Abstract
LINE-1 expression damages host DNA via insertions and endonuclease-dependent DNA double-strand breaks (DSBs) that are highly toxic and mutagenic. The predominant tissue of LINE-1 expression has been considered to be the germ line. We show that both full-length and processed L1 transcripts are widespread in human somatic tissues and transformed cells, with significant variation in both L1 expression and L1 mRNA processing. This is the first demonstration that RNA processing is a major regulator of L1 activity. Many tissues also produce translatable spliced transcript (SpORF2). An Alu retrotransposition assay, COMET assays and 53BP1 foci staining show that the SpORF2 product can support functional ORF2 protein expression and can induce DNA damage in normal cells. Tests of the senescence-associated β-galactosidase expression suggest that expression of exogenous full-length L1, or the SpORF2 mRNA alone in human fibroblasts and adult stem cells triggers a senescence-like phenotype, which is one of the reported responses to DNA damage. In contrast to previous assumptions that L1 expression is germ line specific, the increased spectrum of tissues exposed to L1-associated damage suggests a role for L1 as an endogenous mutagen in somatic tissues. These findings have potential consequences for the whole organism in the form of cancer and mammalian aging.
INTRODUCTION
Long interspersed element-1, LINE-1 or L1, is an autonomous family of retroelements that is currently active in mammalian genomes (1). The human genome has accumulated about 500 000 L1 copies, amounting to 17% of genomic content (2). The majority of L1 inserts are 5′-truncated or rearranged (2); and as a result they are retrotranspositionally inactive. Approximately 3000 L1s in the human genome are full-length (i.e. they contain 5′- and 3′-UTRs and sequences in between without major rearrangement), with about 150 containing both intact open reading frames (ORF) 1 and 2, and about 100 additional elements maintaining only intact ORF2 (3). Both ORFs are required for L1 retrotransposition in cultured cells (4).
L1 expression in the germ line and cells that are closely associated with the germ line has been previously reported (5–7). It has been suggested that full-length L1 mRNA is expressed little, if at all, in somatic tissues (8,9), although it has generally been detected in somatic cells that underwent malignant transformation (10). Recent reports have shown, in addition to the germ line, some L1 protein expression in vascular endothelial cells of human male gonads, L1 RNA expression in lymphoblastoid cell lines, and L1 mobilization in the brains of L1-transgenic mice (7,11,12). Unmethylated L1 loci and L1 mobilization has been reported in normal human brain (13). Because the vast majority of the L1 RNA is spliced and/or prematurely polyadenylated (14,15), detection of L1 proteins in a cell is not a reliable indicator of the retrotransposition potential. Endogenous L1 elements (16), L1 elements transiently expressed in primary cells (17,18), and in somatic cells of transgenic mice (11,19–21), are capable of retrotransposition indicating that there are no intrinsic molecular limitations for L1 protein activity specific to somatic cells.
There are a broad range of factors that lead to DNA damage, both in the germ line and somatic cells. There has been a significant focus on exogenous (i.e. radiation and chemicals), as well as endogenous [replication errors and reactive oxygen species (ROS)], sources of somatic DNA damage. As demonstrated by the disease causing in vivo integration events and tissue culture experiments, expression of the functional L1 elements in human cells results in integration events of L1 as well as its parasites, short interspersed elements (SINEs) and presumably SVA elements. While retrotransposition of L1 elements requires production of the full-length L1 mRNA that contains both functional ORF1 and ORF2 proteins (4), SINE retrotransposons (such as Alu elements) rely only on the production of the functional L1 ORF2 protein in tissue culture-based assays (22). Alu retrotransposons have been much more successful than L1 in occupying the human genome (accumulating to over 1 000 000 copies) and causing over twice the number of diseases originated by L1 elements (23,24). This difference in the total genomic copy number of L1 and Alu elements may come from the variation in the retrotransposition efficiency, post-insertional selection, or both.
In addition to insertional mutagenesis, expression of the functional wild-type (wt) full-length L1, or L1 ORF2 protein alone, in human cancer cells induces DNA double-strand breaks (DSBs) (25–27) in great excess relative to the integration events detected under the same conditions (27). DNA DSBs are known to be highly toxic and mutagenic even when repaired by the wt DNA repair machinery in mammals [reviewed in ref. (28)]. The cellular response to DNA damage usually manifests itself in cell cycle arrest, cell death (apoptosis or necrosis), or senescence. L1 expression in human cancer cells has been reported to induce cell cycle arrest (27) and apoptosis (25,29).
Because L1 expression can contribute to DNA damage not only through insertional mutagenesis but also via generation of DSBs, a better understanding of the expression patterns of endogenous L1 elements, particularly because of the complex processing of their mRNA (14,15,30), would provide a more complete picture of the potential sites of L1-related damage. Our data demonstrate ongoing L1 expression in a broad spectrum of normal human tissues including adult stem cells. Both the expression levels and the L1 RNA processing vary dramatically among the tissues tested in this study. We provide experimental support that L1-related DNA damage in human somatic cells is not limited to the production of the full-length L1 mRNA because of the expression of the L1 splice transcript that likely contributes to the translation of the L1 ORF2 protein. The SpORF2 transcript is detected in numerous human tissues and the expression of SpORF2 mRNA exhibits tissues-specific variation. We also demonstrate that transient expression of the wt L1 or the splice SpORF2 product in normal human fibroblasts and adult stem cells leads to a senescence-like phenotype. Because L1 activity has been well established to contribute to germ line mutagenesis [reviewed in ref. (23)], our finding of somatic L1 expression in a number of human tissues suggests that L1 elements need to be considered as an endogenous mutagen not only in germ line but also in somatic tissues for humans. Our findings suggest that even low levels of somatic DNA damage due to L1 activity have the potential to contribute to genetic instability, aging, and age-related diseases, such as cancer.
MATERIALS AND METHODS
Cell culture
NIH 3T3 (ATCC CRL-1658), MCF7 (ATCC HTB-22), HeLa (ATCC CCL2) cells were maintained as described (14). Human fibroblasts BJ (HCA2) immortalized with human TERT (31) (a generous gift from Dr J. Campisi) were passaged as described (31) and cultured in 5% CO2 and 20 or 5% O2 for DNA damaging assay. Human mesenchymal stem cells (MSCs) were cultured and maintained as described (32). Frozen vials of extensively characterized human MSCs were used at passage 2–4. MSCs were obtained from the Tulane Center for Preparation and Distribution of Adult Stem Cells (http://www.som.tulane.edu/gene_therapy/distribute.shtml). The cells were recovered from the frozen vials as passage 2 cells after they reached ∼80% confluence in 8–9 days and replated at 100 cells/cm2 with a change in medium every 2–3 days for further analysis.
Alu retrotransposition assay
HeLa cells (500 000) per T75 (Corning) were transfected with 2 µg of tagged Alu expression vector (22) and 1 µg of human ORF2 LIS expression vector (see plasmid construction in Materials and methods in Supplementary Data) with 6 µl of Plus reagent and 6 µl of Lipofectamine (Invitrogen). Cells were selected with culture media containing geneticin (G418) at 400 µg/ml (GIBCO). After 12 days under selection cell colonies were fixed and stained. To test the activity of endogenously expressed L1 elements, HeLa cells were transfected with the Alu reporter gene and empty pCEP4 vector as described above. Two independent CsCl preparations of AluNeo and pCEP expression vectors were used for the experiments testing endogenous L1 activity in HeLa cells. Where standard deviations are stated at least an n of 3 was used to determine the indicated value.
Northern blots
RNA extractions and northern blot analysis was performed as previously described (14,15). Two confluent T75 flasks were used for cancer cell lines for the analysis of endogenous L1 expression. PolyA-selected RNA (Ambion) from various human tissues [human testicle catalog #7973, prostate #7989, liver #7961, lung #7969, thymus #7965, ovary (one of the samples was pooled from three different individuals) #7975, brain #7963, placenta #7951, spleen #7971, pancreas #7955, kidney #7977, adrenal #7995, cervix #7993 (pooled from five different individuals), heart #7967, skeletal muscle #7983, colon #7987, stomach #7997, esophagus #6843] was used for the analysis of endogenous L1 expression in human tissues. All tissues were listed as normal. More than one sample was tested for esophagus, ovaries, testis, prostate and brain. Where standard deviations are stated for cancer cell lines at least an n of 3 was used to determine the indicated value. L1 RNA profiles shown in Supplementary Figure S1A are from different individuals than the RNA used for northern blots shown in Figure 1, one of the lanes for testis RNA in Supplementary Figure S1A contains RNA from the same individual shown in Figure 1.
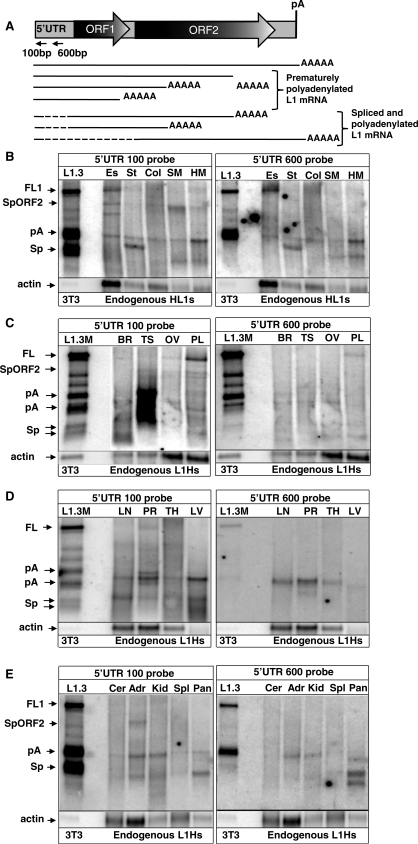
Figure 1.
Endogenous L1 expression in normal human tissues. (A) A schematic representation of the L1 structure and some transcription products detected in normal tissues and cancer cells. Full-length L1 (FL1) element contains a 5′-UTR, two open reading frames 1 and 2 (ORF1 and 2), 3′-UTR and a polyadenylation signal (pA). L1 transcription results in the production of the full-length L1 (FL1) mRNA, prematurely polyadenylated mRNAs (pA), and spliced and polyadenylated mRNAs (Sp), one of which has the potential to express L1 ORF2 protein alone (SpORF2) as confirmed by its capability to mobilize Alu in a tissue culture assay (see Figure 4B). Dashed lines correspond to the L1 sequences that are removed by splicing. Horizontal arrows indicate relative positions of the strand-specific 5′-UTR 100 bp and 5′-UTR 600 bp probes used for northern blot analysis in this study. (B–E) Northern blot analysis of the endogenous L1 expression in various adult human tissues. Es, esophagus; St, stomach; Col, colon; SM, skeletal muscle; HM, heart muscle; Cer, cervix; Adr, adrenal gland; Kid, kidney; Spl, spleen; Pan, pancreas; Br, brain; Ts, testis; Ov, ovaries; Pl, placenta; LN, lung; PR, prostate; TH, thymus; LV, liver. The very left lane shows L1.3 wt (L1.3) and mutant (L1.3 M) that contains a mutation in the strongest polyA site (15) expression profiles in NIH 3T3 cells transiently transfected with these human L1 expression vectors. L1 transcription results in the production of the full-length L1 (FL1) mRNA, prematurely polyadenylated mRNAs (pA), and spliced and polyadenylated mRNAs (Sp) one of which has the potential to express L1 ORF2 protein (SpORF2). Actin denotes β-actin mRNA detected with a strand-specific probe.
RT–PCR
A reverse transcription kit (Promega) was used for reverse transcriptase (RT)–PCR amplification of RNA samples from different tissues according to the manufacture’s protocol. PolyA-selected RNA, 2 µl (1 µg), from testis, ovary, placenta and adrenal gland (Ambion) was used for each reverse transcriptase reaction performed with ORF2(−) primer (all primer sequences are listed in Supplementary Figure S5). RNA from the same individuals was used for RT–PCR and northern blot analysis. Ten micro liters of the diluted RT reaction was used in the total volume of 100 µl with the forward HindIII (+) primer complementary to the first 22 bp of the L1.3 5′-UTR and nORF2(−) primer. The resulting PCR reaction, 2 µl, was used for the second round PCR performed with nested forward and nORF2(−) primers. PCR products were analyzed by gel electrophoresis. Bands were excised, gel purified by QIAquick Gel extraction kit (QIAGEN), cloned into TOPO-2.1 vector (Invitrogen) and sequenced (TGEN, AZ). Identification of the matching genomic L1 loci was done by BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat).
Plasmid construction
See ‘Materials and Methods’ section in Supplementary Data for details.
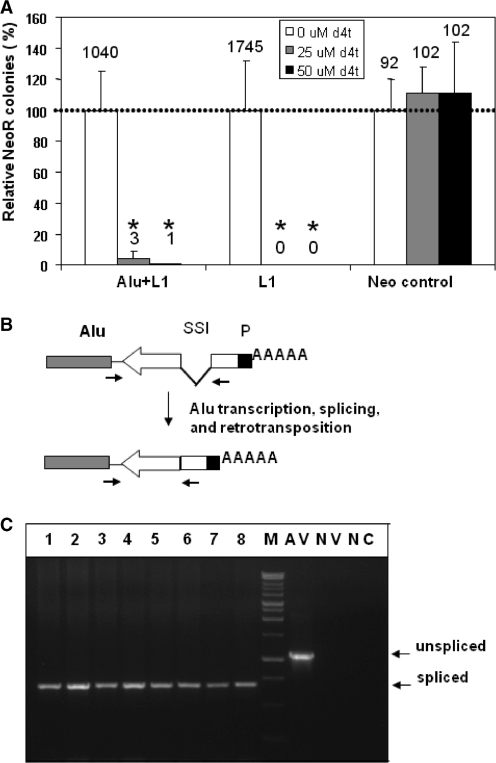
d4T treatment
In order to determine the effective dose of the d4T treatment HeLa cells transiently transfected with a neomycin tagged L1 (L1), with a neomycin tagged Alu (Alu) and a non-tagged L1 expression vector (Alu+L1), or an unrelated plasmid containing the neomycin resistance gene (Neo control) were treated with different doses of 2′,3′-didehydro-3′-deoxy-thymidine, d4T, (0–50 µM) for 7 days as previously reported (33). Following the treatment, cells were grown in the presence of G418 for 14 days, after which the NeoR colonies were stained and counted (Figure 5A). A dose of 50 µM of d4T was used for 7 days in the follow up experiment to determine the dependence of the Alu NeoR generated colonies on the functional endogenous L1 reverse transcriptase activity.
Figure 5.
Endogenously expressed L1 ORF2 supports exogenous Alu retroposition in HeLa cells. (A) Effect of d4T on the neomycin resistant colony formation. HeLa cells transiently transfected with a neomycin tagged Alu supplemented with non-tagged L1 (Alu+L1), a tagged L1 (L1) or an unrelated plasmid containing neomycin resistance gene (Neo control) were treated with different doses of 2′,3′-didehydro-3′-deoxy-thymidine, d4T, (0–50 µM) for 7 days. Following the treatment, cells were grown in the presence of G418 for 14 days, after which the NeoR colonies were stained and counted. The results from the untreated (0 µM) cells were arbitrarily assigned as 100% (dashed line). The relative percentage means ± SD are shown. The total number of colonies observed for the different dose treatments are indicated above each column. No significant effect on the colony formation was detected for the d4T treated control (neo control) when compared to the untreated sample (P > 0.5, paired t-test). Both tested d4T doses significantly affected the ability of the tagged L1 and Alu vectors to generate Neo resistant colonies through the retrotransposition process (P ≤ 0.03, paired t-test astersik). (B) Schematic representation of the Alu expression vector (gray rectangle) tagged with neomycin gene (white arrow) interrupted by a self-spliced intron (SSI). P designates promoter that drives functional Neo gene expression only after successful retroposition. Black arrows indicate relative positions of the primers used in PCR amplification specifically designed to distinguish between a spliced retrotransposed insert and the original unspliced vector sequence [(AluNeo(+) and AluNeoEx1(−)]. Primer sequences are shown in Supplementary Figure S5. To increase specificity, the upstream primer is specific to the Alu construct, so that this pair of primers will not amplify NeoR gene present in other expression vectors to eliminate detection of any potential contamination from the neo/kan-expressing plasmids (Supplementary Figure S4). (C) PCR analysis of DNA recovered from the neomycin resistance (NeoR) colonies generated in HeLa cells without supplementation with the exogenous L1 expression vector. The size of the generated PCR product corresponds to the spliced NeoR gene in the Alu expression vector. AV is Alu expression plasmid (positive control for unspliced product). NV is non-Alu neomycin expression vector (negative control for specific Alu amplification). NC is negative control (PCR reaction without DNA template). M is DNA marker.
Neutral COMET assay
HeLa cells, 106, were plated per T25 18–20 h prior transfection with 3 µg of respective constructs by Lipofectamin reagent (Invitrogen). Cells were trypsinized 24 h post-transfection and resuspended in Ca- and Mg-free PBS (105 cells/ml). One half of the untreated cells was subjected to 5 Gy of ionizing radiation. Neutral Comet Assay was performed according to the directions supplied with Trevigen Reagent Kit for Higher throughput single cell gel electrophoresis assay (Trevigen, catalog #4252-040-K). For more details see ‘Materials and Methods’ section in Supplementary Data.
DNA damage assay
Normal human fibroblasts at 2500 per well (96-well plate) immortalized with Htert (31) were plated and transiently transfected the next day with 0.03 µg/well of the empty (pCEP4), L1wt or SpORF2 vectors by lipofectamine 2000 following the manufacturer’s protocol. Cells were grown in 5% CO2 and 5% O2. The transfection cocktail was left on the cells for 20 h after which the cells were fixed and probed with 53BP1 specific rabbit polyclonal antibody (Novus Biologicals) and Alexa 488 secondary anti-rabbit antibody (Invitrogen) according to the BD Biosciences protocol (http://www.bdbiosciences.com/cgi-bin/literature/archive?type=8&brand=5). Hoechst (Invitrogen) staining was done during the incubation with secondary Ab. Treatment with 1.85 mM of H2O2 for 10 min was used as positive control. Pictures of random fields were taken by a BDPathway 855 at 40× magnification.
Senescence assay
Fifty thousand BJ (HCA2) or human MSCs per well in the six-well plates were transfected with 0.1 µg of the respective DNA by Lipofectamine 2000 (Invitrogen) in 1 ml of media supplemented with serum for 5 h. Zeocin treatment included exposure to 400 µg/ml in serum-supplemented media for 5 h. Forty-eight hours post-transfection cells were fixed and stained for detection of senescence-associated β-galactosidase (SA-β−Gal) activity with Senescence Detection Kit (BioVision) according to the manufacturer’s protocol. The time of incubation at 37°C was adjusted to 6–10 h instead of overnight. Cells were analyzed under LEICA DMIRB microscope at 200× total magnification. Six random pictures were taken for each well and blue cells were scored in Microsoft Photodraw V2 program and expressed as a percentage of the total cells present in each field. Data were normalized to the percentage of the blue cells detected in the non-transfected control. Each experiment was repeated independently three times for each construct and cell type. MSCs from two independent donors were used.
RESULTS
L1 mRNAs are expressed in somatic human tissues
Previous reports that mouse L1 mRNA and protein expression is detected primarily in the germ line (5,6) lead to extrapolation that the human L1s follow the same pattern. We performed northern blot analysis of RNA extracts from normal human tissues with a strand-specific RNA probe complementary to the first hundred base pairs of the L1 sequence (Figure 1A, 5′-UTR 100 bp probe) that detects all known L1 mRNAs (14). We found highly variable expression of endogenous L1 among somatic human tissues, with the amount of the full-length (FL) L1 levels in some tissues comparable to those detected in cancer cells. Northern blot analysis demonstrated that relative expression (normalized to actin mRNA levels) of the FL L1 in esophagus, prostate, stomach and heart muscle is at about 80, 50, 150 and 200%, respectively, of the levels detected in HeLa cells (Figure 1B–D). The levels of the FL L1 in adrenal gland, spleen, kidney, cervix and several other tissues are below the sensitivity of the assay, and thus cannot be accurately assessed. The sum of all L1-related transcripts in testis is as much as an order of magnitude higher than in other tissues. This observation is consistent with the previous report of the 10-fold transcriptional activation of the L1 promoter by testis-determining factor gene SRY (34). Despite having the highest total L1 mRNA levels, testis FL L1 RNA production appears to be severely restricted by RNA processing. This result was confirmed by northern blot analysis of testis RNA extracts obtained from different individuals (Supplementary Figure S1A, out of the two lanes for testis RNA in this figure one is from the same individual as in Figure 1). In comparison, the proportion of the FL L1 mRNA relative to the total L1 RNA production in other tissues varied from almost undetectable levels of FL L1 RNA, as in the testis, to 16.6% (SD = 6.7) in the ovary (Supplementary Table S1).
All human somatic tissues evaluated in this study express L1-related mRNAs with varying degrees of processing. Some of the smaller L1-derived RNAs are ubiquitously expressed and comigrate with the previously identified processed L1 mRNAs produced by transiently transfected human wild-type and mutant L1.3 elements (L1.3 and L1.3M) (Figure 1) (14,15,30). Other transcripts exhibited tissue-specific expression patterns. Although it is possible that some of the bands that do not comigrate with the previously characterized L1 RNAs could represent hybrid transcripts between L1 and cellular genes, this seems unlikely given the low levels detected for such hybrid transcripts (14). To distinguish between spliced and polyadenylated L1 mRNAs, the same RNA blots were evaluated with the 5′-UTR 600 bp probe complementary to the L1 5′-UTR region often removed by splicing (14) (Figure 1). Our results demonstrate that L1 elements undergo splicing and polyadenylation in most of the tested human tissues (Figure 1). However, in some tissues, such as testis, splicing is more dominant than in others, such as stomach, that produces almost exclusively prematurely polyadenylated products.
Endogenous full-length L1 mRNA expression inversely correlates with the production of the spliced and polyadenylated L1 species
We have previously reported that splicing and premature polyadenylation attenuate full-length L1 mRNA production (14,15). The observation of differential L1 processing in normal human tissues suggests that the production of the full-length L1 mRNA in various tissues may depend not only on the L1 promoter strength but also on the amount of the RNA processing supported by a specific cell type. A primary example is testis where overall high expression of the L1-related mRNAs indicates efficient L1 transcription (Figure 1 and Supplementary Figure S1A), whereas the full-length L1 transcripts remain below the detection sensitivity of the assay.
To determine whether premature polyadenylation and splicing play a similar role in malignant cells, endogenous L1 RNA profiles were analyzed in breast and cervical cancer cell lines. Northern blot analysis demonstrated variation in the quantity and pattern of the truncated endogenous L1 products between the transformed cell lines in a manner similar to that observed in somatic tissues. The size and processing of the truncated L1 RNAs in HeLa cells correlates well with the transcripts detected for transiently transfected L1 elements [Supplementary Figure S1B and (14)]. The faster migrating bands in HeLa cells represent spliced L1 RNA as confirmed by northern blot analysis with the strand-specific 5′-UTR 600 bp probe. Similar to human somatic tissues, some of the processed L1 products were detected in all tested cell lines (Figure 2, pA).
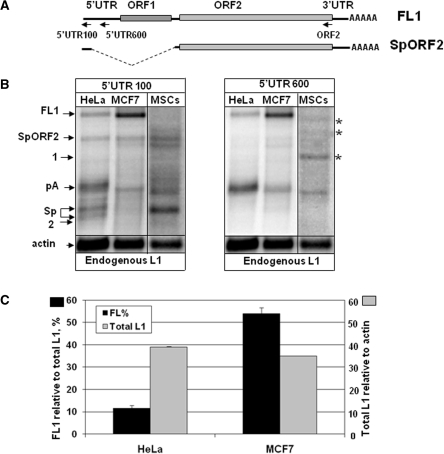
Figure 2.
Endogenous L1 expression in human adult stem cells and cancer cell lines. (A) Schematic of the full-length L1 element (FL1) and the ORF2 splice product (SpORF2). Arrows indicate the positions of the RNA strand-specific probes. (B) The left panel represents northern blot analysis of the RNA profiles of endogenously expressed L1 elements in human cervical (HeLa) and breast (MCF7) cell lines and human mesenchymal stem cells (MSCs) using the 5′-UTR 100 bp RNA strand-specific probe. The right panel is the same northern blots probed with the 5′-UTR 600 bp strand-specific RNA probe that detects polyadenylated, but not spliced, L1 mRNA. See Supplementary Figure S1B for the northern blot with the ORF2 probe. FL1, pA, Sp, SpORF2 and actin are marked as described in Figure 1. Asterisks mark bands specific to human MSCs. (C) Relative L1 expression and processing in MCF7 and HeLa cells. Black bars indicate the amount of the full-length L1 mRNA as a percentage of the total L1-related products detected in each cell line with the 5′-UTR 100 bp probe (scale shown at the left y-axis). Gray bars represent expression of all L1-related transcripts detected in each cell type relative to actin mRNA expression (scale shown at the right y-axis). Note that even though both cell lines express similar steady-state levels of the total L1 mRNAs, the full-length L1 mRNA composes only 10% of the total L1 products in HeLa cells compared to over 50% in MCF7 cells.
Quantitative evaluation of the L1-related products detected by northern blot analysis in human cancer cells shows that the MCF7 breast cancer cell line supports the highest full-length L1 expression. On the other end of the spectrum are HeLa cells that produce ∼15% of the full-length L1 relative to the L1 expression levels in MCF7 cells. Despite the over 5-fold difference in the relative full-length L1 mRNA production between MCF7 and HeLa cells, the expression of the total L1-related products are roughly the same in the two cell lines (Figure 2C, total L1, gray bars), suggesting relatively equal steady-state presence of L1 mRNAs in these cell lines. In part, the observed discrepancy is due to the fact that ∼50% of the mRNA products in MCF7 cells are full-length, while only 10–15% of the L1-related species are full-length in HeLa cells (Figure 2C, FL%, black bars), indicating that there is an inverse correlation between the amount of the truncated L1 species and the FL RNA. These results are comparable to those in human somatic tissues where placenta and ovary generated the lowest amount of the truncated L1 mRNAs with the highest FL L1 expression relative to the total L1 transcripts [16.5 and 16.6% (SD 6.7), respectively] (Supplementary Table S1).
These data demonstrate that even though L1 promoter strength is an imperative determinant of the L1 expression, post-transcriptional L1 mRNA processing is a powerful mechanism that can ultimately control the full-length L1 expression not only in normal but also in malignant human cells.
Human adult stem cells support endogenous L1 expression
Because differentiated somatic cells originate from adult stem cells, we wished to determine whether adult stem cells support endogenous L1 expression. Northern blot analysis of the L1 expression and RNA profiles was carried out for human bone marrow-derived MSCs (32). These cells produce endogenous L1 transcripts with very extensive RNA processing (Figure 2B). Northern blot analysis with the 5′-UTR 600 bp strand-specific probe demonstrated that MSCs support efficient production of the processed mRNAs consistent with the characterized in somatic and cancer cells (Figure 2B, 5′-UTR600 bp probe). They also produce processed L1 mRNAs that are specific to MSCs (Figure 2, bands marked with asterisks). Quantitative analysis of the northern blot results demonstrated that expression of the L1-related species in MSCs is at 20% of the total steady-state levels of the L1 expression detected in MCF7 cells. However, due to the extensive RNA processing, the amount of the full-length L1 transcript is below the sensitivity of the assay in adult stem cells and, therefore, cannot be quantitatively evaluated. These data indicate that efficient L1 expression is supported in a broad spectrum of human somatic tissues including adult stem cells, with highly variable post-transcriptional processing. This processing appears to represent one of the major variations in full-length L1 mRNA expression and presumably activity.
Different somatic tissues express endogenous L1 splice variants with potential biological relevance
We have previously reported functional splice sites within L1 sequence usage of which can remove the entire ORF1 sequence but leave ORF2 intact (14) generating the SpORF2 product. The resulting mRNA would have the potential to make functional L1 ORF2 protein independent of the FL L1 mRNA (Figures 1 and 2). One of the products detected in some normal human tissues, adult stem cells and human cancer cell lines was consistent with the SpORF2 RNA species based on its size and detection with the 5′-UTR 100 bp and ORF2 probes but not with the 5′-UTR 600 bp probe (Supplementary Figure S1B). The amount of this mRNA relative to the amount of the FL L1 mRNA or the total L1-related products varies significantly among human tissues (Supplementary Table S1).
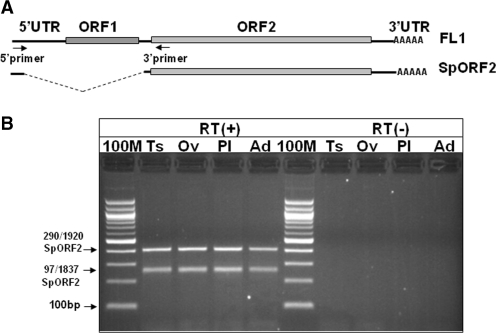
Usage of the splice sites to generate SpORF2 mRNA in normal tissues was confirmed by sequencing of the cDNA obtained by RT–PCR amplification using primers complementary to the beginning of the L1.3 5′-UTR and L1 ORF2 (Figure 3A). RT–PCR produced two L1-specific bands in testis, ovary, placenta and adrenal gland (Figure 3B), one of which was of the expected size for the previously reported SpORF2 identified in HeLa cells (14). Sequence analysis of independent clones confirmed that the faster migrating band is a product of L1 splicing from position 97 to 1837 (coordinates are given relative to the L1.3 accession L19088) as documented previously (14). The slower migrating band is a product of splicing from position 290 to 1920. This second L1 mRNA is also predicted to support translation of ORF2. The same bands were detected by RT–PCR in human MSCs and MCF7 cells (Supplementary Figure S2A). Recovered splice junction sequences matched active (L1Hs) and inactive L1 subfamilies consistent with the previous reports of multiple L1 loci expressed in human cancer and embryonic stem cells (10,35) (Supplementary Figure S2B and C). In addition to the youngest L1 loci that can generate full-length L1 ORF2 this limited cDNA analysis identified L1 loci that can express 3′-truncated ORF2 proteins. Even though retrotranspositionally incompetent, these proteins retain the endonuclease (EN) domain of the L1 ORF2. L1 EN expressed as a truncated fragment of the L1 ORF2 has been shown to possess DNA damaging activity in vitro (36). These data indicate that splice variants that have a potential to express L1 ORF2 independent of ORF1 are likely to be commonly expressed in human somatic tissues including those that do not demonstrate SpORF2 expression by northern blot analysis (i.e. adrenal gland).
Figure 3.
Various adult human tissues support production of the spliced L1 transcript that has a potential to produce ORF2 protein. (A) Schematic of the full-length L1 element (FL1) and the spliced ORF2 transcript (SpORF2). Arrows indicate the positions of the RT–PCR primers. (B) RT–PCR analysis of the L1 splice products in human Ts, testis; Ov, ovary; Pl, placenta; Ad, adrenal gland. RT(+) and RT(−) panels indicate reactions with and without RT, respectively. 100 M, 100 bp marker. The two differentially migrating bands represent L1 splice products utilizing various L1 splice donor and acceptor sites (indicated on the left).
Northern blot analysis with the strand-specific 100 bp 5′-UTR RNA probe was used to determine the relative expression of the SpORF2 product among human somatic tissues as a function of the actin mRNA expression (Figure 1). This quantitative evaluation determined that esophagus, prostate and lung support expression of the L1 SpORF2 mRNA at the levels very similar to those detected in HeLa cells (90, 70 and 80% respectively). In contrast, heart muscle and testis produce 5- 6-fold more SpORF2 mRNA than HeLa cells, while thymus, placenta and stomach generate intermediate levels of this product. As much of the SpORF2 product as the FL L1 mRNA is detected in HeLa cells and heart muscle, while testis, lung and adrenal gland support significantly higher steady-state levels of the SpORF2 mRNA than the FL L1 transcript (Figure 1 and Supplementary Figure S1). The SpORF2 product in the human MCF7 cancer cell line is produced at 26 ± 3.4% of the endogenous FL L1 mRNA detected in these cells. Relative SpORF2 expression in HeLa and MCF7 cells is very similar despite the significant difference in the full-length L1 mRNA expression between the two cell lines. These data suggest potential tissue-specific variation in the cellular exposure to L1-induced damage, depending on the amount of the FL L1 and SpORF2 products in both normal and cancer cells.
Expression of SpORF2 splice product can support Alu mobilization
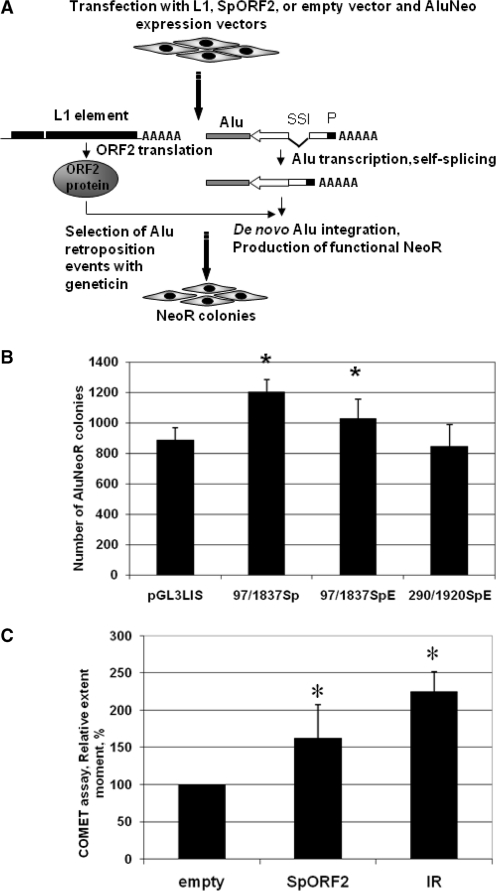
Because the SpORF2 RNA 5′-UTR contains a potential alternative AUG, expression of this RNA may not lead to the efficient L1 ORF2 translation. To confirm that identified spliced variants can effectively translate L1 ORF2 protein when they precede functional L1 ORF2 sequence, recovered cDNAs of spliced 5′-UTRs were subcloned upstream of an active L1 ORF2 inserted into a mammalian expression vector driven by the SV40 promoter (Supplementary Figure S3A). Sp97/1837 (recovered from NIH 3T3 cells transiently transfected with L1.3), Sp97/1837E and Sp290/1920E (recovered from endogenously expressed L1s in somatic human tissues) constructs reconstitute the spliced 5′-UTR of the SpORF2 mRNA products detected in vivo. We used an Alu retrotransposition assay, which requires only the L1 ORF2 protein in cultured human cells (22) to generate Alu retrotransposition-dependent neomycin-resistant (NeoR) colonies (Figure 4A), as an indicator of the functional L1 ORF2 translation from these constructs (22). Alu retrotransposition assays in HeLa cells demonstrated that splice variants support Alu mobilization in HeLa cells as well as the expression vector containing no L1 sequences upstream of the L1 ORF2 (pGL3LIS) (Figure 4B and Supplementary Figure S3B). Although FL L1 mRNAs are almost certainly crucial for L1 retrotransposition, our data demonstrating the ability of the spliced product, SpORF2, to make functional ORF2 protein indicate that damage from Alu insertion may occur independently of the FL L1 RNA expression and L1 retrotransposition. This discovery contrasts with previous assumptions that the only source of functional ORF2 is the FL L1 mRNA (37,38). A potential of the functional L1 ORF2 production by the retrotranspositionally incompetent full-length L1 mRNA independent of the ORF1 translation has been previously suggested (22). The presence of the L1 splice product, SpORF2, in a variety of somatic tissues and cancer cells provides and alternative mechanism for the L1 ORF2 production by both retrotranspositionally competent and incompetent transcriptionally active L1 loci containing wt ORF2 sequence.
Figure 4.
SpORF2 splice product can produce functional L1 ORF2 protein. (A) Experimental approach for L1 ORF2-dependent Alu retroposition. An Alu element (gray box) tagged with a backwards neomycin resistance (NeoR) gene (22) (white arrow indicating the orientation of NeoR transcription) expression of which is driven by a promoter (black box marked with P) in the orientation opposite of the Alu transcription. NeoR gene ORF is interrupted by a self-splicing intron that is removed upon Alu transcription and a functional NeoR gene can be expressed upon tagged Alu retroposition. The tagged Alu expression cassette is transfected into HeLa cells with the wt L1, SpORF2 expression vectors or tagged Alu vector alone, which detects ‘background activity’ generated by the endogenously expressed L1 ORF2. (B) SpORF2 L1 RNA products can produce functional ORF2 protein to drive Alu retroposition in tissue culture. A mean± SD of NeoR colonies for each construct is shown (see Supplementary Figure S3A for construct design and Supplemenatry Figure S3B for additional controls). (C) Transient expression of SpORF2 splice product in HeLa cells leads to DNA damage. HeLa cells were transiently transfected with the empty or SpORF2 expression vector, or subjected to 5 Gy of IR. Neutral COMET assay was performed and a mean± SD of the tail extent moment from three independent experiments is shown. Asterisks indicate statistically significant difference between the treatments and the control empty vector (t-test, P < 0.05).
Endogenously expressed L1s have a potential to produce functional ORF2 protein
Because many FL genomic L1 elements have mutations that inactivate their reading frames (2), having L1-related RNAs does not guarantee that they are making functional products. We hypothesized that HeLa cells can support exogenously expressed Alu retroposition if the functional L1 ORF2 protein is generated in these cells by either the full-length L1 mRNA or the SpORF2 product. HeLa cells transiently transfected with the Alu retrotransposition cassette described in Figure 4A (22) in combination with 1 µg of the vector expressing an exogenous L1 element (39) or the empty vector resulted on average in 1249 ± 69.3 and 52.6 ± 17.34 NeoR colonies, respectively.
To determine that generation of these NeoR colonies depends on the functional L1 RT activity, we used a HIV RT inhibitor 2′,3′-didehydro-3′-deoxy-thymidine, d4T or stavudine, which has been shown to inhibit L1 RT function in a competitive manner (33,40). These previous studies demonstrated that these levels of inhibitor do not have any adverse impact on cell growth or colony formation. Figure 5A demonstrates that treatment of HeLa cells with increasing doses of d4T resulted in the dramatic reduction of both L1, as well as Alu retrotransposition driven by exogenous L1 (Alu+L1), while the same treatment of the HeLa cells transiently transfected with a neomycin (Neo) expression vector had very little effect on the formation of the NeoR colonies through random integration. L1 and Alu element retrotransposition requires target-primed reverse transcription (TPRT) (41), the control transfection with unrelated Neo expression vector relies on the plasmid integration into the host DNA and does not depend on the presence of the functional RT. Equal numbers of colonies formed in HeLa cells following transient transfection with the Neo expression vector with, or without, d4T treatment indicating that the exposure to this chemical has no detectable toxic effect on the cells. Exposure to d4T of HeLa cells transiently transfected with the tagged Alu expression vector alone resulted in the disappearance of the NeoR colonies. The apparent inability of the exogenous tagged Alu to form NeoR colonies in the presence of the inhibitor of the L1 reverse transcriptase activity indicates that these Alu-generated colonies depend on the endogenous L1 RT activity, supporting endogenous expression of functional L1 ORF2 protein in HeLa cells.
To further characterize NeoR colonies formed by Alu without any exogenous source of L1, DNA from eight colonies generated in HeLa cells transfected with Alu and the empty vector was recovered and analyzed by PCR to evaluate the absence/presence of the self-splicing intron (Figure 5B and C, and Supplementary Figure S4). All evaluated colonies were confirmed as bonafide retrotransposition events. These data support endogenous expression of functional L1 ORF2 in HeLa cells and are consistent with the previous reports of the endogenous expression of the functional L1 elements in human (10,35) and rat (42) cells as well as mobilization of exogenously expressed Alu by the functional endogenous L1 proteins in mouse and human cells (22,43,44).
SpORF2 expression causes DNA damage
Previous reports demonstrated that expression of the wt L1 element, or L1 ORF2 alone, in cancer cells leads to DNA damage in the form of DSBs (25–27) that can trigger cell cycle arrest (27) or apoptosis (25,29). Because multiple somatic tissues and tumor cell lines support SpORF2 formation without, or in parallel with, the production of the full-length L1 mRNA, we wished to determine whether SpORF2 could produce DNA damage in these cells independent of the full-length L1. HeLa cells were transiently transfected with the SpORF2 construct and DNA damage was assessed by a neutral COMET assay that detects DNA DSBs (45). Gamma irradiation was used as a positive control. Figure 4C demonstrates that transient expression of the SpORF2 splice product resulted in an increase in the average tail moment that corresponds to DNA damage compared to the cells transfected with the empty vector. This result is consistent with the previous observation of γ-H2AX foci formation after L1 ORF2 expression in HeLa cells (27).
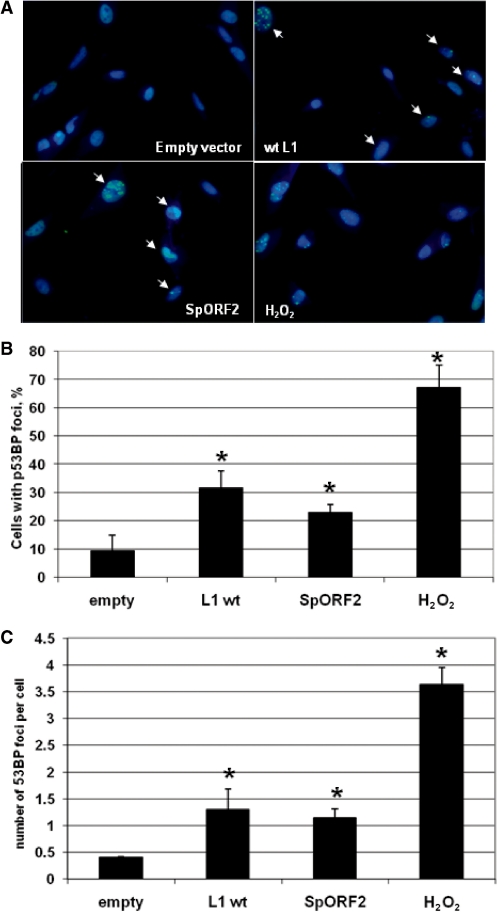
DNA damage was also assessed in normal human fibroblasts transiently transfected with empty vector or vectors expressing wt L1 or SpORF2 splice product by detecting p53-binding protein 1 (53BP1) foci in response to DNA DSBs with antibody specific to this protein. 53BP1 is one of the characterized responders recruited to the sites of DNA DSBs (46). Detection of 53BP1 foci with 53BP1 specific antibody demonstrates a significant increase in the number of cells with DNA DSBs as well as in the average number of breaks per cell in normal human fibroblasts transiently transfected with L1 and SpORF2 expression vectors relative to the empty vector control (Figure 6A–C). These data are consistent with the previous reports of L1-induced DNA damage in cancer cells and support the idea that SpORF2 expression in normal cells may contribute to genomic instability independent or in addition to the full-length L1-associated insult to DNA integrity.
Figure 6.
L1 and SpORF2 expression in normal human cells results in DNA damage. (A) Normal human fibroblasts transiently transfected with the empty vector or vectors expressing L1 or SpORF2 were analyzed for 53BP1 foci formation 21 h post-transfection. 53BP1 is one of the characterized responders recruited to the sites of DNA DSBs and it is commonly used as a marker for DNA DSBs. Treatment with 1.85 mM of H2O2 for 10 min was used as positive control. Overlay of Hoechst and antip53BP antibodies is shown. Arrows indicate cells with distinct staining foci. (B) Quantitation of the 53BP1 foci positive cells as a percent of the total cells (mean ± SD). Asterisks indicate significant difference determined by paired t-test with P = 0.024, 0.014, 0.001 for L1, spORF2 and H2O2, respectively. (C) Quantitative assessment of the average number of 53BP1 foci per cell (mean ± SD). Asterisks indicate significant difference determined by paired t-test with P = 0.003, 0.017 and 0.006 for L1, spORF2 and H2O2, respectively.
Expression of exogenous full-length L1 and SpORF2 in normal human fibroblasts, and adult stem cells results in the senescence-like phenotype
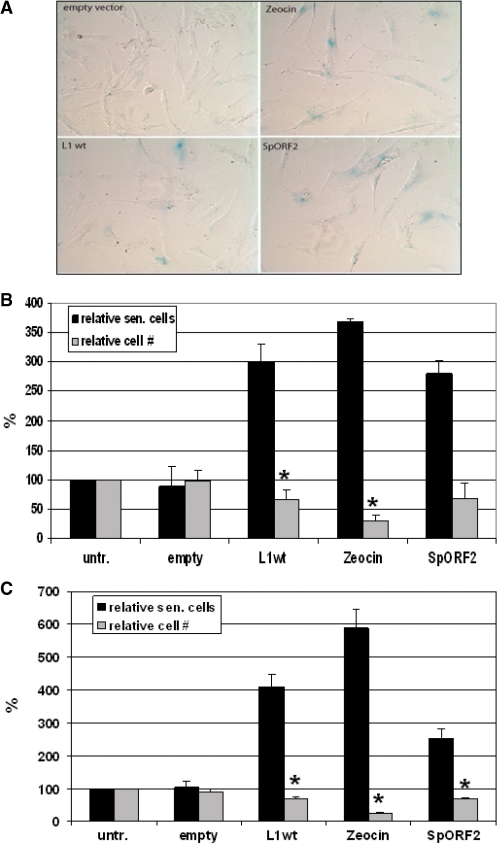
L1 elements have been reported to compromise the integrity of the cellular DNA via the introduction of DSBs (25–27). DSBs are normally highly toxic and mutagenic to mammalian cells and have previously been associated with apoptosis triggered by L1 expression (25,27). Another common cellular response to DNA damage often occurring in normal cells is senescence. Senescent cells express senescence-associated β-galactosidase detection of which is routinely used as a marker of cellular senescence (47). Exposure to a number of DNA damaging agents leads to cellular senescence (48). To determine whether creation of DNA damage in mammalian cells by L1 elements can also result in cellular senescence, normal human fibroblasts immortalized with human telomerase (31), and MSCs were transiently transfected with either empty vector, a vector carrying wt L1.3 element or an SpORF2 expression vector. For the positive control, cells were treated with a DNA damaging agent zeocin. Both cell types exhibited a senescence-like phenotype in response to the FL L1 expression and L1ORF2 as supported by the expression of senescence-associated β-galactosidase (Figure 7) (47). L1 transfected cells also showed some morphological characteristics of senescent cells much like fibroblasts treated with zeocin (49) (Figure 7A) and consistent with the data obtained for cancer cells (29). These data establish that expression of retrotranspositionally active L1s or ORF2 protein alone can compromise the integrity of the normal cell function and lead to a senescence-like phenotype reminiscent of the stress-induced premature senescence triggered by other DNA damaging agents (50).
Figure 7.
Full-length L1 and SpORF2 expression triggers senescence-like phenotype in normal human fibroblasts and human adult stem cells. (A) Normal human fibroblasts transfected with the empty vector, vectors expressing wt L1, or SpORF2, or treated with a DNA damaging agent zeocin. Blue color represents cells that express senescence-associated β-galactosidase. (B) Quantitation of the L1-induced senescence in normal human fibroblasts, BJ, immortalized with human telomerase. Black bars represent relative amount of senescent cells among human fibroblasts treated as described in (A). Grey bars represent relative amount of cells observed after each treatment in any given field. Asterisks indicate statistically significant difference between the cell numbers of cells transfected with the empty vector and cells transfected with the wt L1 expression cassette or zeocin treatment (t-test, P-value 0.014 and 0.00025, respectively). Untransfected (untr.) is mock-transfected cells. (C) Quantitation of the L1-induced senescence in human adult stem cells. Black and gray bars represent relative amount of senescent cells and total cells as described in (B). Asterisks indicate statistically significant difference between the cell numbers of cells transfected with the empty vector and cells transfected with the wt L1 expression cassette, zeocin treatment, or SpORF2 vectors (t-test, P-value 0.01, 0.0002 and 0.013, respectively).
DISCUSSION
The field of human mobile elements has often focused on their mutagenic effect within the germ line (5,6) because transposable element amplification to high copy number in the genome is dependent on germ line insertions. Detection of endogenous FL or SpORF2 L1 transcripts in this study in many tested adult tissues broadens the potential impact of mobile elements on human health and development. Our findings of modest levels of the FL L1 expression in human ovary and testis, and expression of only processed L1 mRNAs in kidney, lung, liver and brain are in agreement with the reported expression in a transgenic mouse model (8). However, sporadic tissues, such as esophagus, placenta and prostate demonstrate that some somatic tissues can express L1 elements fairly well. A recent report of L1 mobilization in a transgenic mouse model demonstrates higher L1 retrotransposition rates in somatic tissues than in the germ line (21), suggesting the possibility that expression of endogenous L1 elements in differentiated tissues may result in an appreciable amount of damage over time. Although FL L1 mRNAs are certainly the dominant source of L1 retrotransposition, generation of the SpORF2 transcript that can make functional ORF2 protein indicates that the FL L1 transcripts with both functional ORFs are not absolutely required to cause cellular damage. This contrasts with previous assumptions that functional ORF2 protein can only be produced from the FL mRNAs (37,38). It is apparent that detection of the L1 ORF2 protein does not necessarily guarantee the presence of the full-length L1 mRNA.
Insertion of L1 elements is not the only mechanism by which they may damage genomes. It is well accepted that L1 element expression is a necessary driver for insertion of Alu and presumably SVA elements (22,51), as well as processed pseudogenes (52). More importantly, only expression of the ORF2 of L1 is required to drive Alu mobilization (22). Thus, tissues that make spliced forms of L1 RNA but not the FL L1 transcripts may make functional ORF2 and cause mutations by driving Alu retrotransposition.
There are almost twice as many Alu inserts as L1 integrants that have been characterized as causing disease [reviewed in ref. (23)]. In addition, there are twice as many total Alu elements as L1 copies in the human genome (2). Even though differential post-integration selection against L1 and Alu inserts and a bias in detection of the disease causing integration events can certainly account for some of the disparity in their accumulation rate, it is also possible that the relative paucity of the full-length L1 mRNA in testis and expression of the SpORF2 splice product in both testes and ovaries may promote potentially higher rate of Alu retrotransposition relative to L1. Thus, the presence of the SpORF2 products in germ line may contribute to the higher copy number of Alu elements in the genome (2) and their higher contribution to disease (53) relative to L1 inserts.
Methylation and promoter strength were originally thought to be the only major regulatory mechanisms restricting L1 activity (54,55). Recently cellular factors, such as the APOBEC3 gene family and the nuclear excision repair endonuclease complex ERCC1/XPF (56) and the potential of RNAi (57), were shown to negatively modulate L1 retrotransposition (58–60). Even though transcriptional activity of the L1 promoter is almost certainly a crucial step in ensuring endogenous L1 expression, our data demonstrate that it is not the only mechanism controlling production of the retrotranspositionally active L1 mRNA. Although polyadenylation and splicing were previously shown to limit expression of L1 elements, this is the first demonstration that differential processing of endogenous L1 mRNA can account for considerable differences in the relative abundance of the full-length L1 transcript. Our studies demonstrate that in most tissues and cell lines, post-transcriptional regulation of L1 RNA is a major factor in controlling L1 expression (Figures 1 and 2).
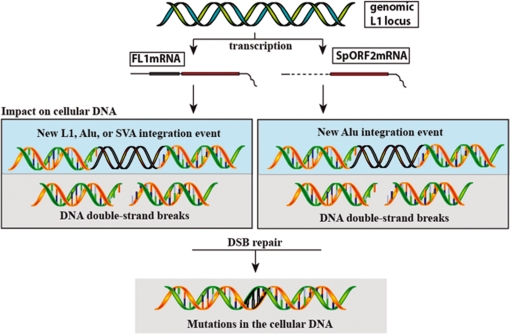
In addition to the mobilization of itself and its parasites, L1 activity also produces DNA DSBs (25–27). Consistent with the previous reports of DNA damage induced by either the wt L1 of ORF2 alone in cancer cells (27), we demonstrate that expression of the SpORF2 splice product in normal human fibroblasts leads to detectable DNA damage. This observation suggests that even very low levels of the L1 ORF2 activity in somatic tissues generated from either the full-length L1 or the SpORF2 splice product may contribute to the gradual accumulation of DNA damage during the lifespan of an individual. This damage can be in the form of de novo integration events of retroelements that can alter gene architecture and expression (14,61,62), point mutations resulting from the error-prone repair of ORF2-induced DNA lesions, or recombination events triggered by the ORF2 nicking activity [reviewed in ref. (63)]. Furthermore, the ability of L1 elements to produce the SpORF2 splice product means that even retrotranspositionaly incompetent L1 elements that maintain intact ORF2 can generate DNA damage (27,36) and mobilize Alu elements (22). As summarized in Figure 8 cells, such as adult stem cells, which predominantly produce the SpORF2 transcript are likely to endure little damage associated with the full-length L1 mRNA. While prostate, esophagus, ovaries and a number of other cell types that produce both the full-length and the SpORF2 mRNAs are possibly exposed to the full arsenal of the L1-related genomic instability. Thus, L1 elements may potentially contribute, in a way similar to ROS, to both the organismal aging process as well as to a number of age-related diseases potentially in a tissue-specific manner. It is even possible that overproduction of L1 elements in cells still expressing p53 (25) could lead to cell death that may serve as a defense mechanism against L1 activity promoting genomic instability that has a potential to contribute to malignant transformation. There is also potential for increased L1-related damage with age because of decreased DNA repair response in aging mammalian cells (64–66) or age-associated hypomethylation of genomic DNA (67).
Figure 8.
A summary of the biologically relevant L1-related mRNA products and their respective impact on the host genome. Transcription of the functional L1 locus results in the production of either the full-length mRNA (FL1mRNA), the splice ORF2 mRNA (SpORF2mRNA) or both. FL1mRNA protein products can mobilize L1, Alu, and SVA elements, while SpORF2mRNA only produces ORF2 protein and as a result can only assist Alu retrotransposition. Expression of either L1 mRNA can generate ORF2, which leads to introduction of DNA DSBs potentially resulting in accumulation of mutations in the cellular genome.
Overall, our data suggest that L1-induced damage to cells is not confined to germ line and it is likely not limited to fully active elements. In addition, there is a potential for the tissue-specific variation in the L1-associated damage depending on the spectrum of the L1-related molecules supported by individual cell types. Our observations combined with the reported variation of the combined L1 activity in the population (68) create the need to reevaluate the potential impact of many of the full-length L1 elements present in the human genome on the human health. Further studies providing a more comprehensive analysis of potential variation in somatic L1 expression among individuals in population will be helpful in ascertaining the impact of these elements on human health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01GM45668 to P.D., P20 RR020152 to P.D., V.P.B. and A.R-E., 5K01AG030074-02 to V.P.B., R01 GM079709-01 to A.M.R.-E.); National Science Foundation (EPS-0346411 to P.D.); State of Louisiana Board of Regents Support Fund (to P.D.); Ellison Medical Foundation New Scholar in Aging award (547305G1 to V.P.B.); Louisiana Cancer Research Consortium (LCRC) Competitive Advantage Award (to A.M.R.-E.); Tulane University Research Enhancement Funds (to A.M.R.-E.). Funding for open access charge: National Institutes of Health (US).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Baddoo and Tulane Proteomics Core for assistance with the COMET assays, Mary Price and LCRC/Tulane Cancer Center Cell Analysis Core for the p53BP foci staining, and Patrice Penfornis for handling of MSCs. We thank C. Bulot for help with CsCl DNA purifications. We are grateful to Dr Judith Campisi for sharing reagents.
REFERENCES
- 1.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Penzkofer T, Dandekar T, Zemojtel T. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 2005;33:D498–D500. doi: 10.1093/nar/gki044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 5.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 8.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH. A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Seluanov A, Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 12.Rangwala SH, Zhang L, Kazazian HH., Jr Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 2009;10:R100. doi: 10.1186/gb-2009-10-9-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 16.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- 17.Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, Seluanov A, Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol. Cell Biol. 2007;27:1264–1270. doi: 10.1128/MCB.01888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl Acad. Sci. USA. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An W, Han JS, Schrum CM, Maitra A, Koentgen F, Boeke JD. Conditional activation of a single-copy L1 transgene in mice by Cre. Genesis. 2008;46:373–383. doi: 10.1002/dvg.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 23.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 24.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum. Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 25.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34:1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11:S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 29.Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio MA, Kim SH, Campisi J. Reversible manipulation of telomerase expression and telomere length. Implications for the ionizing radiation response and replicative senescence of human cells. J. Biol. Chem. 2002;277:28609–28617. doi: 10.1074/jbc.M203747200. [DOI] [PubMed] [Google Scholar]
- 32.Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 33.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 36.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 37.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc. Natl Acad. Sci. USA. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, Ostrowski MA. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS ONE. 2008;3:e1547. doi: 10.1371/journal.pone.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 42.Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Lower R, Buschmann C, Horn AV, Traub P, Schumann GG. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan CR, Sheffield RF, Rudin CM. Human Alu element retrotransposition induced by genotoxic stress. Nat. Genet. 2003;35:219–220. doi: 10.1038/ng1259. [DOI] [PubMed] [Google Scholar]
- 45.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 46.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aoshiba K, Tsuji T, Nagai A. Bleomycin induces cellular senescence in alveolar epithelial cells. Eur. Respir. J. 2003;22:436–443. doi: 10.1183/09031936.03.00011903. [DOI] [PubMed] [Google Scholar]
- 49.Delacote F, Deriano L, Lambert S, Bertrand P, Saintigny Y, Lopez BS. Chronic exposure to sublethal doses of radiation mimetic Zeocin selects for clones deficient in homologous recombination. Mutat. Res. 2007;615:125–133. doi: 10.1016/j.mrfmmm.2006.11.028. [DOI] [PubMed] [Google Scholar]
- 50.Chen QM, Bartholomew JC, Campisi J, Acosta M, Reagan JD, Ames BN. Molecular analysis of H2O2-induced senescent-like growth arrest in normal human fibroblasts: p53 and Rb control G1 arrest but not cell replication. Biochem. J. 1998;332(Pt 1):43–50. doi: 10.1042/bj3320043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 53.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 54.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thayer RE, Singer MF, Fanning T. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–277. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- 56.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair. 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 58.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 60.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 61.Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ustyugova SV, Lebedev YB, Sverdlov ED. Long L1 insertions in human gene introns specifically reduce the content of corresponding primary transcripts. Genetica. 2006;128:261–272. doi: 10.1007/s10709-005-5967-2. [DOI] [PubMed] [Google Scholar]
- 63.Hedges DJ, Deininger PL. Inviting instability: transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 2006;616:46–59. doi: 10.1016/j.mrfmmm.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorbunova V, Seluanov A. Making ends meet in old age: DSB repair and aging. Mech. Ageing Dev. 2005;126:621–628. doi: 10.1016/j.mad.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, Ham YH, Kim TJ, Choi HM, Han GJ, Lee JH, et al. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp. Mol. Med. 2006;38:686–693. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- 66.Mayer PJ, Lange CS, Bradley MO, Nichols WW. Gender differences in age-related decline in DNA double-strand break damage and repair in lymphocytes. Ann. Hum. Biol. 1991;18:405–415. doi: 10.1080/03014469100001702. [DOI] [PubMed] [Google Scholar]
- 67.Mays-Hoopes L, Chao W, Butcher HC, Huang RC. Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Dev. Genet. 1986;7:65–73. doi: 10.1002/dvg.1020070202. [DOI] [PubMed] [Google Scholar]
- 68.Seleme MC, Vetter MR, Cordaux R, Bastone L, Batzer MA, Kazazian HH., Jr Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc. Natl Acad. Sci. USA. 2006;103:6611–6616. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.