Abstract
DNA methylation is an epigenetic modification that plays an important role in gene regulation. It can be influenced by stochastic events, environmental factors and developmental programs. However, little is known about the natural variation of gene-specific methylation patterns. In this study, we performed quantitative methylation analyses of six differentially methylated imprinted genes (H19, MEG3, LIT1, NESP55, PEG3 and SNRPN), one hypermethylated pluripotency gene (OCT4) and one hypomethylated tumor suppressor gene (APC) in chorionic villus, fetal and adult cortex, and adult blood samples. Both average methylation level and range of methylation variation depended on the gene locus, tissue type and/or developmental stage. We found considerable variability of functionally important methylation patterns among unrelated healthy individuals and a trend toward more similar methylation levels in monozygotic twins than in dizygotic twins. Imprinted genes showed relatively little methylation changes associated with aging in individuals who are >25 years. The relative differences in methylation among neighboring CpGs in the generally hypomethylated APC promoter may not only reflect stochastic fluctuations but also depend on the tissue type. Our results are consistent with the view that most methylation variation may arise after fertilization, leading to epigenetic mosaicism.
INTRODUCTION
It is becoming increasingly clear that DNA sequence variations and environmental factors alone cannot account for the phenotypic differences among individuals. Studies in genetically identical organisms, i.e. monozygotic twins, isogenic animal strains and cloned animals implicated non-DNA sequence and non-environment-based effects as a main source of phenotypic variation (1–4). After genes and environment, epigenetic factors, in particular DNA methylation, may form the molecular basis of this ‘third component’. Much of the existing DNA methylation variation may result from stochastic events during epigenetic reprogramming in gametogenesis, early embryogenesis and later developmental processes. This spontaneous epimutation rate can be modified by genetic and environmental factors (5–7). It is estimated that epimutations occur 100 times more frequently than somatic DNA mutations (8,9).
Epigenetic information is not encoded by the DNA sequence itself but by reversible modifications of DNA and/or histones, that can be transmitted from cells to daughter cells and sometimes even from one generation to another (7,10). Methylation of the 5′-cytosine residues in CpG dinucleotides directly targets DNA. It is associated with posttranslational histone modifications that lead to a locally condensed inactive chromatin structure (11,12). One important component of epigenetic gene regulation are CpG islands (CGIs) that are 500–2000 bp long and associated with promoters in most mammalian genes. Most CpGs in promoters are protected from methylation in somatic tissues. Promoter methylation leads to stable gene silencing during development/differentiation and in disease processes (13–15). For example, pluripotency genes switch from a demethylated and transcriptionally active state in embryonic cells to a fully methylated repressed state in somatic cells (16,17). In contrast, tumor suppressor genes are demethylated and active in somatic cells; ectopic methylation begins early in tumorigenesis (18–20). Genomic imprinting is a parent-specific epigenetic modification in which allele-specific expression depends on male versus female germline transmission. The differentially methylated regions (DMRs) of imprinted genes are involved in the control of parental allele-specific gene activity (21,22).
Genomic DNA methylation patterns may exhibit considerable variation among human individuals, as well as intraindividual changes over time (23–26). Large-scale epigenome mapping revealed that CpG-rich regions exhibit substantially less interindividual variation of DNA methylation patterns than CpG-poor regions. Most CGIs showed relatively similar low methylation levels in all individuals, with a few methylated CpGs randomly distributed among the many unmethylated CpGs (27). This is consistent with the view that the density of methylated CpGs in a cis-regulatory region (rather than individual CpGs, i.e. in transcription factor binding sites) turns a gene ‘on’ or ‘off’ (15,28). It is well-known that epigenetic gene silencing during tumor initiation and progression involves methylation of entire CGIs but not single critical CpGs (18–20). It is plausible to assume that epigenetic variation contributes significantly to both phenotypic variation among individuals and human disease (7,14,29). However, in order to associate unexpected methylation levels of developmentally important genes with disease processes, it is crucial to assess the normal range of methylation variation. Using quantitative methylation assays, we measured the methylation levels in cis-regulatory regions of imprinted, pluripotency and tumor suppressor genes in different tissues and/or developmental states.
MATERIALS AND METHODS
Tissue samples and DNA preparation
Blood samples were obtained with informed consent from 12 healthy monozygotic and 14 dizygotic twin pairs as well as from unrelated individuals. Chorionic villus samples (CVS) of 48 term placentas of healthy newborns were obtained with informed consent of the mothers from the Department of Gynecology and Obstetrics, Mainz University. Sperm samples of 45 males with normal spermiograms (according to WHO guidelines) were obtained with informed consent of the donors from the Fertility Center Wiesbaden.
In addition, we analyzed native chorionic villi from 29 first-trimester spontaneous abortions and 12 cultured CVS. Ten fetal brain samples (frontal cortex) were obtained from spontaneous abortions and stillbirths that underwent pathomorphological examination at the Department of Paediatric Pathology, Mainz University. Following diagnostics, the excess samples were made anonymous. Placental and fetal tissues were taken only from pregnancies without detectable chromosome and fetal abnormalities. Tissues were dissected within 24 h after abortion/stillbirth and stored at −80°C until further analysis. Adult brain samples from 12 humans between 24 and 83 years of age were obtained from the Institute of Legal Medicine and the Department of Neuropathology, Mainz University. Causes of death reflected accidents and a wide spectrum of common diseases (e.g. cardiovascular disease). Medical history and histological examination revealed no evidence for hereditary brain disorders. After removal of meninges frontal cortex tissue (area A10) was dissected between 1–2 days postmortem and immediately frozen and stored at −80°C. Use of anonymized excess tissue materials (from pathomorphological diagnostics) for scientific analyses was approved by the local ethics committee [Aerztekammer Rheinland-Pfalz, Decisions No. 837.103.04 (4261) and 837.073.07 (5608)].
The DNeasy Blood and Tissue Kit (Qiagen, Hilden Germany) was used for genomic DNA isolation. Sperm samples were first purified with Pure Sperm 40/80 (Nidacon, Molndal, Sweden) and and then treated with 100 mM Tris–Cl, 10 mM EDTA, 500 mM NaCl, 1% SDS and 2% β-mercaptoethanol. After incubation for 2 h at 56°C with proteinase K, DNA was prepared, as described above.
Studied genes
The DMRs of two paternally methylated (H19 and MEG3) and four maternally methylated (LIT1, NESP55, PEG3 and SNRPN) genes as well as the promoters of the pluripotency gene OCT4 and the tumor suppressor gene APC were analyzed by bisulfite pyrosequencing. Amplicons targeting methylation-dependent cis-regulatory regions of these well-studied genes were chosen from the literature (30–35). Although the gene-specific pyrosequencing assays are based on only a small number (2–7) of CpGs per gene, the analyzed sites are thought to be representative of a given DMR or promoter. Individual CpGs in larger CGIs cannot stably maintain methylation states that differ from those of the neighboring CpGs. Usually, the entire CGI is either methylated or demethylated (27,28). PCR and sequencing primers (Table 1) were designed with the Pyrosequencing Assay Design Software (Biotage). A single nucleotide polymorphism (SNP), rs2107425, with approximately 50% heterozygosity is located at the 4th base from 5′-end of the nested reverse primer for H19. In previously published methylation analyses (30) and also in our own experience, this A to G change did not cause a biased amplification of DNA methylation. The binding sites for the other studied genes do not contain known SNPs. Although we cannot entirely exclude that the one or other analyzed sample displays a novel allelic sequence variation, such rare variants, if any, cannot explain the observed methylation variation of imprinted genes.
Table 1.
Genes and primers for pyrosequencing
| Gene | Primer | Sequence (5′–3′) | Amplicon length (bp) | Chromosomal localization (bp) | Number of CpGs | Reference |
|---|---|---|---|---|---|---|
| H19 | Outer forward | TTTTTGGTAGGTATAGAGTT | 231 | Chromosome 11 1 977 647-1 977 878 | 4 | El-Maarri et al. (30) |
| Outer reverse | AAACCATAACACTAAAACCC | |||||
| Nested forward | TGTATAGTATATGGGTATTTTTGGAGGTTT | |||||
| Nested reverse* | TCCTATAAATATCCTATTCCCAAATAACC | |||||
| Sequencing | TGGTTGTAGTTGTGGAAT | |||||
| MEG3 | Forward | GATTTTTTTTATATATTGTGTTTG | 220 | Chromosome 14 100 361 907-100 362 129 | 3 | Zechner et al. (35) |
| Reverse* | CTCATTTCTCTAAAAATAATTAACC | |||||
| Sequencing | GTGTTTGAATTTATTTTGTTTGG | |||||
| LIT1 | Forward | AATTAGTAGGTGGGGGG | 122 | Chromosome 11 2 677 751-2 677 873 | 2 | Mackay et al. (31) |
| Reverse* | CTAAAAAACTCCCTAAAAATC | |||||
| Sequencing | GGGGGTAGTYGGAG | |||||
| NESP55 | Outer forward | TTTTTTATTTTATAGGGTGTATTT | 343 | Chromosome 20 56 848 506-56 848 849 | 3 | El-Maarri et al. (30) |
| Outer reverse | AAAATAAAATACTTAAACACCAC | |||||
| Nested forward | TTTTTGTAGAGTTAGAGGGTAGGT | |||||
| Nested reverse* | AAAAAAAACAACTCAAAATCTACC | |||||
| Sequencing | GTGTTTAAGAGGATGGAT | |||||
| PEG3 | Forward | GGTGTAGAAGTTTGGGTAGTTG | 153 | Chromosome 19 62 043 756-62 043 909 | 4 | Mackay et al. (31) |
| Reverse* | CTCACCTCACCTCAATACTAC | |||||
| Sequencing | TGTTTATTTTGGGTTGGT | |||||
| SNRPN | Forward* | AGGGAGTTGGGATTTTTGTATT | 237 | Chromosome 15 22 751 105-22 751 342 | White et al. (32) | |
| Reverse | CCCAAACTATCTCTTAAAAAAAAC | 2 × 3 = 6 | ||||
| Sequencing 1 | ACACAACTAACCTTACCC | 3 | ||||
| Sequencing 2 | CCAACCTACCTCTAC | 3 | ||||
| OCT4 | Forward | AAGTTTTTGTGGGGGATTTGTAT | 185 | Chromosome 6 31 246 461-31 246 646 | 2 | Deb-Rinker et al. (33) |
| Reverse* | CCACCCACTAACCTTAACCTCTA | |||||
| Sequencing | TGAGGTTTTGGAGGG | |||||
| APC | Forward* | GGTTAGGGTTAGGTAGGTTGT | 193 | Chromosome 5 112 101 274-112 101 467 | 7 | Schatz et al. (34) |
| Reverse | ACTACACCAATACAACCACATATC | |||||
| Sequencing | CCACACCCAACCAA |
This table provides the primer sequences for bisulfite pyrosequencing of our studied genes, the length and chromosomal localization (according to ensembl 54, May 2009) of the amplified segments, and the number of CpG sites in the amplicons. Biotinylated primers are indicated by star symbols.
Imprinting of the paternally expressed IGF2 gene and the maternally expressed non-protein-coding H19 gene on chromosome 11p15 is regulated by differentially methylated CCCTC binding factor sites in the 5′-region of H19 (36). Methylation disturbances in the H19 germline DMR lead to dysregulation of IGF2-H19 imprinting and intrauterine growth retardation (Silver–Russell syndrome) (37) and overgrowth (Beckwith–Wiedemann syndrome) (38), respectively. The germline DMR within intron 10 of the KCNQ1 gene regulates expression of the paternally expressed KCNQ1OT1 (LIT1) transcript and the maternally expressed CDKN1C gene on chromosome 11p15. Epimutations in this LIT1 DMR are also associated with Beckwith–Wiedemann syndrome (39). The methylation status of the germline DMR in the SNRPN promoter/exon 1 is important for imprint establishment/maintenance of genes in the Prader-Willi/Angelman syndrome region (40). The PEG3 gene on chromosome 19q13.4 is paternally expressed in embryo and placenta and can induce apoptosis. It has a DMR in its promoter region (41). The maternally expressed non-coding MEG3 RNA represents a growth suppressor. Hypomethylation of the MEG3 promoter on chromosome 14q32.2 is associated with low-birth weight, muscular hypotonia and various dysmorphisms (35). The GNAS locus on chromosome 20q13.3 has a highly complex imprinting pattern. Its different products (Gsalpha, XLalphas and NESP55) are involved in early postnatal adaptions and neuroendocrine functions. NESP55 is expressed from the maternal chromosome in restricted brain areas (42).
The transcription factor OCT4 (POU5F1) is the key gene for maintaining pluripotency in mammalian cells (43). It is highly expressed in human oocytes, down-regulated in early embryos, and then expressed de novo from the embryonic genome in blastocysts. Differentiation of the somatic tissues is associated with downregulation of OCT4 (44). Germline mutations in the APC tumor suppressor gene cause familial adenomatous polyposis, a hereditary cancer syndrome. Somatic inactivation of APC by promoter hypermethylation is frequently seen in sporadic colorectal cancers (18).
Bisulfite pyrosequencing
Bisulfite treatment of genomic DNA was performed using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany). The PCR reaction mixture consisted of 2.5 µl 10× PCR buffer, 2.5 µl 50 mM MgCl2, 2.5 µl 10 mM dNTP mix, 1.0 µl (100 ng) of each forward and reverse primer, 0.5 µl (2.5 U) FastStart Taq DNA Polymerase (Roche Diagnostics, Mannheim, Germany), 14 µl PCR-grade water and 100 ng template DNA. PCR amplifications were carried out with an initial denaturation step at 94°C for 3 min, 35–45 cycles of 94°C for 30 s, primer-specific annealing temperature for 30 s, 72°C for 60 s, and a final extension step at 72°C for 10 min. Bisulfite pyrosequencing was performed on a PSQ96MA Pyrosequencing System (Biotage, Uppsala, Sweden) with the PyroGold SQA reagent kit (Biotage) (45). The Pyro Q-CpG software (Biotage) was used for data analysis. To demonstrate the reliability of our quantitative methylation assays, we performed duplicate tests for a subset of tissue samples (mainly samples representing outliers in the box plot analysis). The methylation difference between duplicate measurements was 2.7 ± 1.6% for H19 (12 samples tested), 1.4 ± 0.9% for MEG3 (13 samples), 2.5 ± 1.5% for LIT1 (11 samples), 1.8 ± 1.1% for NESP55 (14 samples), 1.9 ± 1.4% for PEG3 (19 samples), 1.1 ± 0.9% for SNRPN (10 samples), 1.1 ± 1.1% for OCT4 (3 samples), and 1.1 ± 0.6% for APC (14 samples).
Data analysis
Quantitative methylation data were analyzed with SPSS version 17.0.1 (http://www.spss.com). Box plots were generated using the default parameters of SPSS. They display the location, dispersion and skewness of a set of data. The bottom and the top of the box indicate the 25th and 75th percentile, respectively. The T bars extend from the boxes to at most 1.5 times the height of the box. Outliers are samples that do not lie within these T bars, extreme outliers have values more extreme than three times the box length away from the median. Samples falling in the T bars were considered to be normally methylated, whereas outliers (and extreme outliers) may represent extreme methylation values. The χ2-test was used to compare the proportion of outliers in different tissues. A P < 5% was considered significant.
Statistical inference concerning the methylation data sets for the eight studied genes between different tissues was done with a multivariate analysis of variance (MANOVA), which is generalization of common analysis of variance. T-test was used to compare the methylation values of a particular gene between two groups.
Scatter plots were used to visualize the relationship between the DNA methylation levels (of a given gene) in unrelated blood samples and the age of the examined individuals. A linear regression was calculated to infer the dependence of methylation percentage on age. The goodness of fit measure (R2) was used to quantify this relation. R2 can range from +1 (perfect linear dependence) to 0 (no correlation). The P-value was calculated by ANOVA. It determines whether or not R2 significantly differs from 0.
We plotted a receiver operating characteristics (ROC) curve for the six studied imprinted genes, comparing the pairwise methylation differences of monozygotic (MZ) and dizygotic (DZ) twins. Using the area under curve as a test statistic for discriminative power, the corresponding P-value is exactly that obtained by a Wilcoxon rank sum test. In addition, a multivariate discrimination analysis (Fisher’s linear discriminant analysis) including the methylation data sets of the six studied genes was performed. The non-parametric Wilcoxon test was then used to compare the pairwise methylation differences of two groups (MZ versus DZ).
PAM analysis (46) was used to assess whether the methylation pattern across seven neighboring CpG sites in the APC promoter is predictive of the tissue type. By pattern we mean the relative difference in the methylation values among the seven CpGs. The input data were the mean-centered methylation data of all samples.
RESULTS
Gene-, tissue- and developmental stage-dependent variation of DNA methylation
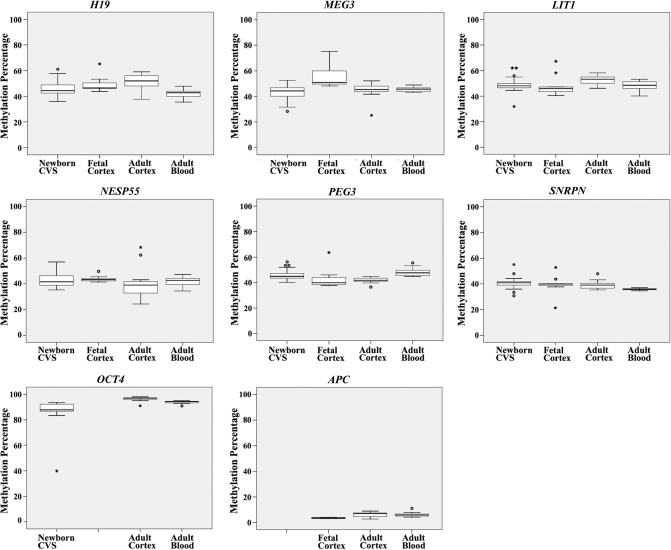
Here, we determined the methylation percentages of six DMRs (H19, MEG3, LIT1, NESP55, PEG3 and SNRPN) and two promoters (OCT4 and APC) in chorionic villi from 48 healthy newborns, in 10 fetal and 12 adult cortices, and 26 adult blood samples. As expected, imprinted genes were differentially methylated with median methylation values ranging from 37% to 53%, OCT4 was hypermethylated with median methylation values from 88% to 97%, and APC was hypomethylated with median methylation values from 4% to 7% (Table 2). The box plots in Figure 1 demonstrate that the median methylation of DMRs and promoters as well as the range of methylation variation are dependent on the gene, tissue type and developmental stage. For example, the median H19 DMR methylation ranged from 43% in adult blood to 52% in adult brain, whereas the median SNRPN methylation ranged from 36% in adult blood to 41% in CVS. Compared to the developing fetal brain, H19 and LIT1 were >5% upmethylated, whereas MEG3 was >5% downmethylated in adult brain.
Table 2.
Median methylation and number of extreme methylation values in different tissues
| Gene | Fetal cortex |
Adult cortex |
Newborn CVS |
Adult blood |
||||
|---|---|---|---|---|---|---|---|---|
| Median methylation | Number of outliers | Median methylation | Number of outliers | Median methylation | Number of outliers | Median methylation | Number of outliers | |
| H19 | 47% | 1/10 | 52% | 0/13 | 44% | 1/45 | 43% | 0/22 |
| MEG3 | 51% | 0/9 | 45% | 1/10 | 44% | 1/47 | 46% | 0/25 |
| LIT1 | 46% | 2/10 | 53% | 0/13 | 48% | 4/40 | 49% | 0/20 |
| NESP55 | 43% | 1/10 | 39% | 2/13 | 42% | 0/33 | 43% | 0/24 |
| PEG3 | 40% | 1/10 | 42% | 1/13 | 45% | 3/48 | 48% | 1/25 |
| SNRPN | 40% | 2/9 | 40% | 1/13 | 41% | 4/34 | 36% | 0/25 |
| OCT4 | ND | ND | 97% | 1/13 | 88% | 1/12 | 94% | 1/12 |
| APC | 4% | 0/10 | 7% | 0/8 | ND | ND | 6% | 1/26 |
| Imprinted genes | 7/58 (17%) | 5/75 (7%) | 13/247 (5%) | 1/141 (1%) | ||||
| Samples with outliers | 3/10 (30%) | 3/13 (23%) | 10/48 (21%) | 3/25 (12%) | ||||
| Samples with outliers in multiple genes | 1/10 (10%) | 2/13 (15%) | 3/48 (6%) | 0/25 (0%) | ||||
This table presents the median methylation (of all analyzed CpGs) and the number of outliers (including extreme outliers) in the studied DMRs and promoters in chorionic villi (of healthy newborns), fetal and adult cortex, and adult blood. ND, not done.
Figure 1.
Median methylation and range of methylation variation of two paternally methylated DMRs, H19 and MEG3, and four maternally methylated DMRs, LIT1, NESP55, SNRPN and PEG3, of imprinted genes as well as of promoters of one pluripotency gene, OCT4, and one tumor suppressor gene, APC. The box plots of a given gene show the distribution of DMR or promoter methylation values in 48 CVS from healthy newborns, 10 fetal and 12 adult brain samples, and 26 adult blood samples from unrelated individuals. The median is represented by a horizontal line. The bottom of the box indicates the 25th percentile, the top the 75th percentile. Outliers are shown as open circles, extreme outliers as stars.
When only looking at the six imprinted genes, which have been studied in all tissues, CVS displayed 5%, fetal brain 17%, adult brain 7% and adult blood samples 1% extreme DMR methylation values (Table 2). The number of outliers (including extreme outliers) in blood samples was significantly lower (χ2-test, P < 0.05) than in any other studied tissue. The difference between CVS and fetal brain was also significant. Three of 48 (6%) CVS, one of 10 (10%) fetal cortices, two of 13 (15%) adult cortices, but none (0%) of 25 adult blood samples displayed extreme DMR methylation values in two or more imprinted genes. In one fetal cortex 5 of the 6 studied DMRs were hypermethylated. Collectively, these data suggest that methylation imprints are relatively tightly regulated in blood, whereas CVS and brain exhibit considerably more methylation variation, consistent with a relaxation of imprinting (DMR methylation). With the notable exception of one CVS with extreme (40%) OCT4 hypomethylation, the pluripotency gene OCT4 was hypermethylated (83–98%) and the tumor suppressor gene APC was hypomethylated (2–11%) in all analyzed tissue samples.
In addition, we performed a MANOVA for each studied gene to test for differences between the methylation data sets of CVS, fetal brain, adult brain and adult blood. The null hypothesis that there are no differences between tissues was clearly rejected (Wilks’ Lambda test, P < 0.001) for H19, MEG3, PEG3, SNRPN and APC. There was a trend (P = 0.05) for LIT1. Group-wise comparisons revealed significant(t-test, P < 0.05) H19 methylation differences between CVS and adult blood, between fetal cortex and adult blood, as well as between adult cortex and adult blood. MEG3 methylation differences were found between CVS and fetal cortex, between fetal and adult cortex, and between fetal cortex and adult blood; PEG3 methylation differences between CVS and adult cortex, between CVS and adult blood, and between adult cortex and blood; SNRPN methylation differences between CVS and adult blood and between adult cortex and blood; APC methylation differed between fetal and adult cortex and between fetal cortex and adult blood.
Methylation changes with age
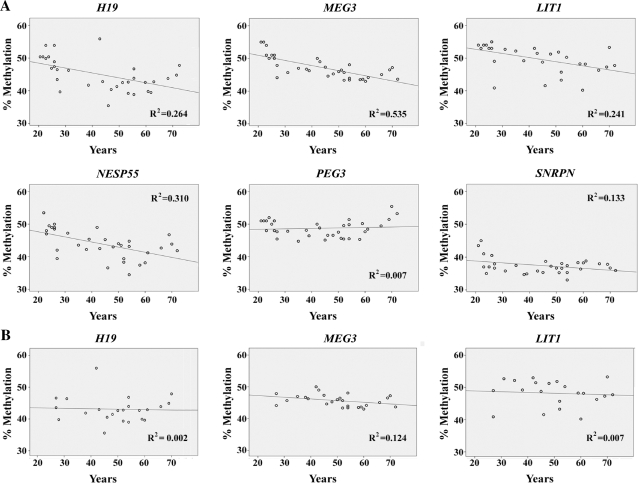
A general decrease in DNA methylation with age has been reported in blood cells (25). When we plotted the measured methylation values of six imprinted genes in blood samples against age (between 21 and 72 years) of the donor and calculated the linear regression (Figure 2A), there was a significant decrease of methylation with age for H19 (R2 = 0.264; P = 0.003 that there is no correlation between methylation and age), MEG3 (R2 = 0.535; P < 0.001), LIT1 (R2 = 0.241; P = 0.006), NESP55 (R2 = 0.310; P = 0.001) and SNRPN (R2 = 0.133; P = 0.037). However, this age effect was completely lost when removing all data points between 21 and 25 years (Figure 2B). T-test revealed highly significant (P < 0.001) differences in H19, MEG3, LIT1, NESP55 and SNRPN methylation between younger individuals (up to 25 years) and those >25 years.
Figure 2.
Association between age and H19, MEG3, LIT1, NESP55, SNRPN and PEG3 methylation. (A) Regression lines and data points represent measurements in blood samples of unrelated healthy individuals between 21 and 72 years. R2 is the linear correlation coefficient. (B) Regression lines when removing all samples from individuals up to 25 years.
Methylation variation in monozygotic and dizygotic twins
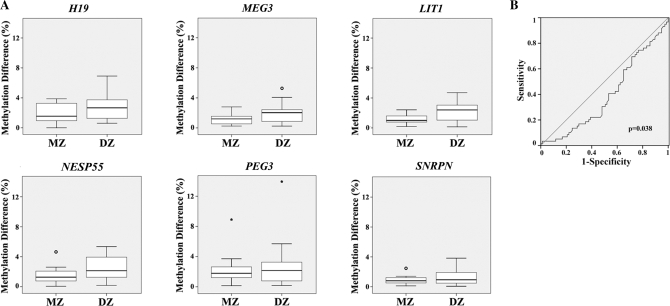
Figure 3A displays the differences in DMR methylation in blood samples of 12 MZ and 14 DZ pairs. For all six studied imprinted genes, the (median) pairwise methylation difference between MZ was somewhat smaller than between DZ. Because of the small number of studied twin pairs, there was no significant difference between the MZ and DZ groups at the single gene level. However, when a ROC curve (Figure 3B) was calculated with the methylation data sets of all six studied genes (including outliers), there was a significant (Wilcoxon test, P = 0.038) difference between MZ and DZ. A multivariate discrimination analysis based on the pairwise methylation differences of the six studied genes correctly classified 86% of the MZ and 88% of the DZ. This supports the assumption that the methylation differences of imprinted genes between pairs are predictive of the mode of twinning. The age of the MZ ranged from 27 to 70 years (mean 43 years), whereas the age of the DZ ranged from 38 to 72 years (mean 55 years). Although we cannot exclude age as a confounding variable, the observation that DMR methylation is rather stable between 27 and 72 years (Figure 2B) argues against this possibility.
Figure 3.
Differences in gene-specific methylation levels between pairs of monozygotic (MZ) and dizygotic twins (DZ). (A) The box plots display the pairwise difference in DMR methylation of H19, MEG3, LIT1, NESP55, SNRPN and PEG3 between MZ (12 pairs) and DZ (14 pairs). The median methylation difference is represented by a horizontal line. (B) ROC curve comparing the pairwise methylation differences of MZ and DZ. The area under the curve indicates the discriminative power of pairwise methylation differences in six studied genes between the MZ and DZ groups.
Methylation patterns of neighboring CpG sites
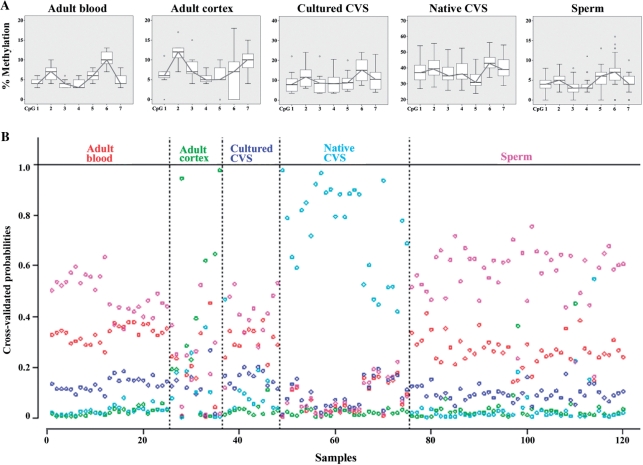
Our pyrosequencing assay for the tumor suppressor APC targets seven neighboring CpGs. With the notable exception of placenta (native CVS), the analyzed region of the APC promoter was hypomethylated in all tissues (Figure 4A). Native placental villi (n = 29) displayed a much higher CGI methylation (median 37%) than cultured CVS (n = 12, median 10%). The maternal APC allele has been reported to be methylated in the placenta (47), implying that cell culture leads to a loss of APC methylation (maternal imprinting). Adult cortex (n = 12), blood (n = 26), and sperm (n = 45) samples displayed median methylation values of 8%, 6% and 4%, respectively. By visual inspection of the fluctuation of methylation levels across the seven studied CpG sites (Figure 4A), CpGs 2, 3, 6 and 7 appeared to be most predictive of the tissue type. When we used Fisher’s discriminant analysis including the methylation data sets of these four CpGs 80% of the adult blood, 82% of adult cortex, 58% of cultured CVS, 100% of native CVS and 76% of sperm samples were classified correctly from the other tissues. In addition, we performed a PAM analysis (Figure 4B) to assess whether the methylation pattern across the seven analyzed CpGs (relative to each other) is predictive of the tissue type. It turned out that native CVS can be distinguished properly from the rest of the tissues. Any further distinction based upon the shrunken centroid classifier was not possible, because in the remaining cases the classifier mostly decided for ‘sperm’, irrespective of the true tissue origin.
Figure 4.
(A) Methylation patterns of seven neighboring CpG sites in the APC promoter region in adult blood (n = 26), adult cortex (n = 12), cultured (n = 12) and native (n = 29) CVS from first trimester abortions, and sperm (n = 45) samples. The box plots display the distribution of methylation values at each CpG site. Please note the different graduations of scale on the y-axis. (B) PAM analysis of methylation patterns across the seven CpGs. Each sample is represented by one column. PAM uses cross-validation to calculate posterior class probabilities for each sample. A classifier which is based upon this would vote for the class with the highest posterior in a given sample. Native CVS and sperm samples would be classified correctly to a large extent, whereas almost all other samples would be misclassified as sperm.
DISCUSSION
In contrast to the genome of an individual which is determined for all cells of the body at the time of conception, the epigenome is highly dynamic and differs from tissue to tissue. The most dramatic epigenetic changes occur during genome-wide reprogramming in gametogenesis and early embryogenesis (48,49). Genome-wide demethylation in the primordial germ cells erases essentially all methylation patterns to ensure an equivalent epigenetic state in germ cells of both sexes. Parent-specific methylation patterns according to the sex or the germline are then established during germ-cell differention (50). Genome-wide demethylation waves in the early embryo erase most germline methylation patterns, followed by de novo methylation and establishment of somatic methylation patterns (48,51). In addition to the carefully directed epigenetic reprogramming processes during development, epigenetic modifications may occur randomly or in response to environmental influences and ageing (5–7,23–25,29). Epigenetic variation may not only contribute substantially to phenotypic differences among individuals but also play an important role in the etiology of common (complex) human disease (52–54). This is obvious in most cancers which display dramatically increased methylation of tumor suppressor genes (18–20).
Although there is no simple linear relationship between DNA methylation and gene activity and the regulatory mechanisms may differ from gene to gene, in general the level and/or pattern of methylation in cis-regulatory regions is thought to control chromatin conformation and transcriptional potential. Threshold models suggest that changes shifting CpG methylation above a critical density lead to gene silencing (27,55). Our quantitative study revealed considerable variation in DNA methylation in hypomethylated, differentially methylated or hypermethylated cis-regulatory regions that are directly or indirectly involved in the control of gene activity. Both the average methylation level and the range of methylation variation depend on gene locus, tissue type and development stage, and age. It is noteworthy that the analyzed CVS and blood samples were from healthy newborns and adults; the brain samples were from fetuses and adults without detectable brain pathology. The lowest and the highest DMR methylation values in individual samples were 36% (in an adult blood) and 65% (in a fetal brain) for H19, 25% (in an adult brain) and 75% (in a fetal brain) for MEG3, 32% (in a CVS) and 67% (in a fetal brain) for LIT1, 24% and 68% (both in adult brain) for NESP55, 37% (in an adult brain) and 64% (in a fetal brain) for PEG3, and 21% (in a fetal brain) and 55% (in a CVS) for SNRPN. These functionally important methylation patterns exhibit a high degree of interindividual variability that by far exceeds DNA sequence variation. This enormous epigenetic variability cannot be explained by experimental noise, which is in the order of 1–2% discrepancy between the measured methylation levels of technical replicates.
Most epimutations underlying the variation in DNA methylation may occur after fertilization and probably represent stochastic errors in the establishment or maintenance of an epigenetic state. If an epimutation occurs in the germline, all cells of the individual should be affected, leading to clear DMR hypomethylation (<10%) or hypermethylation (>90%) in our pyrosequencing assay. In contrast, if an epimutation occurs during embryogenesis or later in life, it affects only a subset of cells, resulting in somatic mosaicism. The wide range of methylation variation in cis-regulatory regions suggests the possibility that basically each cell in a given tissue may be unique in its epigenomic profile. Thus, epigenetic analyses are not only hampered by the fact that only few tissues such as blood are readily accessible, but also by the high degree of epigenetic mosaicism. Because bisulfite treatment heavily degrades DNA, methylation analyses of single cells are both technically challenging and time-consuming (56,57), and therefore not particularly suitable for studying cell-to-cell variability in larger cell populations. Currently available quantitative methylation assays such as bisulfite pyrosequencing provide only a rough estimate on the percentage of normally versus extremely (hypo- or hyper)methylated cells in a tissue. In addition, when interpreting the medical relevance of our data, i.e. whether or not the observed methylation differences influence gene expression, we have to take into account that the analyzed tissues consist of different cell types and heterogeneous cell populations. From single cell expression analyses it becomes increasingly clear that the average cell does not exist (58,59).
With the notable exception of native chorionic villi, which show differential methylation of the APC promoter (47), several hundred analyzed CpGs in cultured CVS, adult brain, blood and sperm all displayed <20% methylation. Average promoter methylation ranged from 4% in sperm to 10% in cultured CVS. Interestingly, the methylation patterns, that is the relative differences in methylation among seven analyzed neighboring CpGs also appeared to differ among tissues. For example, in adult blood with an average CGI methylation of 6% the median methylation decreased from 10% at CpG 6 to 4% at CpG 7, whereas in adult cortex with an average CGI methylation of 7% it increased from 7% at CpG 6 to 10% at CpG 7. This tissue-specific fluctuation between neighboring CpGs is much higher than the technical noise (1.1 ± 0.6% between replicates) of our APC pyrosequencing assay and, therefore may represent a true biological phenomenon, i.e. tissue- and developmental stage-dependent differences in nucleosome positioning (60) and/or binding of specific transcription factors (61). By discriminant analysis based on only a few CpG sites in the APC promoter, we can correctly predict the tissue type in most analyzed samples. More sophisticated algorithms including multiple genes might considerably improve discriminative power.
Collectively, our results highlight substantial variation of DNA methylation at cis-regulatory regions among apparently healthy individuals. Although the observed hypo- and hypermethylation values may represent normal variation, this does not necessarily imply that they are biologically irrelevant. In a conceptually related study on imprinted gene methylation in human abortions and stillbirths (fetal muscle samples), we found that MEG3 hypermethylation was associated with downregulation of this gene and H19 hypermethylation increased the likelihood for biallelic expression of the autocrine growth factor IGF2 (62). In this context, it is noteworthy that altered DMR methylation patterns and relaxation of imprinting have been described in normal populations. For example, about 10% of blood samples from healthy individuals showed biallelic IGF2 expression (63). Loss of IGF2 imprinting occurs in colorectal cancer and many other tumor types. The observation that IGF2 was also dysregulated in normal mucosa and peripheral blood lymphocytes of these cancer patients (64) promotes the idea that relaxation of imprinting may predispose to tumorigenesis and, by extrapolation, other complex diseases. Most likely, the role of epigenetic changes for phenotypic variation and disease susceptibility is largely underestimated. Further research is needed to determine the functional implications of the enormous DNA methylation variation for the fine tuning of gene regulation.
FUNDING
Funding for open access charge: German Research Foundation (HA 1374/8-1).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Daniela Weise for excellent technical assistance.
REFERENCES
- 1.Gärtner K, Baunack E. Is the similarity of monozygotic twins due to genetic factors alone? Nature. 1981;292:646–764. doi: 10.1038/292646a0. [DOI] [PubMed] [Google Scholar]
- 2.Gärtner K. A third component causing random variability beside environment and genotype.. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 3.Yanagimachi R. Cloning: experience from the mouse and other animals. Mol. Cell. Endocrinol. 2002;187:241–248. doi: 10.1016/s0303-7207(01)00697-9. [DOI] [PubMed] [Google Scholar]
- 4.Wong AH, Gottesman II, Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Hum. Mol. Genet. 2005;14:R11–18. doi: 10.1093/hmg/ddi116. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland JE, Costa M. Epigenetics and the environment. Ann. NY Acad. Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 6.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horsthemke B. Epimutation in human disease. Curr. Top. Microbiol. Immunol. 2006;310:45–59. doi: 10.1007/3-540-31181-5_4. [DOI] [PubMed] [Google Scholar]
- 8.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyal R, Reinhardt R, Jeltsch A. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 2006;34:1182–1188. doi: 10.1093/nar/gkl002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 12.Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 15.Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 16.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 17.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 19.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome - components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 20.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 21.Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu. Rev. Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 23.Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. J. Am. Med. Assoc. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech. Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farcas R, Schneider E, Frauenknecht K, Kondova I, Bontrop R, Bohl J, Navarro B, Metzler M, Zischler H, Zechner U, et al. Differences in DNA methylation patterns and expression of the CCRK gene in human and non-human primate cortices. Mol. Biol. Evol. 2009;26:1379–1389. doi: 10.1093/molbev/msp046. [DOI] [PubMed] [Google Scholar]
- 27.Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sontag LB, Lorincz MC, Luebeck EG. Dynamics, stability and inheritance of somatic DNA methylation imprints. J. Theor. Biol. 2006;242:890–899. doi: 10.1016/j.jtbi.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Foley DL, Craig JM, Morley R, Olsson CA, Dwyer T, Smith K, Saffery R. Prospects for epigenetic epidemiology. Am. J. Epidemiol. 2009;169:389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 31.Mackay DJ, Boonen SE, Clayton-Smith J, Goodship J, Hahnemann JM, Kant SG, Njølstad PR, Robin NH, Robinson DO, Siebert R, et al. A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum. Genet. 2006;120:262–269. doi: 10.1007/s00439-006-0205-2. [DOI] [PubMed] [Google Scholar]
- 32.White HE, Durston VJ, Harvey JF, Cross NC. Quantitative analysis of SNRPN gene methylation by pyrosequencing as a diagnostic test for Prader-Willi syndrome and Angelman syndrome. Clin. Chem. 2006;52:1005–1013. doi: 10.1373/clinchem.2005.065086. [DOI] [PubMed] [Google Scholar]
- 33.Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J. Biol. Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
- 34.Schatz P, Distler J, Berlin K, Schuster M. Novel method for high throughput DNA methylation marker evaluation using PNA-probe library hybridization and MALDI-TOF detection. Nucleic Acids Res. 2006;34:e59. doi: 10.1093/nar/gkl218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zechner U, Kohlschmidt N, Rittner G, Damatova N, Beyer V, Haaf T, Bartsch O. Epimutation at human chromosome 14q32.2 in a boy with a upd(14)mat-like clinical phenotype. Clin. Genet. 2009;75:251–258. doi: 10.1111/j.1399-0004.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000;127:711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 37.Gicquel C, Rossignol S, Cabrol S, Houang M, Steunou V, Barbu V, Danton F, Thibaud N, Le Merrer M, Burglen L, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 38.Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, et al. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 2005;13:1025–1032. doi: 10.1038/sj.ejhg.5201463. [DOI] [PubMed] [Google Scholar]
- 39.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum. Mol. Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 40.Horsthemke B, Buiting K. Imprinting defects on human chromosome 15. Cytogenet. Genome Res. 2006;113:292–299. doi: 10.1159/000090844. [DOI] [PubMed] [Google Scholar]
- 41.Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009;442:18–25. doi: 10.1016/j.gene.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plagge A, Kelsey G. Imprinting at the GNAS locus. Cytogenet. Genome Res. 2006;113:178–187. doi: 10.1159/000090830. [DOI] [PubMed] [Google Scholar]
- 43.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Monk M, Hitchins M, Hawes S. Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol. Hum. Reprod. 2008;14:347–355. doi: 10.1093/molehr/gan025. [DOI] [PubMed] [Google Scholar]
- 45.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 46.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guilleret I, Osterheld MC, Braunschweig R, Gastineau V, Taillens S, Benhattar J. Imprinting of tumor-suppressor genes in human placenta. Epigenetics. 2009;4:62–68. doi: 10.4161/epi.4.1.7471. [DOI] [PubMed] [Google Scholar]
- 48.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 49.Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Curr. Top. Microbiol. Immunol. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- 50.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 51.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 52.Abdolmaleky HM, Smith CL, Faraone SV, Shafa R, Stone W, Glatt SJ, Tsuang MT. Methylomics in psychiatry: modulation of gene-environment interactions may be through DNA methylation. Am. J. Med. Genet. B. 2004;127:51–99. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 53.Hatchwell E, Greally JM. The potential role of epigenomic dysregulation in complex human disease. Trends Genet. 2007;23:588–595. doi: 10.1016/j.tig.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 54.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kerjean A, Vieillefond A, Thiounn N, Sibony M, Jeanpierre M, Jouannet P. Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res. 2001;29:e106. doi: 10.1093/nar/29.21.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geuns E, De Rycke M, van Steirteghem A, Libaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum. Mol. Genet. 2003;12:2873–2879. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- 58.Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;3:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 59.May A, Kirchner R, Müller H, Hartmann P, El Hajj N, Tresch A, Zechner U, Mann W, Haaf T. Multiplex RT-PCR expression analysis of developmentally important genes in individual mouse preimplantation embryos and blastomeres. Biol. Reprod. 2009;80:194–202. doi: 10.1095/biolreprod.107.064691. [DOI] [PubMed] [Google Scholar]
- 60.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 61.Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pliushch G, Schneider E, Weise D, El Hajj N, Tresch A, Seidmann L, Coerdt W, Müller AM, Zechner U, Haaf T. Extreme methylation values of imprinted genes in human abortions and stillbirths. Am. J. Pathol. 2010 doi: 10.2353/ajpath.2010.090764. in press (PMID 20093482) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakatani T, Wei M, Katoh M, Okita C, Wada D, Mitsuya K, Meguro M, Ikeguchi M, Ito H, Tycko B, et al. Epigenetic heterogeneity at imprinted loci in normal populations. Biochem. Biophys. Res. Commun. 2001;283:1124–1130. doi: 10.1006/bbrc.2001.4916. [DOI] [PubMed] [Google Scholar]
- 64.Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat. Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]