based DNA-binding domain. These new sequence-independent catalytic domains can now be attached to zinc finger proteins capable of targeting single sites in a cellular genome (22) to allow for user-specified genome editing. We anticipate that this will lead to similar levels of success as seen with the zinc finger nucleases that are built from the sequence-independent catalytic domain of the FokI restriction endonuclease (24,25). These enzymes have been used extensively for genome editing in a variety of organisms and human cell types, and are currently being tested in a phase I clinical trial (26,27). A remaining challenge is to evolve catalytic domains with novel substrate specificities, as opposed to the relaxed specificity developed in this study. This may require more sophisticated library design and greater understanding of the structural determinants of specificity in the serine recombinases.
We anticipate that this system will also enable the efficient re-engineering of other common SSRs, such as Cre and Flp, which have also been the subject of several directed evolution studies (6–9,12,13,28). Similarly, the evolution of integrases (29) and transposases (30) would benefit from this approach. It is also important to note that our studies have suggested that RecZFs are more easily reengineered to act on new substrates relative to published work with the more commonly used tyrosine recombinases, Cre and Flp, as mentioned above. We hypothesize that this is attributable to the modular structure of RecZFs, in which the catalytic domain and DNA-binding domain are functionally and structurally distinct. This is in contrast to the tyrosine recombinases, in which these domains are overlapping (1). This unique feature of RecZFs may constitute a distinct advantage as the field of SSR engineering progresses. This also suggests that the split gene reassembly system may show even greater benefit to the more challenging task of evolving tyrosine recombinases.
The conserved mutations identified in this study suggest potential residues that may be critical for recombination and sequence recognition. This is strengthened by the conservation of many selected mutations with other serine recombinases. Recently, Olorunniji and Stark evaluated several conserved residues in this family of enzymes for their role in catalysis by the Tn3 resolvase (31). None of the residues analyzed in their study were mutated in our evolved variants. Therefore our work has identified a new set of residues that are clearly important in regulating catalysis and sequence specificity by serine recombinases. Future studies that focus on these new positions may provide greater insight into the regulatory mechanisms of site-specific recombination. Additionally, mutagenesis targeted to these positions may be an effective means of reprogramming activity of many members of the serine recombinase family. This work may ultimately lead to the ability to engineer novel SSRs by rational design.
Finally, the utility of this evolution system is supported by the selection of the specific conserved mutations from a library of random mutations across the whole domain. The Gin catalytic domain used in these enzymes consists of 142 amino acids. Therefore there are ∼2.7 × 103 possible protein variants containing one mutation, 7.3 × 106 variants containing two mutations, and ∼2 × 1010 variants with three mutations, as was the average mutation rate per domain in this study. Importantly, the majority of random mutations are expected to be destabilizing and negatively affect protein function and possibly counteract activating mutations. Consequently the ability to specifically isolate variants that contain mutations corresponding to conserved residues across this class of enzymes suggests an extremely high level of selectivity, especially considering the initial library sizes in this study were ∼5 × 106. This also suggests that the selective pressures applied in this evolution system mirror the natural pressures that these enzymes have encountered while performing their native functions. In summary, the efficient and straightforward selection of these critical mutations is a testament to the robust stringency and high-throughput capacity of the approach.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (CA126664 and GM065059 to C.F.B.); the Skaggs Institute for Chemical Biology and a National Institutes of Health Postdoctoral Fellowship (CA125910 to C.A.G.). Funding for open access charge: Internal funding.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Akopian A, Marshall Stark W. Site-specific DNA recombinases as instruments for genomic surgery. Adv. Genet. 2005;55:1–23. doi: 10.1016/S0065-2660(05)55001-6. [DOI] [PubMed] [Google Scholar]
- 3.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 4.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Lakshmipathy U, Ozgenc A, Thyagarajan B, Lieu P, Fontes A, Xue H, Scheyhing K, MacArthur C, Chesnut JD. hESC engineering by integrase-mediated chromosomal targeting. Methods Mol. Biol. 584:229–268. doi: 10.1007/978-1-60761-369-5_13. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 8.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc. Natl Acad. Sci. USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voziyanov Y, Stewart AF, Jayaram M. A dual reporter screening system identifies the amino acid at position 82 in Flp site-specific recombinase as a determinant for target specificity. Nucleic Acids Res. 2002;30:1656–1663. doi: 10.1093/nar/30.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordley RM, Smith JD, Graslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 12.Bolusani S, Ma CH, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucleic Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J. Mol. Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 14.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandell JG, Barbas CF., 3rd Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbas CF, 3rd, Burton DR, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 18.Zaccolo M, Gherardi E. The effect of high-frequency random mutagenesis on in vitro protein evolution: a study on TEM-1 beta-lactamase. J. Mol. Biol. 1999;285:775–783. doi: 10.1006/jmbi.1998.2262. [DOI] [PubMed] [Google Scholar]
- 19.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 20.Vandevenne M, Filee P, Scarafone N, Cloes B, Gaspard G, Yilmaz N, Dumoulin M, Francois JM, Frere JM, Galleni M. The Bacillus licheniformis BlaP beta-lactamase as a model protein scaffold to study the insertion of protein fragments. Protein Sci. 2007;16:2260–2271. doi: 10.1110/ps.072912407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi AT, Stains CI, Ghosh I, Segal DJ. Sequence-enabled reassembly of beta-lactamase (SEER-LAC): a sensitive method for the detection of double-stranded DNA. Biochemistry. 2006;45:3620–3625. doi: 10.1021/bi0517032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl Acad. Sci. USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 26.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alper J. One-off therapy for HIV. Nat. Biotechnol. 2009;27:300. doi: 10.1038/nbt0409-300. [DOI] [PubMed] [Google Scholar]
- 28.Konieczka JH, Paek A, Jayaram M, Voziyanov Y. Recombination of hybrid target sites by binary combinations of Flp variants: mutations that foster interprotomer collaboration and enlarge substrate tolerance. J. Mol. Biol. 2004;339:365–378. doi: 10.1016/j.jmb.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 29.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29:5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 31.Olorunniji FJ, Stark WM. The catalytic residues of Tn3 resolvase. Nucleic Acids Res. 2009;37:7590–7602. doi: 10.1093/nar/gkp797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.