Abstract
Post-transcriptional nucleoside modifications fine-tune the biophysical and biochemical properties of transfer RNA (tRNA) so that it is optimized for participation in cellular processes. Here we report the crystal structure of unmodified tRNAPhe from Escherichia coli at a resolution of 3 Å. We show that in the absence of modifications the overall fold of the tRNA is essentially the same as that of mature tRNA. However, there are a number of significant structural differences, such as rearrangements in a triplet base pair and a widened angle between the acceptor and anticodon stems. Contrary to previous observations, the anticodon adopts the same conformation as seen in mature tRNA.
INTRODUCTION
Transfer RNA (tRNA) has the vital role of delivering amino acids to the ribosome allowing the synthesis of proteins as coded by mRNA. After synthesis by RNA polymerase, each tRNA transcript is converted to a mature tRNA through processing and nucleoside modification. The mechanisms utilized for processing vary not just between the kingdoms of life but also between species, but nonetheless ensure that excess nucleotides in the form of introns and 5′ and 3′ extensions are removed and that the correct 3′ CCA extension required for aminoacylation is present (1,2). Nucleoside modification is an equally important process, bringing about the modification of around 10% of the nucleosides in a typical tRNA such that the final mature tRNA contains not only the four canonical nucleosides but a number of modified nucleosides (3). While nucleoside modification occurs in other RNAs, it is noteworthy that 92 of the 107 known modified nucleosides exist in tRNA making it the most diversely modified RNA (4).
Since the elucidation of the first crystal structure of yeast tRNAPhe in 1974, several crystal structures of free mature tRNAs in the absence of interaction partners have been determined (5,6). At the time of writing structures were available for: (i) yeast tRNAPhe in monoclinic (7–10) and orthorhombic forms (11), (ii) yeast tRNAAsp in two differing conformations (12), (iii) two crystal forms of Escherichia coli tRNAfMet in differing conformations (13), (iv) yeast  (14) and (v) Bos taurus tRNALys (15). X-ray structures revealed that the predicted cloverleaf structure of the tRNA formed by Watson–Crick base pairs folds into an L-shaped tertiary structure, which is further stabilized by a number of tertiary and triplet base pairs (16) and interactions with Mg2+ ions. The two structures of yeast tRNAPhe determined independently in 2000 benefited from the advances in macromolecular crystallography made since the initial 1970s study. The higher resolution of these structures led to a greater understanding of the roles played by water molecules and divalent metal ions like Mg2+ in stabilizing the tertiary structure of tRNA (7,8).
(14) and (v) Bos taurus tRNALys (15). X-ray structures revealed that the predicted cloverleaf structure of the tRNA formed by Watson–Crick base pairs folds into an L-shaped tertiary structure, which is further stabilized by a number of tertiary and triplet base pairs (16) and interactions with Mg2+ ions. The two structures of yeast tRNAPhe determined independently in 2000 benefited from the advances in macromolecular crystallography made since the initial 1970s study. The higher resolution of these structures led to a greater understanding of the roles played by water molecules and divalent metal ions like Mg2+ in stabilizing the tertiary structure of tRNA (7,8).
Mature E. coli tRNAPhe is 76-nt long and contains 10 modified nucleosides: 4-thiouridine (s4U) at position 8, dihydrouridine (D) at positions 16 and 20, pseudouridine (Ψ) at positions 32, 39 and 55, 2-methylthio-N6-isopentenyladenosine (ms2i6A) at position 37, 7-methylguanosine (m7G) at position 46, 3-(3-amino-3-carboxypropyl) uridine (acp3U) at position 47 and 5-methyluridine (m5U) at position 54 (3). The effects of nucleoside modifications have been deduced from comparisons of the biochemical and biophysical characteristics of mature tRNA with (i) unmodified tRNA transcripts (produced in vitro with T7 RNA polymerase) (17–21) and (ii) partially modified tRNAs (produced in mutant cell strains lacking the tRNA-modifying enzyme required for a particular modification) (22,23). These comparisons showed that nucleoside modifications serve to fine-tune and stabilize the structure of the tRNA such that it is optimized for participation in cellular processes (24). During aminoacylation, for example, nucleoside modifications serve as positive and negative determinants (25), while during translation they stabilize the conformation of the anticodon loop (26).
Initial biophysical studies with unmodified and mature yeast tRNAPhe revealed that nucleoside modifications stabilize the tertiary structure of the tRNA with the transcript displaying a higher degree of conformational flexibility as demonstrated by thermal denaturation, UV crosslinking and Pb(II) cleavage experiments (21,27–29). The same trend has also been observed for other tRNAs (30,31). Imino proton NMR studies revealed that while the transcript of yeast tRNAPhe did not adopt the native conformation in the absence of Mg2+, the spectra of the transcript resembled that of the mature tRNAPhe at Mg2+ concentrations of 5 mM or higher, suggesting that Mg2+ is able to compensate for the lack of modified nucleosides and stabilize the native structure of the tRNA (20). A further study with E. coli tRNAVal and its transcript showed that the two forms have similar tertiary structures and inter-arm angles. The presence of modifications, however, resulted in a 20-fold reduction of the rate of imino proton exchange, an indication that the modifications not only lead to the formation of a particular conformation but also reduce the dynamics of that conformation (32). Modifications also impact upon the biochemical behaviour of tRNA. For example, mature and unmodified E. coli tRNAPhe display differential behaviour during aminoacylation (18) and interactions with EF–Tu and the ribosome, with the modifications acting to increase the accuracy of protein synthesis (19). The effects of modifications on biochemical processes with other tRNAs have been reviewed elsewhere (25,26).
The roles played by individual modifications are best understood for those found in the anticodon stem loop (ASL), which is the most heavily modified region of tRNA. Modifications at positions 34 (the wobble position of the anticodon) and 37 play particularly important roles in maintaining the conformation of the anticodon loop. Insights into the roles played by specific modified bases in E. coli tRNAPhe have been provided by NMR structures of the ASL segment. In the absence of all modifications, or the presence of just Ψ32 or N6-isopentyladenosine (i6A) at position 37, the ASL is comprised of a stem of 7 bp and a loop of 3 nt. This indicated that single, isolated modifications do not significantly alter the conformation of the ASL with respect to that adopted by an unmodified ASL. The data do, however, show that modifications alter the dynamics of the ASL, with Ψ32 stabilizing the stem and i6A destabilizing the 3-nt loop (33,34). The ASL segment of yeast tRNAPhe containing modified bases at four positions has also been studied by NMR and the resulting structure revealed that the more heavily modified ASL has a conformation closer to that seen in the crystal structure of mature yeast tRNAPhe (35). Crystal structures of the ribosome in complex with both tRNA and mRNA show that the ASL of tRNA at the A site forms the U-turn motif when interacting with mRNA (36). As modifications at positions 34 and 37 of the ASL favour the U-turn conformation, the entropic penalty associated with its formation during translation is avoided (26,37).
Our understanding of the structure of unmodified tRNA is largely based upon crystal structures of tRNA transcripts in complex with protein molecules (such as aminoacyl tRNA-synthetases and, editing and modifying enzymes) and ribosomes, which not only stabilize but also alter the conformation of tRNA. Surprisingly, to date there is no structure available for a standalone full-length unmodified tRNAPhe. In the absence of such information, a direct comparison of mature and unmodified tRNA structures is complicated by the fact that differences in unmodified tRNA may be induced by interacting protein partners rather than the lack of modifications. Here, we present the crystal structure of the transcript of E. coli tRNAPhe which permits this issue to be addressed.
MATERIALS AND METHODS
tRNA transcription
Plasmid pCF0 containing the gene for E. coli tRNAPhe (carrying C3–G70 to G3–C70 transversion in the acceptor stem (19) under the T7-RNA polymerase promoter was donated by O.C. Uhlenbeck (Northwestern University, Evanston, IL, USA). Transcripts of tRNAPhe were prepared as described (37) and purified from nucleotides by gel filtration. Transcripts of tRNAPhe were aminoacylated and Phe-tRNAPhe was purified to homogeneity by reverse-phase HPLC. Purified Phe-tRNAPhe was deacylated in 0.5 M Tris–HCl buffer pH 9.0 for 1 h at 37°C. The resulting preparation of E. coli tRNAPhe transcript was ethanol-precipitated, dissolved in water and stored at −80°C in small aliquots.
Crystallization
Crystals of the tRNA transcript were obtained as a result of efforts to crystallize its complex with Bacillus anthracis ThiI (purified as described previously) (38). The complex comprised 70 μM of ThiI and 230 μM of tRNAPhe transcript in 100 mM KCl, 10 mM HEPES–NaOH pH 8.0, 1 mM ATP and 1 mM MgCl2. Crystals were grown by sitting drop vapour diffusion after mixing 150 nl of the protein–RNA mixture with 150 nl of the reservoir solution (0.3 M KCl, 0.1 M HEPES–NaOH pH 7.5 and 35% v/v MPD) and equilibrating this against 54 μl of the reservoir solution. Hexagonal crystals grew after a period of 6 months at 20°C. Prior to data collection, the crystals were vitrified in liquid nitrogen straight from the drop. Diffraction data were collected on beamline I04 at Diamond Light Source (Didcot, UK).
Crystallography
The data set was integrated using iMosflm (39) and then imported into the CCP4 suite for all subsequent steps (40). After scaling the data with Scala (41) it became apparent from calculation of the Matthews’ coefficient (42) that, given the 622 point group symmetry, the asymmetric unit would be unable to contain the entire ThiI–tRNA complex. Phaser (43) was used to perform a number of searches covering all potential combinations of space groups and search models but the only acceptable solution (RFZ = 4.4 and TFZ = 9.9) was obtained in the space group P 6222 using chain A of yeast tRNAPhe (PDB 1EHZ) (7). Assisted by a composite omit map from Phenix (44) to aid rebuilding and overcome bias, the model was improved though iterative cycles of manual rebuilding with Coot (45) and refinement with Refmac (46) using individual isotropic B-factors. In the later stages of refinement, a single TLS group (47) was used for the entire tRNA and in the final stages water molecules and metal ions were added to the model. The identity of the metal ions was based purely upon the coordination environment of the metal and the B-factors after refinement as neither Mg2+ nor K+ have a significant f″ at the wavelength used for data collection. The Mg2+ ion coordinated by U8–A9 and C11–U12 was built octahedrally coordinated by six water molecules and restrained as such during refinement on the basis of the high resolution structures of yeast tRNAPhe (PDB 1EHZ and 1EVV). While there was no clear density for the solvent molecules in the 2m|Fo|−D|Fc| maps contoured at 1σ, refinement without the coordinating water molecules led to a 6σ positive peak in the m|Fo|−D|Fc| map and chemically unreasonable distances between the Mg2+ and the coordinating residues, thus validating the magnesium-solvent cluster. The final model (Rwork = 22.6% and Rfree = 26.1%) contains the nucleotides corresponding to positions 1–73 of the tRNA, nine water molecules, two Mg2+ ions and one K+ ion. Figure generation and least-squares structural alignments were performed with ccp4mg (48). Base pairs were analysed by BP Viewer at the Nucleic Acid Database (49).
RESULTS AND DISCUSSION
Crystal structure
An unmodified transcript of E. coli tRNAPhe has been crystallized and its structure was determined by molecular replacement using the structure of yeast tRNAPhe (PDB 1EHZ). Crystals belong to the space group P 6222 with one molecule per asymmetric unit and the final model was refined at 3 Å resolution (Table 1). Nucleotides 1–73 are clearly defined in the electron density maps except for bases of U16 and U20. The last three nucleotides 74–76 (the 3′ CCA extension) were omitted from the model because of the lack of clear electron density.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Wavelength (Å) | 0.9704 |
| Space group | P 62 2 2 |
| a, b, c (Å) | 99.5, 99.5, 110.3 |
| Resolutiona | 46.5–3.00 (3.16–3.00) |
| Number of reflections | |
| Total | 107 790 (15 965) |
| Unique | 6886 (971) |
| Completeness (%) | 99.8 (100.0) |
| Multiplicity | 15.7 (16.4) |
| <I /σ(I)> | 12.3 (2.3) |
| Rmergeb | 0.126 (1.244) |
| Refinement | |
| Number of atoms | |
| Total | 1572 |
| tRNA | 1561 |
| Metals | 3 |
| Water | 9 |
| Rworkc | 0.226 (0.312) |
| Rfreec | 0.261 (0.236) |
| RMSD bond length (Å) | 0.006 |
| RMSD bond angle (°) | 0.8 |
| RMSD chiral | 0.05 |
| Wilson B-factor (Å2) | 70.9 |
| Mean B-factor (Å2) | |
| Overall | 81.0 |
| tRNA | 88.4 |
| Metals | 66.6 |
| Water | 71.1 |
| ESU coordinate based on ML (Å) | 0.31 |
| ESU B-value based on ML (Å2) | 35.8 |
aFigures in parentheses refer to the highest resolution shell.
bRmerge = ΣhklΣi|Ii − <I>| / ΣhklΣi<I>.
cRwork and Rfree = Σhkl||Fobs|−|Fcalc|| / Σhkl|Fobs|. Rfree was calculated using a set of reflections (5% of total) which were randomly chosen and excluded from the refinement.
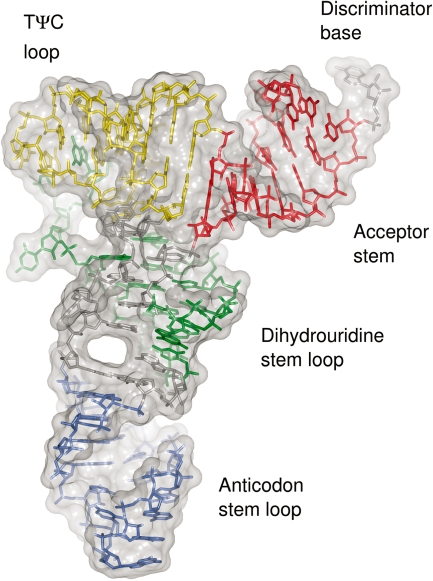
The structure of the tRNA has the conserved L-shape common to all cytoplasmic tRNAs. This architecture is maintained by the canonical Watson–Crick base pairs within the stems of the tRNA (Figure 1). Most of the tertiary interactions observed in previously determined crystal structures of mature tRNAs are observed in the present structure: the trans Watson–Crick Hoogsteen U8–A14 and U54–A58 base pairs, the trans Watson–Crick G15–C48 ‘Levitt’ base pair, the trans Watson–Crick sugar-edge G18–U55 base pair and the cis Watson–Crick G19–C56 base pair. Also preserved are the triplet base pairs U8–A14–A21, A9–U12–A23 and C13–G22–G46. The absence of the U18–C56 and A26–G44 tertiary base pairs and the G10–C25–G44 triplet base pair will be discussed below.
Figure 1.
Structure of unmodified E. coli tRNAPhe. The nucleotides of the acceptor stem (red), D stem loop (green), ASL (blue) and TΨC loop (yellow) are coloured according to the region they belong to. Molecular surface is shown in grey.
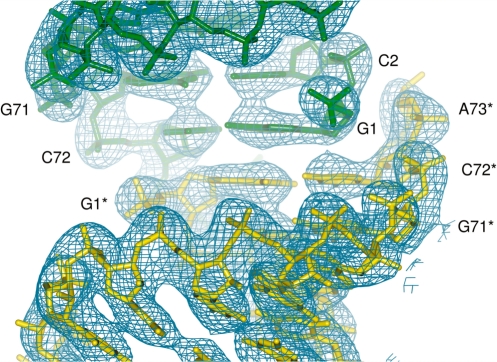
Crystallographic 2-fold symmetry results in the formation of an almost continuous helix comprised of the acceptor stem of one tRNA molecule and the acceptor stem of a symmetry-related tRNA, an arrangement similar to that previously observed in the crystal structure of modified tRNAfMet from E. coli (13). In the present structure, the G1–C72 base pair of one tRNA stacks with its symmetry equivalent (Figure 2). Such packing and absence of a clear electron density for the 3′-terminal CCA nucleotides indicates that this segment is either disordered or has been lost through partial degradation, which might explain why crystals took 6 months to grow.
Figure 2.
Crystal packing of the acceptor stem. G1–C72 base pairs from two tRNA molecules (yellow and green) are related by a crystallographic 2-fold axis. Nucleotides in one of the symmetry mates (yellow) are denoted with a star. Corresponding 2m|Fo|−D|Fc| electron density maps are contoured at 1.5σ.
Conformational differences between modified and unmodified tRNA
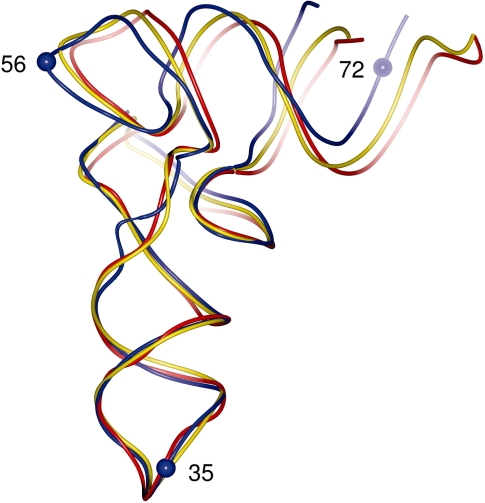
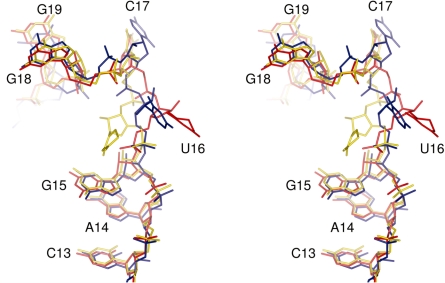
In the absence of the crystal structure of modified E. coli tRNAPhe, we compared the present structure with the available structure of the mature yeast tRNAPhe which has been refined at 1.93 Å (monoclinic crystal form, PDB 1EHZ) (7). This tRNA shares 63% sequence identity with the E. coli tRNAPhe prior to the nucleoside modifications made to the yeast tRNAPhe. Least-square superposition using the backbone P and C1′ atoms of nucleotides 1–72 resulted in an RMSD of 2.0 Å. While the overall conformation and fold of the two structures are similar, there are also differences which are distributed along the tRNA chain. When the superposition was repeated using either the acceptor stem (nucleotides 1–9 and 66–72) or the ASL (positions 28–42) the RMSD values decreased to 1.1 Å and 0.7 Å, respectively (Figure 3). Inspection of aligned structures revealed that while both regions are structurally similar, the relative orientations of the two arms of the tRNA differ between the two structures, with the angle defined by atoms P35, P56, P72 being 73° in the present structure and 67° in mature yeast tRNAPhe in both the monoclinc (PDB 1EHZ) and orthorhombic (PDB 4TRA) crystal forms. The analysis was extended to include all eight structures of yeast tRNAPhe available in the PDB and this resulted in a mean angle of 68° and a standard deviation of 1°, indicating that the difference between the transcript E. coli tRNAPhe and yeast tRNAPhe is significant (Supplementary Table S1). We also assessed the variation in the angle within E. coli tRNAPhe transcripts crystallized in complex with other macromolecules (four structures in total). This resulted in a mean angle of 58° and a standard deviation of 1° indicating the degree to which transcripts are remodelled, relative to the present structure, during their interactions with other macromolecules (Supplementary Table S2).
Figure 3.
Difference in conformation between unmodified and modified tRNAs. Superposition of unmodified E. coli tRNAPhe (blue) and yeast tRNAPhe in its monoclinic form (yellow, PDB 1EHZ) and orthorhombic form (red, PDB 4TRA). The P atoms of residues 35, 56 and 72 (blue spheres) were used for the calculation of inter-arm angles.
Base pairing
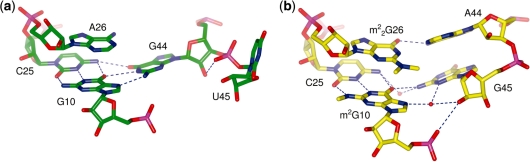
Comparison with the structure of modified yeast tRNAPhe also shows that differences in base pairing occur in the core of the tRNA where the most striking feature is the formation of the triplet base pair comprised of G10, C25 and G44 while A26 is left unpaired (Figure 4a). In yeast tRNAPhe, the equivalent triplet is formed by m2G10 (N2-methylguanosine), C25 and G45, and a cis Watson–Crick base pair is formed between 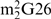 G26 (N2, N2-dimethylguanosine) and A44 (Figure 4b). Other conformational changes in the transcript tRNA are seen in the vicinity of this triplet—the pyrimidine ring of U45 is flipped out of the tRNA core such that it interacts with the core region of a symmetry-related tRNA. The ribose O4′ of the same nucleotide interacts with the exocyclic N2 amine of a symmetry-related G27 and the N3 amine of U45 interacts with the N1 imine of a symmetry-related A25. In the absence of additional structural information, we cannot rule out the possibility that differences between the present structure and mature yeast tRNAPhe are a consequence of crystal packing and/or differences in the nucleotide sequences.
G26 (N2, N2-dimethylguanosine) and A44 (Figure 4b). Other conformational changes in the transcript tRNA are seen in the vicinity of this triplet—the pyrimidine ring of U45 is flipped out of the tRNA core such that it interacts with the core region of a symmetry-related tRNA. The ribose O4′ of the same nucleotide interacts with the exocyclic N2 amine of a symmetry-related G27 and the N3 amine of U45 interacts with the N1 imine of a symmetry-related A25. In the absence of additional structural information, we cannot rule out the possibility that differences between the present structure and mature yeast tRNAPhe are a consequence of crystal packing and/or differences in the nucleotide sequences.
Figure 4.
The rearrangement of triplet base pairs. (a) The triplet base pair in the unmodified E. coli tRNAPhe compared with (b) the triplet base pair in yeast tRNAPhe (PDB 1EHZ). Nucleotides are shown as sticks with oxygen atoms in red, nitrogen atoms in blue and phosphorus atoms in magenta. Water molecules are shown as red spheres and hydrogen bonds are depicted as blue dashed lines.
Unmodified E. coli tRNAPhe has also been crystallized in complex with the Thermus thermophilus 70S ribosome (36) and E.coli MiaA (50,51). In both the 70S ribosome (PDB 2J02) and one structure of MiaA (PDB 3FOZ), the base pairing in the core of the tRNA molecule is the same as observed in modified yeast tRNAPhe. In these structures, the triplet base pair is formed between G10, C25 and G45 while a further base pair is formed between A26 and G44. Different base pairing is however observed in other structures of unmodified tRNAPhe bound to MiaA (PDBs 2ZM5 and 2ZXU). In these structures, a triplet base pair is formed between C11, G24 and G44, a second base pair is formed between G10 and C25 while A26 is left unpaired. Neither arrangement of base pairs is identical to that observed in the present structure, suggesting that the remodelling of the tRNA by proteins or ribosomes results in conformational changes which are facilitated by plasticity in the core of the tRNA.
D-loop
The electron density for the D-loop, in particular positions 16–20, and the B-factors of nucleotides in this region indicate that this is one of the most flexible regions of the tRNA. It has been reported that flexibility of the D-loop in mature tRNA is increased by the presence of dihydrouridine (8). What the present structure shows is that the D-loop is flexible even without dihydrouridine at positions 16 and 20, although the lack of crystal contacts may also contribute to its flexibility. Comparisons of the present structure with yeast tRNA in its monoclinic (PDB 1EHZ) and orthorhombic (PDB 4TRA) crystal forms indicate that the major differences in the loop are the conformations of nucleotides 16 and 17 (Figure 5). Shi and Moore (7) noted that the conformation of D16 in yeast tRNAPhe was dependent on the presence or absence of particular divalent cations. In the presence of only Mg2+ (equivalent to the orthorhombic crystal form) the pyrimidine ring of D16 pointed away from U59. In the presence of Mg2+ and either Mn2+ or Co2+ (monoclinic crystal form), however, the pyrimidine ring pointed towards U59. We calculated omit maps through refinement of the structure without U16. The resulting m|Fo|−D|Fc| map had a positive density peak which could not be explained by just one of the two conformations of D16 described above (Supplementary Figure S1). Given the resolution, we modelled a single conformer in which U16 points away from U59 as this gave the best overall fit to the electron density (Supplementary Figure S1).
Figure 5.
Conformation of the D-loop. Stereo view of the structures of unmodified E. coli tRNAPhe (blue) and yeast tRNAPhe in its monoclinic (yellow, PDB 1EHZ) and orthorhombic (red, PDB 4TRA) forms. In yeast tRNAPhe dihydrouridine is present at positions 16 and 17.
ASL
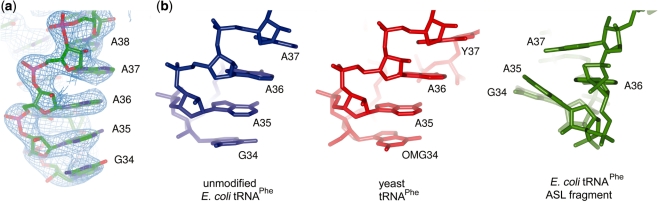
The electron density maps for the ASL are well defined allowing the accurate positioning of all nucleotides (Figure 6a). When the structure was re-refined with residues 28–42 omitted from the model, clear density appeared at a 2σ contour level in the difference maps calculated with coefficients m|Fo|−D|Fc|, indicating that the positions are correct and not a consequence of model bias (Supplementary Figure S2). In the final model, the ASL adopts the U-turn motif in which the anticodon loop contains 7 nt. This is the same conformation observed in mature tRNAs and also in unmodified tRNAs when it is bound at the A site of the ribosome (36). Superposition of nucleotides 31–39 of the transcript and mature yeast tRNAPhe (PDB 1EHZ) results in an RMSD of 0.7 Å, illustrating the similarity between the two structures (Figure 6b). In contrast to mature tRNA, the anticodon loop in the NMR structure of unmodified ASL of E. coli tRNAPhe (lowest energy conformer; PDB 1KKA) does not adopt the U-turn motif but instead has a non-standard conformation in which the anticodon loop contains 3 nt. Superposition of that structure with the transcript resulted in an RMSD of 5.3 Å (Figure 6b) reflecting the difference. Thus, the isolated ASL and ASL within the tRNA molecule have different conformations, even though both have the same sequence and lack nucleoside modifications.
Figure 6.
Anticodon stem loop conformation. (a) Electron density map for nucleotides 34–38. The 2m|Fo|−D|Fc| map is contoured at 1.5σ. (b) The conformation of these nucleotides (left) is essentially the same as in modified tRNA (middle) but differs from the conformation observed in the lowest energy conformer of the unmodified ASL fragment (right). All three ASLs are shown in the same orientation after superposition using the backbone P and C1′ atoms of nucleotides 31–39.
Of the 15 nucleotides that make up the ASL, those at positions 34, 36, 37 and 38 are involved in crystal packing interactions. In A36, the 2′OH makes hydrogen bonds with the backbone O3′ and O2′ atoms of a symmetry-related G71, while the N3 atom of the base accepts a hydrogen bond from the same O2′. In A37, the 2′OH can make hydrogen bonds with the ribose O2′ and base O2 of a symmetry-related C70. A single hydrogen bond occurs between the 2′OH of A38 and the 2′OH of C4. The purine ring of G34 is stabilized through stacking with the purine ring of a symmetry-related A73.
While crystal packing interactions may stabilize the conformation of the anticodon loop, it is more likely that the U-turn motif was induced by other factors. These might include the torsional and positional restraints imposed by the core of the tRNA molecule and/or the presence of monovalent and divalent cations. The concentrations of monovalent and divalent cations present during crystallization have been demonstrated to have a considerable influence on the folding of tRNA (52,53) and are comparable to the intracellular concentrations of K+ and Mg2+ reported for E. coli (54). In contrast, the NMR study of the ASL was performed at low concentrations of Na+ and K+ and in the absence of Mg2+ (10 mM NaCl, 10 mM potassium phosphate pH 6.8 and 0.05 mM EDTA) (33).
It is pertinent that substantial differences also exist between the structural information obtained for tRNALys,3 by NMR and X-ray crystallography. The NMR structure of an ASL lacking modifications except Ψ39 showed that the stem was extended by the base pairs C32–A38 and U33–A37 and that the loop was comprised of only three nucleotides (U34–U36) (55). The crystal structure of the full-length mature tRNALys,3, however, revealed that the ASL adopted the U-turn motif. Differences between the two structures were attributed to the absence of modifications in the ASL segment used in the NMR study. However, our data on unmodified E. coli tRNAPhe show that the ASL adopts the U-turn motif and this is in agreement with the observation that this tRNA is functional even in an unmodified state (18,19). In view of these results, it could be thought that the full structure of the tRNA may be necessary to stabilize a canonical ASL structure (with or without modifications), while a more relaxed isolated ASL favours a non-canonical structure. It was however shown that full-length unmodified tRNALys,3 cannot bind a ribosome programmed with codons AAA or AAG and that it is only by the conversion of U34 to 2-thiouridine (s2U) that ribosome binding is restored (56).
Metal ions
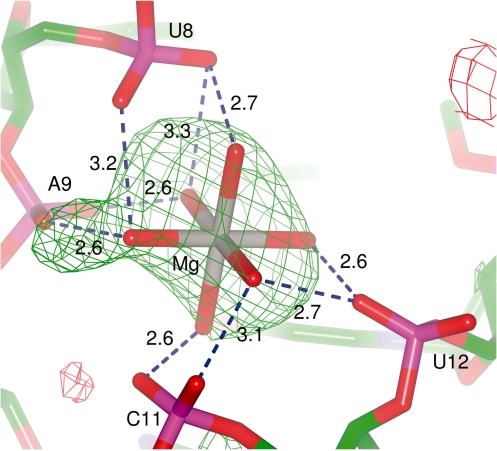
Three peaks in the electron density map were modelled as metals. A peak located between the phosphate groups of U8, A9, C11 and U12 was modelled as a Mg2+ ion coordinated by six water molecules based upon the presence of a similarly coordinated Mg2+ ion in two high resolution structures of mature yeast tRNAPhe (PDB 1EHZ and 1EVV) (7,8) (Figure 7). Consistent with the other structures, the magnesium ion in this structure is >4.5 Å away from each phosphate and it is through the water molecules that the Mg2+ is coordinated by the phosphates (water–oxygen distances of 2.5–3.0 Å). While there is no clear density for the coordinating water molecules in the 2m|Fo|−D|Fc| map contoured at 1σ (Supplementary Figure S3), their removal leads to a 6σ positive peak in the m|Fo|−D|Fc| difference map indicating that in the absence of coordinating water molecules the model is lacking electron density in this region (Figure 7). This suggests that the solvent molecules are bridging the interaction between the Mg2+ and the phosphates. A second Mg2+ ion is located between the phosphates of A7 and A14. In this position, the Mg2+ ion is close enough to the oxygen atoms of the phosphates to interact without bound water molecules (the closest Mg2+-O distances being 2.9 Å to O1P of A7 and 2.5 Å to O2P of A14). A third metal was located in the core of the tRNA between the bases of A9, A23, G24, C35 and G44. Given the reagents present during the crystallization experiment and the chemical environment of this region, this peak was modelled as a K+ ion. A search of MERNA (a database of metal ion binding sites in RNA: http://merna.lbl.gov/) reveals a number of high resolution crystal structures in which K+ ions are coordinated by carbonyls, amines and imines with typical coordination distances of 2.5–3.2 Å. In the present structure, the K+ ion is within 2.6 Å of O6 of G44 and 3.0 Å of N7 of G24.
Figure 7.
The coordination of the hydrated Mg2+ ion. The m|Fo|−D|Fc| electron density map contoured at 3σ was calculated after omitting the coordinating water molecules. The hydrogen bonds between the phosphate groups and the water molecules are depicted by dashed blue lines, along with the distances in Angstroms.
CONCLUSIONS
We have determined the crystal structure of a transcript of E. coli tRNAPhe. As expected, the structure adopts the same overall fold as mature tRNAs. Comparisons of the present structure with previously determined structures of mature yeast tRNAPhe revealed a number of significant differences, such as the increased angle between the acceptor and anticodon stems, and the rearrangement of a triplet base within the core of the tRNA. These differences could be attributed to the absence of modifications or variations between the nucleotide sequences. In contrast to previous studies performed with an isolated ASL, the anticodon loop in the present structure adopts the U-turn motif comprised of a 5-bp stem and a 7-nt loop. The conformation of the anticodon loop is thus the same as seen in mature tRNAs and also in unmodified tRNAs bound at the A site of the ribosome.
ACCESSION NUMBER
The coordinates and structure factors have been deposited with the Protein Data Bank under the accession code 3L0U.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council PhD studentship (to R.T.B.); Deutsche Forschungsgemeinschaft (to M.V.R.); Wellcome Trust (to A.A.A.). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Eleanor Dodson, Garib Murshudov, Jennifer Potts, Johan Turkenburg and Callum Smits for useful discussions, Michael Mrosek (Diamond Light Source) for providing assistance during data collection and Simone Möbitz for expert technical assistance.
REFERENCES
- 1.Schurer H, Schiffer S, Marchfelder A, Morl M. This is the end: processing, editing and repair at the tRNA 3′-terminus. Biol. Chem. 2001;382:1147–1156. doi: 10.1515/BC.2001.144. [DOI] [PubMed] [Google Scholar]
- 2.Morl M, Marchfelder A. The final cut - The importance of tRNA 3′-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jühling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozenski J, Crain PF, McCloskey JA. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BFC, Klug A. Structure of yeast phenylalanine transfer RNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Suddath FL, Quigley GJ, McPherso A, Sussman JL, Wang AHJ, Seeman NC, Rich A. Three dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 7.Shi HJ, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jovine L, Djordjevic S, Rhodes D. The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J. Mol. Biol. 2000;303:113–113. doi: 10.1006/jmbi.2000.3950. [DOI] [PubMed] [Google Scholar]
- 9.Jack A, Landner JE, Rhodes D, Brown RS, Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J. Mol. Biol. 1977;111:315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- 10.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sussman JL, Holbrook SR, Warrant RW, Church GM, Kim S-H. Crystal structure of yeast phenylalanine transfer RNA: I. Crystallographic refinement. J. Mol. Biol. 1978;123:607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- 12.Westhof E, Dumas P, Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Cryst. A. 1988;44:112–124. [PubMed] [Google Scholar]
- 13.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisne C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008;36:4894–4901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basavappa R, Sigler PB. The 3 Å crystal structure of yeast initiator transfer RNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benas P, Bec G, Keith G, Marquet R, Ehresmann C, Ehresmann B, Dumas P. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000;6:1347–1355. doi: 10.1017/s1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serebrov V, Vassilenko K, Kholod N, Gross HJ, Kisselev L. Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Escherichia coli tRNAPhe. Nucleic Acids Res. 1998;26:2723–2728. doi: 10.1093/nar/26.11.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson ET, Uhlenbeck OC. Determination of recognition nucleotides for Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1992;31:10380–10389. doi: 10.1021/bi00157a028. [DOI] [PubMed] [Google Scholar]
- 19.Harrington KM, Nazarenko IA, Dix DB, Thompson RC, Uhlenbeck OC. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993;32:7617–7622. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- 20.Hall KB, Sampson JR, Uhlenbeck OC, Redfield AG. Structure of an unmodified tRNA molecule. Biochemistry. 1989;28:5794–5801. doi: 10.1021/bi00440a014. [DOI] [PubMed] [Google Scholar]
- 21.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urbonavicius J, Qian O, Durand JMB, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979;6:1571–1581. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin: Landes Biosciences; 2009. [Google Scholar]
- 25.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maglott EJ, Deo SS, Przykorska A, Glick GD. Conformational transitions of an unmodified tRNA: implications for RNA folding. Biochemistry. 1998;37:16349–16359. doi: 10.1021/bi981722u. [DOI] [PubMed] [Google Scholar]
- 28.Behlen LS, Sampson JR, Uhlenbeck OC. An ultraviolet light-induced cross-link in yeast transfer RNAPhe. Nucleic Acids Res. 1992;20:4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behlen LS, Sampson JR, Direnzo AB, Uhlenbeck OC. Lead-catalyzed cleavage of yeast transfer RNAPhe mutants. Biochemistry. 1990;29:2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- 30.Derrick WB, Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA - evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perret V, Garcia A, Puglisi J, Grosjean H, Ebel JP, Florentz C, Giege R. Conformation in solution of yeast transfer RNAAsp transcripts deprived of modified nucleotides. Biochimie. 1990;72:735–744. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen A, McCallum SA, Pardi A. Comparison of the global structure and dynamics of native and unmodified tRNA. Biochemistry. 2005;44:6024–6033. doi: 10.1021/bi0473399. [DOI] [PubMed] [Google Scholar]
- 33.Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- 34.Cabello-Villegas J, Nikonowicz EP. Solution structure of Ψ32-modified anticodon stem-loop of Escherichia coli tRNAPhe. Nucleic Acids Res. 2005;33:6961–6971. doi: 10.1093/nar/gki1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring Modification Restricts the Anticodon Domain Conformational Space of tRNAPhe. J. Mol. Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 36.Selmer M, Dunham CM, Murphy FVIV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 37.Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterman DG, Ortiz-Lombardia M, Fogg MJ, Koonin EV, Antson AA. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Leslie AGW. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallgraphy, No 20. 1992. [Google Scholar]
- 40.CollaborativeComputational Project. The CCP4 suite: programs for protein crystallography. Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Evans P. Scaling and assessment of data quality. Acta Cryst. D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 42.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 48.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Cryst. D. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 49.Yang HW, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seif E, Hallberg BM. RNA-Protein mutually induced fit structure of Escherichia Coli isopentyl-tRNA transferase in complex with tRNAPhe. J. Biol. Chem. 2009;284:6600–6604. doi: 10.1074/jbc.C800235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chimnaronk S, Forouhar F, Sakai J, Yao M, Tron CM, Atta M, Fontecave M, Hunt JF, Tanaka I. Snapshots of Dynamics in Synthesizing N-6-Isopentenyladenosine at the tRNA Anticodon. Biochemistry. 2009;48:5057–5065. doi: 10.1021/bi900337d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhodes D. Initial stages of the thermal unfolding of yeast phenylalanine transfer RNA as studied by chemical modification: the effect of magnesium. Eur. J Biochem. 1977;81:91–101. doi: 10.1111/j.1432-1033.1977.tb11930.x. [DOI] [PubMed] [Google Scholar]
- 53.Shelton VM, Sosnick TR, Pan T. Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations. Biochemistry. 2001;40:3629–3638. doi: 10.1021/bi002646+. [DOI] [PubMed] [Google Scholar]
- 54.Moncany MLJ, Kellenberger E. High magnesium content of Escherichia coli B. Experientia. 1981;37:846–847. doi: 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 55.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 56.Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF. Single atom modification (O -> S) of tRNA confers ribosome binding. RNA. 1999;5:188–194. doi: 10.1017/s1355838299981529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.