Abstract
Trinucleotide repeats (TNRs) are of interest in genetics because they are used as markers for tracing genotype–phenotype relations and because they are directly involved in numerous human genetic diseases. In this study, we searched the human genome reference sequence and annotated exons (exome) for the presence of uninterrupted triplet repeat tracts composed of six or more repeated units. A list of 32 448 TNRs and 878 TNR-containing genes was generated and is provided herein. We found that some triplet repeats, specifically CNG, are overrepresented, while CTT, ATC, AAC and AAT are underrepresented in exons. This observation suggests that the occurrence of TNRs in exons is not random, but undergoes positive or negative selective pressure. Additionally, TNR types strongly determine their localization in mRNA sections (ORF, UTRs). Most genes containing exon-overrepresented TNRs are associated with gene ontology-defined functions. Surprisingly, many groups of genes that contain TNR types coding for different homo-amino acid tracts associate with the same transcription-related GO categories. We propose that TNRs have potential to be functional genetic elements and that their variation may be involved in the regulation of many common phenotypes; as such, TNR polymorphisms should be considered a priority in association studies.
INTRODUCTION
Microsatellites, known also as short tandem repeats (STR) or simple sequence repeats (SSRs), are tracts of tandemly repeated short (1–6 bp) DNA sequence motifs. These sequences are abundant in prokaryotic (1) and eukaryotic (2) genomes and occur in both inter- and intragenic regions, including open reading frames (ORFs). Estimates from the human genome reference sequence indicate that microsatellites may account for ∼3% of the genome. This contribution, however, is highly approximate and depends strongly on how repeat length and sequence purity thresholds are defined. An immanent feature of microsatellites is their high mutability, which leads to both sequence and length polymorphism (3–5), the latter being at least one order of magnitude greater than the former (3,6). The length polymorphism of microsatellites makes them very informative genetic markers; they are used as such in population genetics, genetic mapping and linkage analysis (7–9). Microsatellite polymorphisms are also, next to single nucleotide polymorphisms (SNPs) and copy number polymorphisms (CNPs), very significant components of human genetic variation capable of modifying many common phenotypes.
Trinucleotide repeats (TNRs) are a special class of microsatellites. These sequences have received special attention, primarily because some are known to undergo pathogenic expansions that cause triplet repeat expansion diseases (TREDs). More than 20 genetic disorders belong to this group; they are mostly neurodegenerative and neuromuscular (10,11) disorders. In several TREDs, stable RNA structures formed by triplet repeats present in untranslated regions of the responsible genes are implicated in pathogenesis (12–15); in some other TREDs, CAG repeats expressed as homo-Gln tracts in proteins give rise to pathogenesis (16–18).
The great majority of TNRs do not undergo pathogenic expansion and little is known about their normal function in human genes and transcripts. The features of TNRs that suggest their functionality include: (i) widespread occurrence in exons, (ii) formation of stable hairpin or quadruplex structures by some TNRs and (iii) coding for homo-amino acid (AA) tracts. In this article, we address the question of whether the occurrence of TNRs in human exome is random (null hypothesis) or subject to positive or negative selective pressure (alternative hypothesis). To test the above hypotheses, we compared the frequency of all TNR types in exons with their frequencies in the entire genome. The high overrepresentation of some TNR types and underrepresentation of others in exons favor the alternative hypothesis. To further characterize TNRs localized in exons, we have classified all exonic TNRs with regard to their orientation (sense/antisense), localization in the mRNA (5′-UTR/ORF/3′-UTR) and coded AA. Using the groups of genes defined by the above criteria, we performed gene functional association analysis. We show that most groups of genes containing TNR types overrepresented in exons are strongly associated with function as defined by gene ontology (GO) terms. The above results suggest that TNRs have high potential to be important functional elements in human genes and argue against the common notion that microsatellites are ‘genetic junk’. This functionality can be expressed at the protein, RNA or DNA (genetic) level. We propose that polymorphic TNRs, especially those localized in or close to exons or genetic regulatory elements (promoters, enhancers, microRNA genes, etc.) have considerable phenotype-modifying potential and should be considered high priority genetic variants in genotype–phenotype association studies.
MATERIALS AND METHODS
Identification of TNRs
To identify all TNRs [≥6 repeated units (U)] present in the reference sequence of the human genome NCBI build 36.1, March 2006 Assembly (hg18), we used the BLASTn program available on the webpage of Ensembl Genome Browser—http://www.ensembl.org. The reference human genome sequence was searched in both directions against 10 sequences [(AAC)6, (AAG)6, (AAT)6, (ACC)6, (GAC)6, (ACT)6, (CAG)6, (AGG)6, (ATC)6 and (CGG)6] representing all combinations of nucleotide triplets. We excluded from the analysis triplets composed of homonucleotides, as they actually represent mononucleotide tracts. The BLASTn parameters were as follows: -filter, none; -RepeatMasker, no; -W (word size), 2; -wink (step size), 1; -E (expectancy) was adjusted to obtain only perfect match hits; other parameters, default). The TNRs localized in exons annotated by RefSeq or UCSC were defined as exonic and were characterized according to their mRNA localization (5′-UTR, ORF, 3′-UTR) and encoded AA.
Functional association analysis
We performed a functional association analysis for groups of genes defined by TNR type, mRNA localization and encoded AA. Only groups with ≥20 genes were taken for analysis. We compared these groups of genes with Gene Ontology categories [biological process (BP), cellular compartment (CC) and molecular function (MF)] using the database for annotation, visualization and integrated discovery, DAVID—http://david.abcc.ncifcrf.gov/ (19,20). The DAVID program calculated fold enrichments, fractions of involved genes, appropriate P-values and correction for multiple tests. A Bonferroni corrected P < 0.01 was considered significant unless otherwise stated.
Statistical methods
All statistical analyses were performed using Statistica (StatSoft, Tulsa, OK, USA) or Prism v. 4.0 (GraphPad Software, San Diego, CA, USA). The K–S graphs were created using an online tool available on the webpage of College of Saint Benedict and Saint John’s University, Collegeville, MN—http://www.physics.csbsju.edu/stats/KS-test.html.
Secondary-structure prediction
The secondary structures of the multi-TNR-mRNAs were predicted using the Mfold program (21). The predicted structures of the lowest free-energy conformations were taken for visualization.
RESULTS AND DISCUSSION
Some TNR types are strongly overrepresented and others are underrepresented in human exons
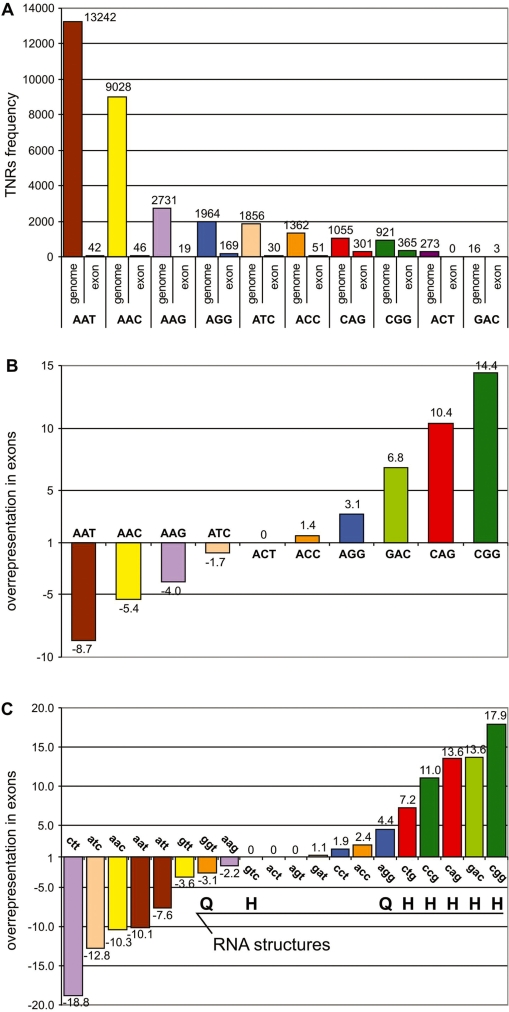
To determine the frequencies of all types of TNRs in the human genome, all uninterrupted TNR tracts composed of six or more repeated units were identified using the BLASTn algorithm. The human genome reference sequence (assembly March 2006) was searched in both directions for each of the 10 non-redundant TNR sequences distinguished by the criteria of combined frames and complementarity: AAC representing (AAC/GTT, ACA/TGT and CAA/TTG), AAG (AAG/CTT, AGA/TCT, GAA/TTC), AAT (AAT/ATT, ATA/TAT, TAA/TTA), ACC (ACC/GGT, CAC/GTG, CCA/TGG), GAC (GAC/GTC, ACG/CGT, CGA/TCG), ACT (ACT/AGT, CTA/TAG, TAC/GTA), CAG (CAG/CTG, AGC/GCA, GCA/TGC), AGG (AGG/CCT, GGA/TCC, GAG/CTC), ATC (ATC/GAT, TCA/TGA, CAT/ATG), CGG (CGG/CCG, GGC/GCC, GCG/CGC). Lowercase letters will be used to distinguish the orientations of TNRs localized in exons (e.g. cag or ctg instead of CAG). By applying the above criteria, we identified 32 448 TNRs in the entire human genome (Supplementary Table S1). The most frequent repeats were AAT, AAC and AAG with a frequency of 13 242, 9028 and 2731 occurrences, respectively (Figure 1A). The least frequent were GAC with 16 and ACT with 273 tracts identified. The frequency of the other TNRs ranged from 921 CGG to 1964 ATC occurrences. The relative frequencies of different TNR types observed in our study is similar to that found in earlier genome-wide surveys that used different repeats length and purity thresholds (22–26). In one of these studies, the genomic frequency of various microsatellite types was shown to correlate inversely with their back-folding annealing temperature, i.e. the tendency of their single strands to form stable hairpin or quadruplex structures (22). This observation well explains the high frequency of AT-rich TNRs having no or low structure-forming potential and the lower frequency of GC-rich TNRs capable of forming stable hairpin or quadruplex structures. However, when we formally compared the frequency of the TNR types observed in our study with the annealing temperatures of specific TNR sequences determined earlier in DNA (22), we found only a modest (r2 = 0.44), marginally significant correlation. This moderate correlation suggests that other factors may also influence the occurrence of various TNR types in the human genome.
Figure 1.
Frequency of TNRs in the human genome and in exons. (A) The total number of TNRs (≥6 U) identified in the reference sequence of the human genome and in human exons. Different colors represent different TNR types and are used consistently throughout the article. (B) Representation rates of the 10 TNR types in human exons. Positive and negative values represent the fold over- and under-representation, respectively. The representation rate was calculated as the ratio of a TNR’s density (number of TNRs per 1 Mbp) in exons to its density in the entire genome including exons. For calculation, we have taken into account the total genome size and the fraction of the genome covered by exons annotated by RefSeq and/or UCSC nomenclature (2.75%). (C) Representation rates calculated separately for each orientation of each TNR type. In calculating these ratios, we assumed that both TNR orientations are represented equally in the genome (e.g. to calculate the representation rate for ccg, we divided the density of ccg in exons by the density of CGG in the genome divided by two). The symbols H and Q indicate TNRs for which RNA strands are capable of forming stable hairpin and quadruplex structures, respectively.
In the next step of the study, we extracted 1030 TNRs localized in the exonic sequences of 878 genes (TNR-containing genes) (Supplementary Table S2). Exons were defined by RefSeq (27) and UCSC nomenclatures (28,29) and accounted for 2.75% of the total human genome sequence. As much as 93% of identified TNR-containing genes belong to the well-validated classes of genes (RefSeq status: validated, reviewed) (Supplementary Table S2), whereas only 80% of all non-redundant human genes (RefSeq defined) belong to this class. This result is consistent with the observation showing that functionally unclassified genes are significantly underrepresented among TNR-containing genes (PANTHER Classification System http://www.pantherdb.org).
The most frequent TNRs in the exonic sequences were CGG with 365 occurrences, CAG with 301 and AGG with 169. The frequencies of other TNRs ranged from 0 (ACT) to 51 occurrences (ACC). Comparing the TNR frequency in the genome with that in exons, we found that all types of TNRs taken together are only slightly (1.16 times) overrepresented in exons [the overall TNR density (coverage) in genomic and exonic sequences is 11 TNR/Mbp (0.0273%) and 13 TNR/Mbp (0.0287%), respectively]. As the frequencies of different types of TNRs in exons differ significantly and there is no correlation between the frequency of TNRs in the genome and in exons (r2 = 0.05), we calculated the over-/under-representation ratio for each TNR type (Figure 1B) as well as for each TNR orientation in exons (Figure 1C). It is apparent that some TNRs are strongly overrepresented in exons and others are underrepresented (Figure 1B and C). The most overrepresented are two CNG-type TNRs: CGG: 14.4× (cgg: 17.9×, ccg: 11.0×) and CAG: 10.4× (cag: 13.6×, ctg: 7.2×), while the most underrepresented are AT-rich TNRs: AAT: −8.7× (aat: −10.1×, att: −7.6×), AAC: −5.4× (aac: −10.3×, gtt: −3.6×) and AAG: −3.8× (aag: −2.2×, ctt: −18.8×). Based on the formally calculated representation factors for all TNR types (not only those related to TREDs), we propose that the observed over- and under-representation of specific TNR types in exons may result from positive and negative selective pressure, respectively. The factor that can also influence over-/under-representation of TNR types in exons is the nucleotide composition of the sequences being compared. It was shown for example that median GC content in human exons (0.51) is higher than in genome (0.41) (30). Although this difference is relatively low when compared to the differences in TNR frequencies it may partially explain the overrepresentation of GC-rich TNRs in exons and opposite trend for AT-rich TNRs. The different nucleotide composition observed in first (coding), internal and last exons (31) can also influence a biased distribution of homo-AA tracts in these exons (e.g. homo-Ala, -Leu, -Gly and -Pro are overrepresented in the first exons whereas homo-Gln, -Glu and -Ser are overrepresented in internal exons) (22).
Length distribution differs significantly between specific TNR types but does not differ between TNRs localized inside and outside exon sequences
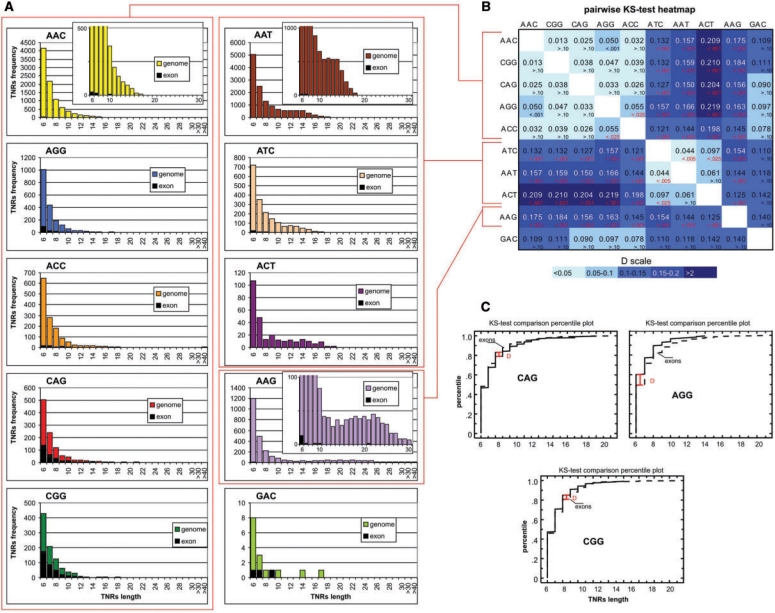
It was recently shown that TNR lengths present in the reference human genome sequence can be used as proxies for the most frequent or average allele lengths (32). We also found a good correlation (R = 0.8; P-value <0.0001) between TNR lengths in the reference sequence and the same TNRs recently genotyped in a Polish population (33). Therefore, the next feature of TNRs that we analyzed was their length distribution.
As shown in Figure 2A, the general trend in length distribution is similar in all TNR types studied. As expected, the shortest tracts are always the most frequent and the frequency of others decreases roughly exponentially with TNR length. For the majority of TNR types, the longest tracts are shorter than 20 U. However, for some TNR types, tracts longer than 20 or even 30 U were identified. Extreme examples are 210 ACC repeats, 123 and 79 ATC repeats, as well as 60 and 61 AAG repeats (see Supplementary Table S1 for details).
Figure 2.
Length distributions of TNR types in the human genome and in exons. (A) For each type, the graph shows the number (y-axis) of TNRs of a given length (x-axis) identified in the human genome (color bars) and in exons (black bars). Bars indicated on the x-axis as >30 and 40 show the combined number of TNRs 31–40 and >40 U in length, respectively. An inset, shown in some graphs, was scaled up to emphasize the length distribution details specific for longer (less frequent) TNRs. (B) Heatmap graph showing the Kolmogorov–Smirnov statistic (D) for the pairwise length distribution comparisons of all TNR types. The color legend is shown next to the graph. The P-values for individual comparisons are also indicated on the graph (in each cell the value of the D statistic is given below). The P < 0.05 are indicated in red. The cluster of light blue squares represents groups of TNR types with similar length distributions. (C) Cumulative fraction plots comparing the TNR length distribution in exons (dashed line) and in non-exon sequences (genome sequence not covered by exons) (solid lines). The y-axis indicates the cumulative fraction of TNRs for certain TNR lengths (x-axis). The maximum distance between fraction plots (K–S-test, D statistic) and appropriate P-values are indicated on the plots.
To formally compare the length distributions of different TNR types, we used the Kolmogorov–Smirnov (K–S) test. Pairwise comparison of the length distributions of all TNR types (Figure 2B) shows that these fall into three distinct groups: 1 contains AAC, CGG, CAG, AGG and ACC repeats, 2 contains ATC, AAT and ACT, and 3 contains AAG. The low number of identified GAC tracts (N = 16) did not allow for their reliable classification.
A more detailed analysis of TNR length distributions within the individual groups distinguishes groups 2 and 3 as having an additional mode with a maximum at 13 and 20 U, respectively (Figure 2A). This is the first formal comparison and classification of TNR types into length-defined groups, although the existence of extra modes in the length distribution of some TNR types was noticed earlier (22–24). The presence of the extra mode increases the fraction of longer TNRs that are more prone to undergo expansions. However, expansions of ACT and AAT TNRs have not yet been detected nor shown to cause human disease. On the other hand, the expansion of AAG repeats in intron 1 of the FXN gene is known to cause the recessive disorder Friedreich ataxia (MIM #229300). The AAG repeat, which in our analysis shows the most distinct length distribution, was analyzed earlier in 20 different genomes (23). It was shown that the unusual length distribution of AAG TNRs and the high fraction of tracts longer than 10 U are specific to mammals. It was also demonstrated that long AAG tracts are highly polymorphic and that there are several AAG loci in the human genome (some of them localized in introns) that contain alleles longer than 65 U, analogous to those causing Friedreich ataxia (23).
Using the K–S test, we also compared the lengths of TNRs localized in exons and outside of exonic sequences. Because different TNR types showed different length distributions, the analysis was performed separately for the CGG, CAG and AGG TNRs for which sufficient numbers of tracts were identified in exons (Figure 2C). The results obtained show that the lengths of TNRs localized to exons do not differ from the lengths of TNRs located outside exon sequences (K–S-test D = 0.04, P = 0.87; D = 0.05, P = 0.77 and D = 0.11, P = 0.08 for CGG, CAG and AGG, respectively). The length of a TNR is probably a compromise between the tendency of TNRs to expand and selective pressure acting against excessive TNR length. Excessively long TNRs can be a source of unnecessary polymorphism or even pathogenic expansions. On the other hand, some level of polymorphism can be beneficial by facilitating adaptive evolution (34).
A similar analysis as above was conducted for all TNR types comparing their length distribution in sequences covered by RefSeq-defined genes (including exons and introns) and sequences out of these regions (data not shown). All TNRs except for AAG showed no differences in length distributions. In the case of AAG TNRs, tracts located in genes were on average 1 U shorter than those located in intergenic regions (average length: 8.9 and 10.1 U, respectively; K–S-test; P < 0.001). This difference may result from natural selection acting against the occurrence of the easily expandable sequences of long AAG tracts in the transcribed regions of protein-coding genes (Figure 2A). As mentioned above Friedreich ataxia is an example of a pathogenic effect caused by expanded AAG.
The localization of TNRs in mRNA strongly depends on their type
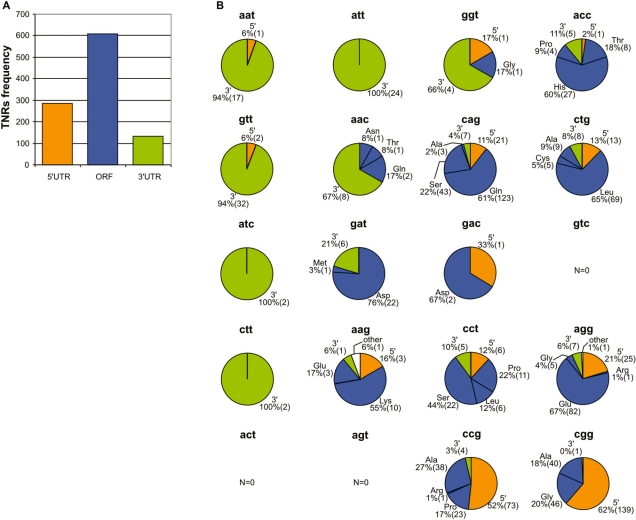
As TNRs occurring in different mRNA regions may have different functions, we classified each TNR present in an exon as belonging to the 5′-UTR, 3′-UTR or ORF. TNRs localized in ORFs were further divided into subgroups according to the coded AA. Figure 3A shows that out of the 1030 TNRs identified in exons, 609 (59%) are localized in the ORF (average TNR density 18 TNR/Mbp), 286 (28%) in the 5′-UTR (average TNR density 16 TNR/Mbp) and 133 (13%) in the 3′-UTR (average TNR density 4 TNR/Mbp). The remaining two TNRs could not be unambiguously assigned to any of these locations.
Figure 3.
Localization of TNRs in mRNA regions. (A) Bar-plot showing the number of TNRs in 5′-UTRs, ORFs and 3′-UTRs. (B) Pie plots showing the distribution of TNRs among mRNA regions separately for each TNR type and orientation. Subfractions of TNRs localized in ORFs coding specific AAs are also indicated. The percentage and the number (in brackets) of TNRs in each fraction are indicated.
As each of the 10 distinct TNR types can occur in two orientations, we analyzed the distribution of 20 possible single-stranded repeated motifs between mRNA sections (Figure 3B) and found that it is not random. The TNRs acc, cag, ctg, cct, agg, aag and gat occur most frequently in the ORF (∼80%). AT-rich TNRs are generally more frequent in the 3′-UTR. Extreme examples are att and aat, which occur almost exclusively in the 3′-UTR (100 and 94%, respectively). On the other hand, ccg and cgg repeats are most frequent in the 5′-UTR (52 and 62%, respectively). This finding may be associated with the fact that repeats harboring CpG dinucleotides are often present in promoter regions and are involved in the regulation of transcription. CG-rich repeats also have the potential to regulate the initiation step of the translation process (35–37). The observed overrepresentation of CG-rich repeats in 5′-UTRs and AT-rich repeats in 3′-UTRs can be partially explained by the well-known higher AT and GC content of 3′- and 5′-UTRs, respectively.
Homo-Gln is the most prevalent homo-AA tract encoded by TNRs
The uninterrupted TNR sequences identified in our study code for 15 of the 20 possible homo-AA tracts. The most frequent are Gln, Ala, Glu and Leu tracts with 125, 90, 85 and 75 occurrences, respectively. Cys, Arg, Met and Asn tracts are very rare (≤5), and Tyr, Trp, Val, Ile and Phe tracts do not occur at all. This distribution corresponds generally to the distribution of homo-AA tracts identified in the human proteome (38,39), despite the fact that analyses conducted by others also include homo-AA tracts encoded by interrupted TNRs (mixes of synonymous codons) that are significantly longer than the same pure codon tracts (6). An exception is the homo-Gln tract, which accounts for 19% of all homo-AA tracts identified in this study but is only the seventh most frequent (5%) among homo-AA tracts detected in the human proteome (39). This discordance can be explained by lower number of interruptions in the TNRs coding for homo-Gln tracts comparing to TNRs coding for other homo-AA tracts. The least frequent (or absent) tracts are those of hydrophobic or highly hydrophobic AAs. Their lower frequency, which was found in this and other studies (38,39), may be explained by the higher toxicity of such tracts (40,41).
Most TNR types localized in ORFs are associated with transcription-related functions (GO terms)
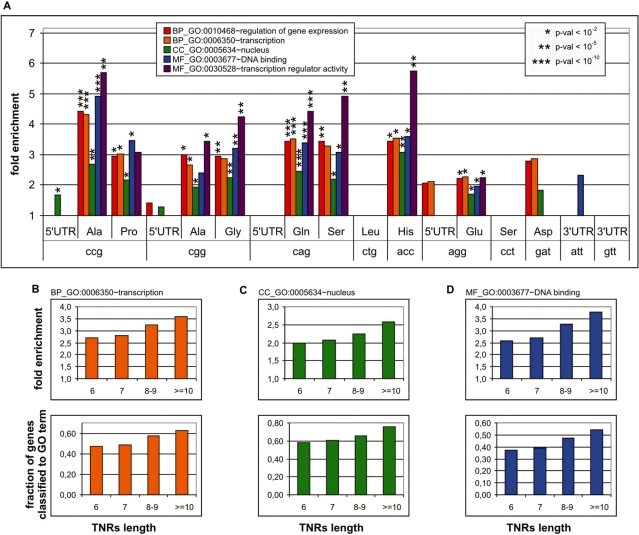
To gain insight into the potential functions of TNRs in exons, we conducted a functional association analysis that compared the list of TNR-containing genes with BP, CC and MF terms defined by the GO classification (42). We assumed that the functions associated with TNRs may be specific for their type, orientation, localization and coded AA tract. Therefore, prior to GO analysis, we classified all TNR-containing genes (Supplementary Table S2) into groups defined by the above criteria (Supplementary Table S3). Only groups composed of 20 or more genes were taken for GO-association analysis. These groups included genes with: (i) gtt in the 3′-UTR (N = 31), (ii) att in the 3′-UTR (24), (iii) acc coding His (26), (iv) agg in the 5′-UTR (22), (v) agg coding Glu (79), (vi) cct coding Ser (20), (vii) gat coding Asp (21), (viii) cag in the 5′-UTR (20), (ix) cag coding Gln (94), (x) cag coding Ser (35), (xi) ctg coding Leu (69), (xii) cgg in the 5′-UTR (134), (xiii) cgg coding Gly (42), (xiv) cgg coding Ala (40), (xv) ccg in the 5′-UTR (72), (xvi) ccg coding Ala (37) and (xvii) ccg coding Pro (21) [note that the differences in the numbers (N’s) indicated here and in Figure 3 are due to the fact that some genes contain more than one TNR of the same type]. The complete results of the GO-association analysis are presented in Supplementary Table S4 and are summarized in Table 1. The most striking result is the association of several groups of TNR-containing genes with transcription-related GO terms [e.g. GO:0006350, transcription (BP); GO:0030528, transcription regulator activity (MF); GO:0005634, nucleus (CC)]. These groups include genes with different TNR types coding for different homo-AA tracts localized in ORFs [acc coding (His), cag (Gln), cag (Ser), cgg (Gly) and ccg (Ala)]. As association with transcription-related functions seems to be common for TNR-containing genes, we reanalyzed all TNR-containing groups of genes for their association with just five representative (arbitrarily selected) transcription-related GO terms, assuming a nominal P < 0.01 as significant (Figure 4A). This step led to the identification of additional groups of genes with transcription-related functions harboring the coding TNRs ccg (Pro), cgg (Ala) and agg (Glu) (Figure 4A). In the case of genes containing gat repeats coding for Asp, enrichment in some of the transcription-related terms is also observed but is statistically not significant. The results obtained for cct coding Ser and gat coding Asp were considered non-informative rather than negative due to the low number of genes present in these groups. Although the five analyzed GO terms were selected arbitrarily, similar results were obtained for other transcription-related GO terms (Supplementary Table S4). Altogether, we identified eight groups of genes containing different types of TNRs located in ORFs that show significant enrichment (2–5×) in transcription-related GO terms. We have further shown that both the enrichment factor and the fraction of genes classified to specific transcription-related GO terms increase with TNR length (Figure 4B). This association with transcription-related GO terms generally was not observed for genes containing TNRs localized in the untranslated regions of mRNA. This result further confirms the observation that TNRs coding for AA tracts are responsible for the observed associations and that increased TNR length (in the analyzed range) enhances this association. The observation that TNR-containing genes are associated with transcription-related functions was reported earlier; however, these reports were either limited to a specific TNR type (CAG coding homo-Gln) (32) or extended to all TNR-containing genes not distinguished by type, location or encoded AA tract (22). Here for the first time, we have shown that several different types of TNRs coding for different AAs are responsible for this association. The only AA characteristic that is overrepresented in the group of AAs tracts associated with transcription-related functions is polarity (we have analyzed many chemical and physical properties of AAs, e.g. those characterized in CHIP Bioinformatics Tools http://snpper.chip.org/bio/showamino). However, the excess of polar AAs among those associated with transcription is probably merely due to the general excess of polar homo-AA tracts in human proteins observed in this study and earlier (38,39).
Table 1.
Functional associations of TNR-containing genes
| TNR | Location | N | Effect | The representative GO-term | Fraction of genes | Fold enrich. | P-value | Bon ferroni |
|---|---|---|---|---|---|---|---|---|
| gtt | 3′ UTR | 31 | No | |||||
| att | 3′ UTR | 24 | No | |||||
| acc | His | 26 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.50 | 3.9 | 8.7E−06 | 4.5E−02 |
| GO:0005634∼nucleus | 0.73 | 3.1 | 2.6E−08 | 2.3E−05 | ||||
| agg | 5′ UTR | 22 | No | |||||
| Glu | 79 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.25 | 2.1 | 2.3E−03 | ns | |
| GO:0005634∼nucleus | 0.76 | 1.4 | 5.6E−06 | 4.8E−03 | ||||
| cct | Ser | 20 | No | |||||
| gat | Asp | 21 | transcription/nucleic acids binding/nucleus locationa | GO:0006355∼regulation of transcription. DNA-dependent | 0.19 | 2.5 | ns | ns |
| GO:0005634∼nucleus | 0.13 | 1.8 | ns | ns | ||||
| cag | 5′ UTR | 20 | No | |||||
| Gln | 94 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.48 | 3.8 | 2.3E−17 | 1.2E−13 | |
| GO:0005634∼nucleus | 0.66 | 2.5 | 1.9E−15 | 1.7E−12 | ||||
| Ser | 35 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.44 | 3.6 | 4.7E−06 | 2.4E−02 | |
| GO:0005634∼nucleus | 0.53 | 2.2 | 3.0E−04 | 2.3E−01 | ||||
| ctg | Leu | 69 | membrane location | GO:0031224∼intrinsic to membrane | 0.63 | 2.1 | 5.0E−08 | 4.3E−05 |
| cgg | 5′ UTR | 134 | protein ST kinase activity | GO:0004674∼protein serine/threonine kinase activity | 0.11 | 4.8 | 2.3E−06 | 6.7E−03 |
| Gly | 42 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.44 | 3.2 | 6.0E−06 | 3.1E−02 | |
| GO:0005634∼nucleus | 0.61 | 2.2 | 8.4E−06 | 7.2E−03 | ||||
| Ala | 40 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.28 | 2.5 | 6.1E−03 | ns | |
| GO:0005634∼nucleus | 0.43 | 1.9 | 3.3E−03 | ns | ||||
| ccg | 5′ UTR | 72 | No | |||||
| Ala | 37 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.61 | 4.8 | 5.5E−12 | 2.9E−08 | |
| GO:0005634∼nucleus | 0.72 | 2.7 | 1.6E−08 | 1.4E−05 | ||||
| Pro | 21 | transcription/nucleic acids binding/nucleus location | GO:0006355∼regulation of transcription. DNA-dependent | 0.45 | 3.3 | 1.5E−03 | ns | |
| GO:0005634∼nucleus | 0.60 | 2.1 | 5.1E−03 | ns |
anot significant.
Figure 4.
Many different groups of TNR-containing genes are associated with transcription-related functions. (A) Bar plot showing the overrepresentation (y-axis) of representative transcription-related GO terms in all analyzed groups of TNR-containing genes. TNR type, mRNA localization and coded AA are indicated on the x-axis. The type and number of GO terms are indicated in the graph legend. The P-value for individual associations is indicated on the graph. (B–D) To test whether transcription-related association depends on TNR length, we divided all genes belonging to the transcription-associated group into four length-defined classes [6 U (N = 167), 7 U (64), 8–9 U (66) and ≥10 U (47)]. The classes were so defined to obtain comparable class sizes sufficient for GO analysis. The bar plots show the association of genes containing increasingly long TNRs with GO:0006350, transcription; GO:0005634, nucleus and GO:0003677, DNA binding terms. The upper and lower panels show fold enrichment and the fraction of genes classified as related to the various GO terms, respectively.
As the properties of AAs do not explain the association of TNR-containing genes with transcription, we hypothesize that this association is related not to a simple excess of a specific type of AA but rather to properties shared by different (but not all; e.g. homo-Leu tracts) homo-AA tracts. It was shown earlier that the presence of such tracts may influence protein localization (40), interactions and aggregation (43), structure (38,44) and toxicity (45). It was also shown that the presence of homo-AA tracts in some proteins is highly conserved across eukaryotes (38). Analysis of the Protein Data Bank (PDB) showed a significant underrepresentation of homo-AA tract-containing proteins among proteins with solved structures (44,46,47). Even proteins with known structures have, in most cases, an unsolved region with homo-AA tracts. This suggests that homo-AA tracts in most proteins form unstable or disordered structures that can serve as flexible linkers or hinges (44) modulating structure and facilitating interactions with other macromolecules (38,44). Another feature of homo-AA tracts that may be implicated in transcription-related functions is the formation of charge clusters that lead to unusual charge distribution in proteins (39). Such charge clusters can be elements facilitating recognition of and interaction with other molecules. It has been shown that charge clusters are associated with transcriptional activation, membrane receptor activity and developmental regulation (39).
The only association of genes containing TNRs in ORFs that does not relate to transcription is the association of ctg TNRs coding homo-Leu tracts with membrane localization [Table 1, Supplementary Table S4 (ctg_ORF_L)]. The overrepresentation of homo-Leu tracts in membrane-associated proteins is most likely related to the high hydrophobicity of Leu. Hydrophobic AA runs are commonly found in transmembrane segments of receptors and other membrane-attached proteins.
We did not find any functional association with genes containing TNRs (att and gtt) in the 3′-UTR. This lack of association is in agreement with the fact that both att and gtt are underrepresented in exons (Figure 1). On the other hand, one TNR localized in the untranslated region that shows a functional association is cgg localized in the 5′-UTR [Supplementary Table S4 (cgg_5UTR)]. Genes containing cgg TNRs in the 5′-UTR are overrepresented in GO terms related to protein phosphorylation and kinase activity. This association shows that the function of TNRs can be expressed not only at the protein level but also at the level of RNA or DNA.
Some mRNAs contain multiple TNRs in various combinations
About 11% (96/878) of TNR-containing genes harbor more than one TNR (Supplementary Table S5). The number of genes containing multiple TNRs in exons (multi-TNR-genes) is shown in Figure 5A (prior to counting the multi-TNR-genes, we merged TNRs that apparently belong to longer TNR-tracts separated by interruptions; such TNRs represent ∼3% of all TNRs). Extreme examples of such genes are presented in Figure 5. The general distribution of TNR types in multi-TNR-genes is similar to the distribution of all TNRs in exons, with the most frequent being cgg, cag and ccg. In most cases (81/96), the TNRs co-occurring in multi-TNR-genes are of different types. The formal analysis of TNR co-occurrence did not show any significantly overrepresented TNR pair (pairs composed of TNRs of the same type were only slightly overrepresented). The functional classification (GO) of multi-TNR-genes also did not reveal significant disparity from the trends observed in other groups of TNR-containing genes. Although of weak power, the above facts suggest that the observed co-occurrence of TNRs in multi-TNR genes is rather random and that gene functionality does not favor specific TNR pairs.
Figure 5.
Genes with multiple TNRs. (A) Graph showing the number of genes with two or more TNRs. (B) The inset table characterizes the TNRs localized to genes containing the highest number (4 and 6) of TNRs. In the table, gene name, TNR type, TNR length, mRNA region, coded AA, genomic localization and genomic distance between successive TNRs are indicated. (C) Secondary structure of POU3F3 mRNA containing six TNRs (the simulation represents the lowest energy structure generated by the Mfold program).
The secondary-structure prediction of mRNAs containing multiple TNR tracts suggests that in specific mRNAs, fully complementary TNR sequences may interact with each other even if they are well separated in the mRNA sequence, thus making the mRNA structures more compact (Figure 5).
Functionality of TNRs can be expressed at the protein, DNA (genetic) and RNA levels
The most important argument for a functional role of homo-AA tracts comes from functional association analyses, both those published earlier (32,38,48) and those presented in our articles. Our results show that almost all groups of genes containing TNRs in the ORF associate with GO-defined terms, and these associations seem to be specific for TNRs localized in ORFs (Figure 4). Potential functions of homo-AA tracts were analyzed in several earlier publications (38,39,48). In this study, we discussed function of homo-AA tracts in the section describing results of functional associations. Although the AA-coding property seems to be the most important factor driving accumulation of TNRs in ORFs, the type of TNR coding for a specific homo-AA tract is not random and probably depends on TNR properties expressed either at the DNA (genetic) or at the RNA level. Similar conclusions can be drawn from results showing that the homogeneity of TNRs coding for AA tracts is higher than that of TNRs localized in the genome (49). Another result suggesting the functionality of TNRs on the DNA and/or RNA level is the functional association of genes containing cgg in the 5′-UTR with GO-defined protein serine/threonine kinase activity.
The function of TNRs at the DNA (genetic) level is probably related mostly to their high mutability. Although mutations are usually associated with deleterious effects, several elegant lines of evidence suggest a beneficial role for the high mutability of microsatellites. A high mutation rate of microsatellites can increase plasticity and facilitate adaptation of certain classes of genes during evolution (49–53). For example, microsatellites located in rapidly evolving developmental genes were shown to differ significantly between morphologically different breeds of dogs (50) and may be considered a major source of phenotypic variation in evolution, facilitating a rapid response to selective pressure (49,50,54). The potential of microsatellites to act as ‘advantageous mutators’ or ‘facilitators of evolution’ was recently discussed in two excellent review articles (34,55).
TNRs located in RNA can also modulate many different functions on the molecular level. It was shown that TNRs and other types of microsatellites in RNA can regulate gene expression (35,56–59), serve as protein binding sites (60,61) and splicing enhancers (62), induce transcription slippage and influence RNA stability (63,64). The above functions are related mainly to microsatellites localized in untranslated portions of transcripts [reviewed in ref. (65)]. The feature that may contribute most to the function of TNRs in RNA is their structure. The functional role of structures formed by TNRs is strongly supported by the correlation of the structure-forming potential of TNRs (66–68) with their overrepresentation in exons. As shown in Figure 1C, the five TNR types most overrepresented in exons form stable hairpin structures in transcripts (66,67). These include four cng (n: any nucleotide) repeats and gac repeats. The latter is of very low frequency in the genome. On the other hand, TNRs that remain single-stranded are strongly underrepresented in exons. This suggests that hairpin-forming repeats have functional roles in the regulation of gene expression (66,69). The situation is less clear for the G-rich repeats agg and tgg (ugg) that form G-quadruplex structures in RNA (68,70). The former is 4.5-fold overrepresented and the latter 3.1-fold underrepresented in human exons. Beside their abundance in ORFs (85%), agg repeats are also frequent in the 5′-UTRs of mRNAs (20%), and they may, like other G-quadruplexes at this location (71), be involved in translational regulation. In contrast, ugg repeats are practically absent in transcripts. It should also be noted that structure-forming properties expressed at the RNA level correlate well with similar properties in DNA single strands (22); thus, it cannot be excluded that DNA structure contributes to TNR functionality.
Although in this study we focused mostly on TNRs localized in exons, TNRs outside exons can also be important functional elements. These TNRs may influence gene expression if localized in promoters, splicing if present in introns, and local chromosome structure, DNA–protein interactions, recombination and other functions.
CONCLUSIONS
In this study, we have shown that the occurrence of TNRs in exons is strongly biased compared with their genomic frequency. Some TNR types are strongly overrepresented (CGG > CAG > GAC > AGG) while others are underrepresented (AAT > AAC > AAG) in exons. This result is a simple and direct argument supporting the notion that TNRs are important functional genetic elements undergoing strong selective pressure (34,49,50,54,55,72). Our results, along with various lines of evidence reported previously [e.g. (38,39,48,50,59,73,74)], allow us to conclude that the functionality of TNRs can be expressed at the protein, DNA (genetic) and RNA levels.
Regardless of the level at which the functionality of TNRs predominates, our results suggest that TNRs are potential functional genetic elements whose polymorphism can modulate many common phenotypes. Therefore, we propose that polymorphic TNRs should be considered as priority variants in association studies. The low number of high-ranked phenotype-TNR associations identified thus far probably results from the fact that most association studies have focused on easily genotyped SNPs. Moreover, little is known about the genome-wide linkage disequilibrium (LD) between TNR polymorphisms and SNP markers. Nevertheless, several reports have shown associations of TNRs with complex human phenotypes/diseases (49,75–77). Among the most striking are the associations of CAG TNRs localized in the androgen receptor (AR) gene with male infertility (78,79) and prostate cancer (80).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Sixth Research Framework Programme of the European Union, Project RIGHT (LSHB-CT-2004-005276); Ministry of Science and Higher Education (Grant No. PBZ-MNiI-2/1/2005, N N302 278937); Operational Programme ‘Innovative economy’ (POIG.01.03.01-00-098/08). Funding for open access charge: Operational Programme ‘Innovative economy’ (POIG.01.03.01-00-098/08).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 2000;10:62–71. [PMC free article] [PubMed] [Google Scholar]
- 2.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pumpernik D, Oblak B, Borstnik B. Replication slippage versus point mutation rates in short tandem repeats of the human genome. Mol. Genet. Genomics. 2008;279:53–61. doi: 10.1007/s00438-007-0294-1. [DOI] [PubMed] [Google Scholar]
- 4.Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madsen BE, Villesen P, Wiuf C. Short tandem repeats in human exons: a target for disease mutations. BMC Genomics. 2008;9:410. doi: 10.1186/1471-2164-9-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borstnik B, Pumpernik D. Tandem repeats in protein coding regions of primate genes. Genome Res. 2002;12:909–915. doi: 10.1101/gr.138802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 8.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 9.Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. The 1993-94 Genethon human genetic linkage map. Nat. Genet. 1994;7:246–339. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- 10.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 11.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 12.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagerman RJ, Leavitt BR, Farzin F, Jacquemont S, Greco CM, Brunberg JA, Tassone F, Hessl D, Harris SW, Zhang L, et al. Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation. Am. J. Hum. Genet. 2004;74:1051–1056. doi: 10.1086/420700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 15.Napierala M, Michalowski D, de Mezer M, Krzyzosiak WJ. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 17.THDCRG. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 18.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 19.Dennis G., Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bacolla A, Larson JE, Collins JR, Li J, Milosavljevic A, Stenson PD, Cooper DN, Wells RD. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008;18:1545–1553. doi: 10.1101/gr.078303.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark RM, Bhaskar SS, Miyahara M, Dalgliesh GL, Bidichandani SI. Expansion of GAA trinucleotide repeats in mammals. Genomics. 2006;87:57–67. doi: 10.1016/j.ygeno.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Clark RM, Dalgliesh GL, Endres D, Gomez M, Taylor J, Bidichandani SI. Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics. 2004;83:373–383. doi: 10.1016/j.ygeno.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Astolfi P, Bellizzi D, Sgaramella V. Frequency and coverage of trinucleotide repeats in eukaryotes. Gene. 2003;317:117–125. doi: 10.1016/s0378-1119(03)00659-0. [DOI] [PubMed] [Google Scholar]
- 27.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 31.Kalari KR, Casavant M, Bair TB, Keen HL, Comeron JM, Casavant TL, Scheetz TE. First exons and introns–a survey of GC content and gene structure in the human genome. In Silico Biol. 2006;6:237–242. [PubMed] [Google Scholar]
- 32.Butland SL, Devon RS, Huang Y, Mead CL, Meynert AM, Neal SJ, Lee SS, Wilkinson A, Yang GS, Yuen MM, et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozanska M, Sobczak K, Jasinska A, Napierala M, Kaczynska D, Czerny A, Koziel M, Kozlowski P, Olejniczak M, Krzyzosiak WJ. CAG and CTG repeat polymorphism in exons of human genes shows distinct features at the expandable loci. Hum. Mutat. 2007;28:451–458. doi: 10.1002/humu.20466. [DOI] [PubMed] [Google Scholar]
- 34.Fondon JW, 3rd, Hammock EA, Hannan AJ, King DG. Simple sequence repeats: genetic modulators of brain function and behavior. Trends Neurosci. 2008;31:328–334. doi: 10.1016/j.tins.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Raca G, Siyanova EY, McMurray CT, Mirkin SM. Expansion of the (CTG)(n) repeat in the 5′-UTR of a reporter gene impedes translation. Nucleic Acids Res. 2000;28:3943–3949. doi: 10.1093/nar/28.20.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 38.Faux NG, Bottomley SP, Lesk AM, Irving JA, Morrison JR, de la Banda MG, Whisstock JC. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlin S, Brocchieri L, Bergman A, Mrazek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl Acad. Sci. USA. 2002;99:333–338. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oma Y, Kino Y, Sasagawa N, Ishiura S. Intracellular localization of homopolymeric amino acid-containing proteins expressed in mammalian cells. J. Biol. Chem. 2004;279:21217–21222. doi: 10.1074/jbc.M309887200. [DOI] [PubMed] [Google Scholar]
- 41.Dorsman JC, Pepers B, Langenberg D, Kerkdijk H, Ijszenga M, den Dunnen JT, Roos RA, van Ommen GJ. Strong aggregation and increased toxicity of polyleucine over polyglutamine stretches in mammalian cells. Hum. Mol. Genet. 2002;11:1487–1496. doi: 10.1093/hmg/11.13.1487. [DOI] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oma Y, Kino Y, Toriumi K, Sasagawa N, Ishiura S. Interactions between homopolymeric amino acids (HPAAs) Protein Sci. 2007;16:2195–2204. doi: 10.1110/ps.072955307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins. 2002;48:134–140. doi: 10.1002/prot.10150. [DOI] [PubMed] [Google Scholar]
- 45.Oma Y, Kino Y, Sasagawa N, Ishiura S. Comparative analysis of the cytotoxicity of homopolymeric amino acids. Biochim. Biophys. Acta. 2005;1748:174–179. doi: 10.1016/j.bbapap.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 46.Saqi M. An analysis of structural instances of low complexity sequence segments. Protein Eng. 1995;8:1069–1073. doi: 10.1093/protein/8.11.1069. [DOI] [PubMed] [Google Scholar]
- 47.Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J. Biomol. Struct. Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 48.Salichs E, Ledda A, Mularoni L, Alba MM, de la Luna S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5:e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wren JD, Forgacs E, Fondon JW, 3rd, Pertsemlidis A, Cheng SY, Gallardo T, Williams RS, Shohet RV, Minna JD, Garner HR. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 2000;67:345–356. doi: 10.1086/303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc. Natl Acad. Sci. USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fondon JW, 3rd, Garner HR. Detection of length-dependent effects of tandem repeat alleles by 3-D geometric decomposition of craniofacial variation. Dev. Genes Evol. 2007;217:79–85. doi: 10.1007/s00427-006-0113-4. [DOI] [PubMed] [Google Scholar]
- 52.Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP. Natural variation in a Drosophila clock gene and temperature compensation. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 53.Zamorzaeva I, Rashkovetsky E, Nevo E, Korol A. Sequence polymorphism of candidate behavioural genes in Drosophila melanogaster flies from ‘Evolution canyon’. Mol. Ecol. 2005;14:3235–3245. doi: 10.1111/j.1365-294X.2005.02616.x. [DOI] [PubMed] [Google Scholar]
- 54.Kashi Y, King D, Soller M. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 55.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Yamada N, Yamaya M, Okinaga S, Nakayama K, Sekizawa K, Shibahara S, Sasaki H. Microsatellite polymorphism in the heme oxygenase-1 gene promoter is associated with susceptibility to emphysema. Am. J. Hum. Genet. 2000;66:187–195. doi: 10.1086/302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, Sasaguri Y. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455:70–74. doi: 10.1016/s0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- 58.Toutenhoofd SL, Garcia F, Zacharias DA, Wilson RA, Strehler EE. Minimum CAG repeat in the human calmodulin-1 gene 5′ untranslated region is required for full expression. Biochim. Biophys. Acta. 1998;1398:315–320. doi: 10.1016/s0167-4781(98)00056-6. [DOI] [PubMed] [Google Scholar]
- 59.Lawson MJ, Zhang L. Housekeeping and tissue-specific genes differ in simple sequence repeats in the 5′-UTR region. Gene. 2008;407:54–62. doi: 10.1016/j.gene.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 60.Stallings RL. Distribution of trinucleotide microsatellites in different categories of mammalian genomic sequence: implications for human genetic diseases. Genomics. 1994;21:116–121. doi: 10.1006/geno.1994.1232. [DOI] [PubMed] [Google Scholar]
- 61.Richards RI, Holman K, Yu S, Sutherland GR. Fragile X syndrome unstable element, p(CCG)n, and other simple tandem repeat sequences are binding sites for specific nuclear proteins. Hum. Mol. Genet. 1993;2:1429–1435. doi: 10.1093/hmg/2.9.1429. [DOI] [PubMed] [Google Scholar]
- 62.Gorbunova V, Seluanov A, Dion V, Sandor Z, Meservy JL, Wilson JH. Selectable system for monitoring the instability of CTG/CAG triplet repeats in mammalian cells. Mol. Cell Biol. 2003;23:4485–4493. doi: 10.1128/MCB.23.13.4485-4493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gay E, Babajko S. AUUUA sequences compromise human insulin-like growth factor binding protein-1 mRNA stability. Biochem. Biophys. Res. Commun. 2000;267:509–515. doi: 10.1006/bbrc.1999.2000. [DOI] [PubMed] [Google Scholar]
- 64.Fabre E, Dujon B, Richard GF. Transcription and nuclear transport of CAG/CTG trinucleotide repeats in yeast. Nucleic Acids Res. 2002;30:3540–3547. doi: 10.1093/nar/gkf483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 66.Krzyzosiak WJ, Sobczak K, Napierala M. In: Genetic Instabilities and Neurological Diseases. Wells RD, Ashizawa T, editors. Burlington, San Diego, London: Academic Press; 2006. pp. 705–713. [Google Scholar]
- 67.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sobczak K, Michlewski G, De Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.078790. doi: 10..1074/jbc.M1109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, Krzyzosiak WJ. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishikawa F, Murakami K, Matsugami A, Katahira M, Nishikawa S. Structural studies of an RNA aptamer containing GGA repeats under ionic conditions using microchip electrophoresis, circular dichroism, and 1D-NMR. Oligonucleotides. 2009;19:179–190. doi: 10.1089/oli.2008.0167. [DOI] [PubMed] [Google Scholar]
- 71.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.King D, Soller M, Kashi Y. Evolutionary tuning knobs. Endeavour. 1997;21:36–40. [Google Scholar]
- 73.Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res. 2008;18:1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc. Natl Acad. Sci. USA. 2009;106:17095–17100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blomberg Jensen M, Leffers H, Petersen JH, Daugaard G, Skakkebaek NE, Rajpert-De Meyts E. Association of the polymorphism of the CAG repeat in the mitochondrial DNA polymerase gamma gene (POLG) with testicular germ-cell cancer. Ann. Oncol. 2008;19:1910–1914. doi: 10.1093/annonc/mdn407. [DOI] [PubMed] [Google Scholar]
- 76.Gysin R, Kraftsik R, Sandell J, Bovet P, Chappuis C, Conus P, Deppen P, Preisig M, Ruiz V, Steullet P, et al. Impaired glutathione synthesis in schizophrenia: convergent genetic and functional evidence. Proc. Natl Acad. Sci. USA. 2007;104:16621–16626. doi: 10.1073/pnas.0706778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Y, Yang Y, Zhang X, Yan C, Xi S, Kang J. Relationship of the CAG repeat polymorphism of the MEF2A gene and coronary artery disease in a Chinese population. Clin. Chem. Lab. Med. 2007;45:987–992. doi: 10.1515/CCLM.2007.159. [DOI] [PubMed] [Google Scholar]
- 78.Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, Yong EL. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J. Clin. Endocrinol. Metab. 1997;82:3777–3782. doi: 10.1210/jcem.82.11.4385. [DOI] [PubMed] [Google Scholar]
- 79.Davis-Dao CA, Tuazon ED, Sokol RZ, Cortessis VK. Male infertility and variation in CAG repeat length in the androgen receptor gene: a meta-analysis. J. Clin. Endocrinol. Metab. 2007;92:4319–4326. doi: 10.1210/jc.2007-1110. [DOI] [PubMed] [Google Scholar]
- 80.Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc. Natl Acad. Sci. USA. 1997;94:3320–3323. doi: 10.1073/pnas.94.7.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.