Abstract
Long-range tertiary interactions determine the three-dimensional structure of a number of metabolite-binding riboswitch RNA elements and were found to be important for their regulatory function. For the guanine-sensing riboswitch of the Bacillus subtilis xpt-pbuX operon, our previous NMR-spectroscopic studies indicated pre-formation of long-range tertiary contacts in the ligand-free state of its aptamer domain. Loss of the structural pre-organization in a mutant of this RNA (G37A/C61U) resulted in the requirement of Mg2+ for ligand binding. Here, we investigate structural and stability aspects of the wild-type aptamer domain (Gsw) and the G37A/C61U-mutant (Gswloop) of the guanine-sensing riboswitch and their Mg2+-induced folding characteristics to dissect the role of long-range tertiary interactions, the link between pre-formation of structural elements and ligand-binding properties and the functional stability. Destabilization of the long-range interactions as a result of the introduced mutations for Gswloop or the increase in temperature for both Gsw and Gswloop involves pronounced alterations of the conformational ensemble characteristics of the ligand-free state of the riboswitch. The increased flexibility of the conformational ensemble can, however, be compensated by Mg2+. We propose that reduction of conformational dynamics in remote regions of the riboswitch aptamer domain is the minimal pre-requisite to pre-organize the core region for specific ligand binding.
INTRODUCTION
The complex three-dimensional structures of RNAs include a large variety of secondary and tertiary structural motifs. Structural motifs defining tertiary folds contribute to local RNA structure formation but can also connect sequentially distant nucleotides and globally constrain RNA conformation. Interestingly, it has been shown that tertiary contacts of structural elements remote from the active center influence the biological functions for a number of different RNAs (1). In addition to the intrinsic properties of an RNA, cofactors including proteins, ions or small ligands mediate RNA structure formation and alter or promote cellular function. Mg2+ ions play a particularly important role since they often enable the formation and functional stabilization of compact RNA structures (2).
Riboswitches represent a class of recently identified RNA regulatory elements. They are generally found in the 5′-untranslated regions of mRNAs. Transcriptional or translational regulation or RNA processing is modulated by binding of a small ligand to an evolutionarily highly conserved ligand-binding domain (aptamer domain). The conformational switch between alternate RNA conformations is supposed to be the structural basis for the regulation of gene expression. According to this model, ligand binding stabilizes one of the alternative conformations of the aptamer domain that further affects formation or destabilization of a 3′-downstream structural element (3). Riboswitches sense various small molecule metabolites, ranging from purine nucleobases to amino acids (4). The high affinity and specificity of riboswitch aptamer domains for their small molecule effectors is coupled to complex tertiary architectures, involving formation of intricate networks of intra- and intermolecular interactions (5). To gain function, a remarkable variety of strategies for ligand recognition and conformational adaptation of riboswitch elements has been discovered. RNA–ligand complex formation can, for example, be critically dependent on cations as e.g. observed for FMN- (6) and TPP-sensing (7,8) riboswitches. A single point mutation in the ligand-binding region of the guanine-sensing riboswitch element (C74U) converts the specificity of the aptamer domain from the cognate ligand guanine to the originally rejected ligand adenine (9). In the case of the metabolite S-adenosylmethionine (SAM), even five structurally different classes of riboswitches with different ligand recognition modes could be identified (10–16). In addition, not only nucleotides that define the ligand-binding pocket but also residues in remote regions are found to be important for ligand binding and/or the regulatory function of various riboswitch elements (17). These residues are involved in the formation of a large number of different long-range tertiary interactions and enable the formation of compact RNA structures (18). Pre-formation of peripheral structural elements already in the ligand-free state could be observed in some riboswitch RNAs; the extent to which secondary and tertiary structure is formed and its Mg2+ dependence are remarkable diverse and associated with different functional implications (19–21).
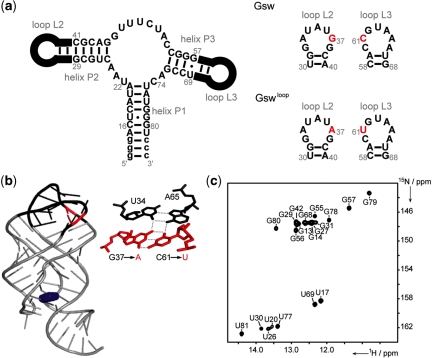
For the adenine- and the guanine-sensing riboswitches, the structure-function relationship is particularly intriguing. The secondary structure of the two aptamer domains (22,23) and the RNA fold of the RNA–ligand complexes (24,25) are highly similar. The secondary structures contain three helical elements (P1, P2 and P3) organized around a three-way junction (22,23) (Figure 1a). Helices P2 and P3 possess capping loops (L2 and L3), whereas one strand of P1 is supposed to be involved in the conformational rearrangement underlying the regulatory function of the riboswitch elements (22,23,26). The RNA fold of the aptamer domains is defined by two regions of complex tertiary interactions. Residues within the central three-way junction are mainly involved in the specific recognition of the cognate ligand; recognition involves the formation of a large number of tertiary interactions that define the ligand-binding pocket (24,25). Specificity is accomplished through formation of an intermolecular Watson–Crick interaction with the complementary RNA residue (C or U) at one specific sequence position (24,25,27). Several mutational studies have identified contributions of RNA residues to ligand-binding ability and affinity (9,22,23,28). The X-ray structures of RNA–ligand complexes of the adenine- and guanine-sensing riboswitch aptamer domains (24,25) strongly indicate the significance of the terminal loop regions (L2 and L3) in the organization of the global RNA fold. Long-range tertiary interactions connect these two loops and induce the compact arrangement of the helical elements, P2 and P3. This inter-helical linkage is mainly defined by the formation of two inter-helical base-quadruples, involving various nucleotides in the loop regions.
Figure 1.
(a) Secondary structure of the guanine-sensing riboswitch aptamer domain (Gsw) of the B. subtilis xpt-pbuX operon and the mutant aptamer domain (G37A/C61U, Gswloop) following the nomenclature of Mandal et al. (22). Nucleotides in the loop regions L2 and L3 that differ in the two constructs are highlighted in red [for further construct details, see refs (27,29)]; (b) X-ray structure of the Gsw-hypoxanthine complex (pdb: 1U8D) (24), loop regions (L2 and L3) are color coded in black, mutated residues at nucleotide positions 37 and 61 in the Gswloop-construct are color coded in red, the ligand hypoxanthine is highlighted in blue; the base-quadruple including the mutation sites for Gswloop is enlarged next to the X-ray structure; (c) 1H,15N-HSQC spectrum of Gswloop with annotated NMR imino proton resonance assignment ([RNA]:[Mg2+] ratio = 1:7, T = 283 K).
However, the purine-sensing riboswitch RNAs show high specificity and affinity for the cognate ligand only, and in addition, pursue different mechanisms to control gene expression (22,23). Ligand binding in the xpt-pbuX guanine-sensing riboswitch results in transcriptional deactivation, in contrast to the adenine-sensing riboswitches reported to date for which ligand binding activates gene expression. Adenine-sensing riboswitch elements were identified that operate either on the level of transcriptional (pbuE) or translational (add) regulation.
In addition, the structural characteristics of the ligand-free states of the aptamer domains and their metal-ion induced structure formation strongly differ (26,28–30). NMR (30) and biochemical (28) studies on the ligand-free state of the adenine-sensing riboswitch aptamer domain indicate conformational heterogeneity. The addition of Mg2+ or small sequence variations in helix P2 result in a homogenously folded RNA along with stabilization of the loop–loop interaction (30). These NMR results are in agreement with fluorescence studies, for which Mg2+ was reported to pre-organize the long-range tertiary interactions (26,28). In contrast, NMR studies on the guanine-sensing riboswitch aptamer domain indicated pre-formation of the long-range tertiary structural element already in the ligand-free state of the RNA (at a temperature of 10°C) irrespective of the presence of Mg2+ ions (29). Despite the differences in the extent of pre-formation of long-range tertiary interactions, ligand binding can be detected for both purine-sensing riboswitches in the absence of Mg2+ even though the tertiary interaction is just partially pre-formed under these conditions in the adenine-sensing riboswitch (30). Thus, although the final RNA-purine complexes show high structural similarity, the conformational RNA ensemble characteristics in the absence of ligand as well as their Mg2+ dependence strongly differ in the adenine- and the guanine-sensing riboswitches. Hence, for a comprehensive understanding of how riboswitches function, it is important to characterize the ligand-free states of the RNA elements, their Mg2+- and temperature-dependent stabilities in solution and how structural properties are coupled to their ligand-binding characteristics.
To address these questions, we extend our previous investigations (29), and present here detailed NMR-spectroscopic studies investigating the influence of RNA–Mg2+ interactions on RNA folding, ligand-binding capability and functional stability of the guanine-sensing riboswitch aptamer domain (Gsw) of the Bacillus subtilis xpt-pbuX operon and its G37A/C61U-mutant (Gswloop) (Figure 1). Applying solution NMR-spectroscopic techniques offers the opportunity to probe potential RNA conformational changes and formation of RNA–ligand complexes at residue-specific resolution under a range of different conditions. The mutations introduced in the loop regions (G37A/C61U) of the riboswitch RNA were chosen in a way that the base-pairing ability of the interacting nucleotides in L2 and L3 is retained and might still enable formation of potential long-range tertiary interactions. The inter-helical tertiary structure, however, is destabilized in the mutant RNA by the formal loss of two hydrogen-bonding interactions. The initial NMR-spectroscopic characterization of the mutant Gswloop revealed that structure formation and ligand-binding characteristics strongly differ from the wild type guanine-sensing riboswitch aptamer domain (29). Most importantly, ligand binding becomes critically dependent on the presence of Mg2+ for the mutant Gswloop.
Here, we characterize the link between existing structural elements for potential formation of low-affinity or specific RNA–ligand complexes and the temperature-dependent functional stability of the mutant Gswloop and the wild-type Gsw. NMR-spectroscopic studies of Gswloop reveal that destabilizing the long-range interactions by the introduced mutations results dramatic alterations of the conformational RNA ensemble characteristics. In the absence of Mg2+, neither loop-loop interactions nor ligand binding can be detected. In addition, a low-affinity encounter complex of the ligand with the RNA as previously reported in case of the wild type Gsw (31) cannot be observed for Gswloop in the absence of Mg2+. However, the ligand-binding characteristics for Gswloop resembling the situation in the wild-type RNA can be recovered at high Mg2+ concentrations. The conformational ensemble defining the free state of Gswloop is biased towards conformations lacking long-range tertiary interactions. The mutant enables us to fine-tune and dissect the formation of the different tertiary structural elements independently through variations of the Mg2+ concentration. Although this mutation is artificial, the detailed analysis of the Mg2+-dependent structure formation in the ligand-free state and the link to its ligand-binding characteristics yields insight into contributions to the function of the wild type guanine-sensing riboswitch element.
MATERIALS AND METHODS
Sample preparation
Guanine-sensing riboswitch aptamer domains [Gsw of the B. subtilis xpt-pbuX operon and Gswloop (G37A/C61U-mutant)] were prepared by in vitro transcription using T7 RNA polymerase (29). Unlabeled model hairpin RNA constructs (helixII and helixIII) were purchased from Dharmacon (Boulder, CO). 15N-labeled rNTPs were purchased from Silantes (Munich). NMR samples were prepared in H2O/D2O (9:1) using the following NMR buffer conditions: 25 mM potassium phosphate, pH = 6.2, 50 mM potassium chloride.
Nuclear Magnetic Resonance (NMR) spectroscopy
NMR experiments were recorded on Bruker NMR spectrometers AV900, AV800, AV700 and AV600 MHz with 5-mm z-axis gradient TXI-HCN or TCI-HCN cryogenic probes and a DRX600 MHz spectrometer equipped with a 5-mm x, y, z-axis gradient TXI-HCN-RT probe at 283 K (if not otherwise stated) using standard pulse sequences [1H,15N-HSQC (32), 2D-1H,1H-NOESY, HNN-COSY (33) and 3D-1H,1H,15N-NOESY-HSQC (34)].
Characterization of Mg2+-dependent effects by NMR spectroscopy
Chemical shift perturbations (CSP) Δδ [Hz] of imino proton resonances were determined from 1H,15N-HSQC spectra at various Mg2+ concentrations. Aliquots of a MgCl2 solution were added increasing the [RNA]:[Mg2+] ratio in steps of equivalents (eq) of Mg2+ compared with the RNA or the RNA–ligand complex ([RNA] = 0.15 mM) ranging from 0 to 33 eq (corresponding to absolute concentrations of [Mg2+] ranging from 0 to 5 mM). CSPs were calculated according to 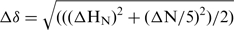 . Non-linear regression of the correlation of CSP with the corresponding [Mg2+]:[RNA] ratio results in estimation of apparent dissociation constants (KD). The KD was analyzed for exemplary imino proton resonances according to the fitting function:
. Non-linear regression of the correlation of CSP with the corresponding [Mg2+]:[RNA] ratio results in estimation of apparent dissociation constants (KD). The KD was analyzed for exemplary imino proton resonances according to the fitting function: 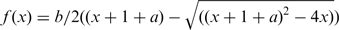 , with f (x) is the CSP at the respective x the [Mg2+]:[RNA] ratio, a the ratio of the dissociation constant to [RNA] and b the CSP for infinite Mg2+ concentrations (35).
, with f (x) is the CSP at the respective x the [Mg2+]:[RNA] ratio, a the ratio of the dissociation constant to [RNA] and b the CSP for infinite Mg2+ concentrations (35).
NMR line width analysis
The NMR studies were performed using unlabeled Gswloop-RNA and 13C,15N-labeled ligands (hypoxanthine or adenine) with [RNA]:[ligand] ratios of ∼5:1 (31). Experiments were performed on a Bruker DRX600 MHz spectrometer equipped with a 5-mm x, y, z-axis gradient TXI-HCN-RT probe at 283 K. The line width values of the respective ligand signals (C2–H2 and C8–H8) were extracted from appropriately zero-filled 1H,13C-HSQC spectra and analyzed by deconvolution using Topspin 2.1.
Circular Dichroism (CD) spectroscopy
CD melting profiles were recorded on a Jasco J-810 instrument within the temperature range of 4–94°C using a quartz cuvette of 1 mm path length. RNA samples were measured in NMR-buffer with final RNA concentrations of 10–15 µM. The melting profiles were followed at a wave length of 264 nm and data were collected with heating rates of 1°C/min. The unfolding transitions (°C) were extracted from the temperature derivative of the fitted CD profile. The heating and cooling profiles could be superimposed indicating reversibility of the transitions.
RESULTS
NMR-spectroscopic characterization of the free (G37A/C61U)-mutant of the guanine-sensing riboswitch aptamer domain
The assignment of the NMR resonances of the solvent exchangeable imino protons is reported for Gswloop as a pre-requisite for its structural analysis. H,N-correlation spectra [1H,15N-HSQC (32)] involving the imino sites of the nucleobases of guanosine and uridine residues in RNA provide two advantages: (i) NMR signals for imino sites are only detectable when protected from exchange with the solvent and give direct evidence for residues involved in stabilizing secondary or tertiary structure. (ii) The chemical shifts of these signals sensitively vary in response to even subtle changes in local structure. They reflect contributions of different potential interactions e.g. with small ligands, proteins, Mg2+ or variations in inter- or intramolecular interaction networks of RNA structural elements (34).
The NMR imino proton resonances of the aptamer domains of wild type and mutant guanine-sensing riboswitches provide sensitive reporters to analyze the differences in tertiary structure formation and ligand-binding characteristics. The formation of long-range loop–loop interactions in Gsw and Gswloop can be followed by four different NMR reporter signals in the 1H,15N-HSQC spectra, respectively. Signals can be monitored for residues U34, G38 and G37 in Gsw and for residues U34, G38 and U61 including the mutation sites (G37A/C61U) in Gswloop. The imino sites of these nucleotides are involved in the base-pairing interactions of the inter-helical base-quadruples. In addition, an imino proton signal is expected for residue G32 in both RNA constructs. Interestingly, this signal is protected from exchange by stacking interactions rather than by hydrogen-bond formation and becomes exclusively detectable when the long-range tertiary interaction is present.
The bound ligand and nucleotides involved in formation of the ligand-binding pocket are locked in place by numerous interactions. As a consequence, additional imino proton signals appear and lead to considerable differences in the NMR spectra (29).
The 1H,15N-HSQC spectrum of the uniformly 15N-labeled 73 nt-Gswloop in its free state indicates a homogeneously folded RNA with conformational dynamics that are fast on the NMR time scale. As typically expected for fast conformational averaging, these dynamics can be detected by increased line widths that are actually larger in the ligand-free conformation of the RNA compared to the ligand-bound state. However, a second set of RNA resonances identifying a second conformation in slow exchange cannot be detected.
In the free state, 20 resolved imino proton resonances are observed (Figure 1). NMR-spectroscopic assignment of the base-pairing patterns and the sequential correlations could be obtained from an analysis of the HNN-COSY experiment (33) and 2D- and 15N-edited 3D-NOESY spectra (Supplementary Figure S1). All imino proton signals could be assigned to residues in the three helical elements P1, P2 and P3. In cases where sequential correlations could not be achieved due to limited spectral dispersion we used model hairpin RNAs representing structural fragments of the full-length RNA construct. The chemical shifts and NOE-connectivities of the model hairpin RNAs were found to be closely similar to the NMR characteristics of the full-length RNA and thus, further confirmed the resonance assignment of Gswloop (Figure 1, Supplementary Figures S1 and S2). No additional imino proton signals besides those of residues in the three helical elements could be observed in the NMR spectra of Gswloop (in the absence of Mg2+). All other imino sites are therefore not involved in hydrogen-bonding interactions but rapidly exchange with the solvent water. This behavior is also observed for imino proton signals of residues that form the closing base pairs of the helical elements (G12, G45, U67, G72 and U75). Thus, our NMR-spectroscopic characterization confirms the proposed secondary structure of the free Gswloop (Figure 1). Additionally, the NMR spectra reveal the absence of the long-range tertiary loop–loop interactions for Gswloop in the absence of Mg2+. This observation is in stark contrast to the wild type guanine-sensing riboswitch aptamer domain (Supplementary Figure S3) for which the loop–loop interaction is detectable in the NMR spectra of the ligand-free state of the RNA even in the absence of Mg2+ (29). The NMR spectra of Gswloop thus show that the mutation destabilizes the long-range loop–loop interaction but does not induce alternative tertiary interactions or conformational heterogeneity indicative of misfolded conformations detectable on the NMR time scale.
Mg2+ dependence of conformational dynamics for Gswloop in the ligand-free state
Mg2+ ions are essential cofactors for the structure and function of various complex RNAs. In particular, formation of tertiary structure motifs often requires Mg2+ ions (1,36).
Analysis of the 1H,15N-HSQC spectra of Gswloop upon titration with Mg2+ revealed strong Mg2+-induced effects on structure formation. We observed chemical shift perturbations (CSP) of various imino proton NMR resonances. Such CSP might either arise when residues are involved in or are in close proximity to Mg2+-binding sites and thus, allow for their localization and the determination of affinities. In addition, CSP might be caused by RNA conformational rearrangements. The Mg2+-induced formation of additional secondary or tertiary structural elements might result in the appearance or disappearance of NMR resonances. These Mg2+-induced differences in RNA structure might also be reflected by changes in NOE-connectivities.
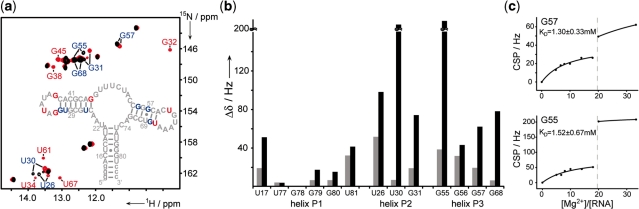
Mg2+-titration of Gswloop from its free state up to an [RNA]:[Mg2+] ratio of ∼1:18 reveals no change in the number of imino sites detectable in the 1H,15N-HSQC spectra. The observable imino proton signals stem from residues in the helical elements P1, P2 and P3. However, significant Mg2+-dependent chemical shift perturbations of a number of imino proton resonances were observed. By analyzing the magnitude of the CSP as a function of the [RNA]:[Mg2+] ratio, apparent KD(Mg2+)-values in the low millimolar range could be derived (Figure 2).
Figure 2.
Mg2+-induced effects on Gswloop structure formation; (a) Overlay of 1H,15N-HSQC spectra of Gswloop (T = 283 K) at [RNA]:[Mg2+] ratios of 1:8 (black) and 1:33 (red). INSET: secondary structure of Gswloop. RNA imino proton resonances that are exclusively detectable at a high [RNA]:[Mg2+] ratio (1:33) are annotated and color coded in red. Imino proton resonances with a chemical shift perturbation Δδ of >40 Hz within [RNA]:[Mg2+] ratios ranging from 1:8 to 1:33 are annotated in blue; (b) chemical shift perturbation (CSP) Δδ [Hz] of resolved imino proton resonances upon Mg2+ titration, gray bar: titration of Gswloop to a final [RNA]:[Mg2+] ratio of 1:8, black bar: titration of Gswloop to a final [RNA]:[Mg2+] ratio of 1:33; (c) CSP [Hz] of two exemplary imino proton resonances (G55 and G57) as a function of the [Mg2+]:[RNA] ratio; the KD-values are determined from the correlation up to a [Mg2+]:[RNA] ratio of ∼18:1. The dashed gray line indicates the minimal [RNA]:[Mg2+] ratio for which all signals of the tertiary loop–loop interaction can be detected in the NMR spectra of Gswloop.
A further increase in Mg2+ concentration leads to drastic changes in the NMR spectra of Gswloop. A number of additional imino proton signals were observed in the 1H,15N-HSQC spectrum. Interestingly, these signals correspond to residues involved in the tertiary loop–loop interaction (U34, G32, G38, U61) or to the closing base pairs in helical elements (G45, U67). These signals are exclusively detected when the long-range tertiary interaction is formed (Figure 2a). Our NMR experiments thus indicate that high concentrations of Mg2+ mediate the formation of the long-range tertiary loop–loop element that is destabilized as a result of the introduced mutations.
Second, at high [RNA]:[Mg2+] ratios the Mg2+-induced formation of the tertiary loop–loop interaction induces sizeable chemical shift changes for signals in the helical elements P2 and P3 (Figure 2). These CSPs might be associated with two effects. They could be caused by local RNA conformational changes induced by interactions with Mg2+ and, in addition, due to changes in inter-helical packing as soon as the formation of the tertiary long-range interaction induces the parallel arrangement of helices P2 and P3. The changes in the CSPs are rather abrupt. These abrupt changes likely reflect a global conformational change taking place in a coupled process of Mg2+ binding and RNA folding due to formation of a stable, Mg2+-mediated long-range tertiary interaction.
NMR spectra of Gswloop were recorded in the presence of cobalt hexamine (37) in order to clarify whether direct inner-sphere metal coordination or non-specific electrostatic interactions and shielding of negative charges are the minimal pre-requisites to mediate the formation of the long-range tertiary RNA–RNA interactions. Titration of Gswloop with this [Mg(H2O)6]2+ analogue leads to cobalt hexamine association determined by CSP on the imino proton resonances. In addition, not only Mg2+ but also the addition of cobalt hexamine induces the appearance of the imino proton reporter signals in 1H,15N-HSQC spectra indicative of the long-range tertiary loop–loop interaction (data not shown). These results indicate that outer-shell metal–ion coordination is already sufficient to facilitate the formation of the long-range tertiary structural element for Gswloop.
Structural characteristics of the free RNA conformation determine properties of Gswloop–ligand complex formation
The titration of Gswloop with Mg2+ in the presence of the ligand hypoxanthine followed by NMR spectroscopy indicates that ligand binding takes place and induces formation of an RNA–ligand complex that resembles the wild type Gsw–hypoxanthine complex (Supplementary Figure S4). The NMR spectra of the Gswloop–ligand complex in the presence of Mg2+ show the expected additional imino proton resonances resulting from signals of the bound ligand and characteristic residues in the ligand-binding core region of the RNA (29). As discussed in the previous paragraphs, the conformation of Gswloop in the ligand-free state is strongly dependent on the Mg2+ concentration. According to the structural characteristics of these Mg2+-dependent ligand-free RNA conformations, three different scenarios for ligand binding to Gswloop were observed dependent on the [RNA]:[Mg2+] ratios.
In the absence of Mg2+, the tertiary loop–loop interaction is not pre-formed and the mutant Gswloop cannot bind hypoxanthine. The NMR spectra show no RNA–ligand complex signals. Also a 10-fold excess of ligand does not result in complex formation suggesting that such missing structural pre-formation may not be compensated at high ligand concentrations.
For low Mg2+ concentrations with [Gswloop]:[Mg2+] ratios up to ∼1:18, characteristic RNA–ligand complex signals can be detected in the NMR spectra. However, the tertiary loop–loop interaction and the ligand-binding pocket are exclusively formed upon simultaneous binding of ligand and Mg2+.
In contrast, for high Mg2+ concentrations with [Gswloop]:[Mg2+] ratios >1:18, the long-range tertiary loop–loop interaction is already detectable in the ligand-free state of the RNA. When hypoxanthine is added, chemical shift changes can be observed for resonances of the loop residues and of residues in helices P2 and P3, in addition to the appearance of the signals of the ligand-binding core region of the RNA. These chemical shift changes indicate that not only ligand binding, but an additional conformational rearrangement in these remote regions is associated with the ligand-binding process (Supplementary Figure S5).
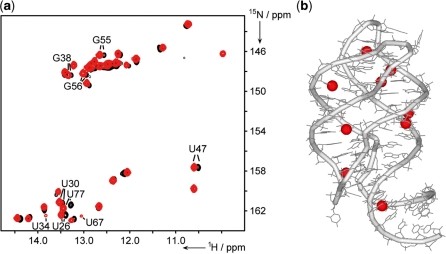
However, comparison of the NMR spectra of the final Gswloop–ligand complexes at (ii) low and (iii) high Mg2+ concentrations reveal CSPs for a number of imino proton resonances (Figure 3). RNA signals that show a CSP of >25 Hz following a Mg2+-titration ([RNA]:[Mg2+] ratio of (ii) ∼1:7 up to (iii) ∼1:33) of the Gswloop–ligand complex are highlighted in Figure 3. At high Mg2+ concentrations ([Gswloop]:[Mg2+] ratio ∼1:33), two additional imino proton signals arising from residues U34 and U67 are detectable. Both residues are located close to the tertiary loop–loop interaction in the structure of the RNA–ligand complex. The observed Mg2+-dependent effects of imino proton resonances in the Gswloop–ligand complex are in good agreement with the Mg2+-binding sites obtained for the wild type Gsw–hypoxanthine complex (29). Thus, although RNA–ligand complex formation can already be detected in NMR spectra at low [Gswloop]:[Mg2+] ratios, the RNA characteristics of the RNA–ligand complexes vary with increasing Mg2+ concentrations.
Figure 3.
Mg2+-induced effects on Gswloop–hypoxanthine complexes; (a) overlay of 1H,15N-HSQC spectra of the Gswloop–hypoxanthine complex with (ii) 7 eq (black) and (iii) 33 eq (red) of Mg2+. Residues that show a chemical shift perturbation Δδ of >25 Hz or can exclusively be detected in the presence of high Mg2+ concentrations are annotated; (b) residues depicted in (a) are highlighted as red spheres on the X-ray structure of the Gsw–hypoxanthine complex [pdb: 1U8D (24)].
Comparison of the NMR spectra of the final RNA–ligand complexes of the wild type and the mutant guanine-sensing riboswitch aptamer domains at elevated Mg2+ concentrations illustrates a high degree of similarity with the exception of imino proton resonances that differ due to the RNA mutations (G37 and U61). As expected for very similar complex structures, the only considerable chemical shift differences in the 1H,15N-HSQC spectra of these RNA–ligand complexes are observed for residues situated in spatial vicinity to the mutation sites in the loop regions of the two RNA constructs (Supplementary Figure S4). In conclusion, the NMR conformational analysis indicates that destabilizing effects of the mutations in the formation of a long-range loop–loop interaction for Gswloop are compensated by the addition of Mg2+ both, in the ligand-free Gswloop and in the RNA–ligand complex; the local stability may, however, be different compared to the wild-type Gsw.
Long-range tertiary interactions influence the RNA conformational state capable of low-affinity ligand binding
For wild-type Gsw, we have previously established that formation of an initial encounter complex between the ligand and the aptamer domain precedes formation of the high-affinity RNA–ligand complex. The encounter complex is characterized by low-affinity and reduced specificity ligand binding. It is detected by increased NMR line widths of the non-cognate ligand adenine in the presence of the guanine-sensing riboswitch aptamer domain and, reciprocally, of the non-cognate ligand hypoxanthine in the presence of the adenine-sensing riboswitch aptamer domain (31). The low-affinity binding may indicate a partial pre-organization of the ligand-binding core of the RNA enabling the transient interaction with the ligand.
In an attempt to unravel the influence of the long-range tertiary structural element on the properties of such an initial encounter complex, we utilized an analogous experimental set-up for Gswloop. The NMR line widths of the non-exchangeable ligand signals (C2–H2 and C8–H8) were determined using isotopically (13C,15N)-labeled ligands, hypoxanthine and adenine, in the presence of unlabeled Gswloop (Supplementary Table S1). For the free ligands in solution, the NMR line widths were determined to be 6.9 ± 0.1 Hz and 7.5 ± 1.0 Hz for adenine and hypoxanthine, respectively. As expected in the presence of Gswloop and Mg2+, the line width of hypoxanthine increases to 33.0 ± 3.2 Hz, reflecting the formation of a specific RNA–ligand complex in slow exchange. A low-affinity complex of Gswloop and adenine with an increase in the ligand line width to 11.3 ± 0.1 Hz was observed at a [Gswloop]:[Mg2+] ratio >1:18. These results resemble the ligand-binding characteristics both, of specific binding of the cognate ligand hypoxanthine and of formation of an initial encounter complex of the non-cognate ligand adenine also detected for the wild-type Gsw (31). Interestingly, the RNA-induced line-broadening effects of ligand NMR signals in the absence of Mg2+ are only minor and equivalent to line widths previously observed in the presence of a control RNA construct lacking the ligand-binding site (31). As Gswloop in the absence of Mg2+ is not capable of low-affinity ligand binding, we conclude that the increased conformational dynamics of the RNA ensemble in the absence of long-range tertiary interactions prevent formation of such an initial encounter complex.
RNA–Mg2+ interactions affect the temperature-dependence of the functional stability for guanine-sensing riboswitch aptamer domains
The ligand-free RNA conformations of Gsw and Gswloop comprise very similar secondary structures but differ in the extent of pre-formed long-range tertiary interactions as well as the variable Mg2+ dependence of their formation. To delineate the contributions of RNA–Mg2+ interactions to RNA structure formation and functional stability we analyzed the temperature-dependent unfolding profiles of Gsw and Gswloop combining global information of CD melting experiments (38) and residue-specific resolution of temperature-dependent NMR studies (Table 1).
Table 1.
Temperature-dependent unfolding transitions of guanine-sensing riboswitch aptamer domains (Gsw and Gswloop)
| RNA | Cofactor | First transition (°C) | Second transition (°C) |
|---|---|---|---|
| Gsw | – | 20.6 | 68.2 |
| Gsw | Mg2+ | 65.9 | 78.9 |
| Gsw | Hypoxanthine | 21.9 | 67.9 |
| Gsw | Mg2+ and hypoxanthine | 69.5 | 80.0 |
| Gswloop | – | – | 67.4 |
| Gswloop | 6 eq Mg2+ | – | 68.1 |
| Gswloop | 33 eq Mg2+ | 36.1 | 73.8 |
| Gswloop | 33 eq Mg2+ and hypoxanthine | 37.0 | 73.5 |
Comparison of thermal denaturation experiments of the two RNA constructs, Gsw and Gswloop, supports the link of RNA–Mg2+ interactions to tertiary RNA structure stability. For RNA constructs (Gsw with and without Mg2+, Gswloop in the presence of high Mg2+ concentrations) or RNA–ligand complexes for which the tertiary loop–loop interaction is pre-formed according to the NMR data, the melting profiles show two transitions (Figure 4c, Supplementary Figure S6). Melting profiles of Gswloop in the absence of Mg2+ or at low Mg2+ concentrations where the long-range loop–loop interaction is not stable formed reveal only a single unfolding transition at 67.4–68.1°C. Accordingly, the first melting transition can be tentatively assigned to unfolding of the tertiary loop–loop structural element, while the second transition (or in case of Gswloop in the absence of Mg2+ or at low Mg2+ concentrations the only transition) can be attributed to unfolding of secondary structures.
Figure 4.
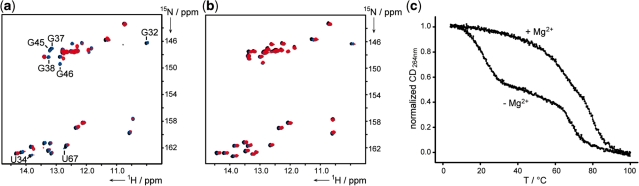
Temperature-dependent characteristics of Gsw–hypoxanthine complexes; overlay of temperature-dependent 1H,15N-HSQC spectra (black: 10°C, blue: 20°C, red: 30°C) of (a) the Gsw–hypoxanthine complex, (b) the Gsw–hypoxanthine complex in the presence of Mg2+ (15N-labeled RNA, unlabeled hypoxanthine). Imino proton signals not detectable at 30°C are annotated; (c) normalized CD melting profiles of the Gsw–hypoxanthine complex in the presence and absence of Mg2+.
Taking advantage of residue-specific resolution that can be obtained by NMR spectroscopy we characterized the first melting transition in temperature-dependent NMR experiments to gain further evidence for the assignment of this transition to the unfolding of specific structural elements (Figure 4). The CD melting experiments reveal a first transition for the Gsw–hypoxanthine complex at 21.9°C, while upon addition of Mg2+ a shift of this first melting transition to 69.5°C could be observed (Figure 4c). Comparison of 1H,15N-HSQC spectra of the Gsw–hypoxanthine complex at different temperatures indicates that a number of imino proton NMR signals disappear at 30°C, while, in contrast, at this temperature all signals of the RNA–ligand complex in the presence of Mg2+ can be observed in the NMR spectrum. In detail, for the Gsw–hypoxanthine complex the signal intensities of nucleotides forming the base-quadruples of the tertiary loop–loop interaction U34, G37 and G38 and of nucleotides G32, G45 and U67 that can only be detected if the long-range tertiary interaction is present, decrease at 30°C. Concerning tertiary interactions constituting the ligand-binding pocket, characteristic imino proton signals broaden but the signal for G46 is the only reporter that diminishes at 30°C. However, a further increase in temperature (35–40°C) then results in NMR spectra characterized by the absence of all other NMR resonances of nucleotides involved in formation of the ligand-binding pocket.
For the ligand-free state of Gsw, no imino proton signals from nucleotides situated in the ligand-binding core can be observed. However, NMR resonances from nucleotides located in the loop regions of the RNA reveal similar temperature-dependent effects upon addition of Mg2+ ions (data not shown). Based on these NMR results, the first temperature-dependent unfolding transitions can reliably be assigned to unfolding of the respective existing tertiary interactions.
The effect of the two cofactors on the thermal stability of the RNA complexes is different. While hypoxanthine does not induce a significant shift of the unfolding transitions neither for Gsw nor for Gswloop, a strong stabilization of tertiary structure could be observed in the presence of Mg2+ (Figure 4c, Table 1). However, while the two unfolding transitions are well separated e.g. for the Gsw–hypoxanthine complex in the absence of Mg2+, at higher Mg2+ concentrations temperature-dependent unfolding of tertiary and secondary structural elements becomes less distinguishable. For Gsw in the ligand-free and the ligand-bound state, the presence of Mg2+ induces a shift of the first unfolding transition of ∼45.3–47.6°C, while the secondary structure unfolding transitions are similar in both states, respectively.
For Gswloop and Gswloop–hypoxanthine complexes at [RNA]:[Mg2+] ratios for which the tertiary loop–loop interaction is formed, two melting transitions are observable. However, while for experimental conditions comparable to Gsw in the presence of Mg2+ the second transition shows a similar stabilization through association of Mg2+ ions, the first unfolding transition can here be determined to be 36.1–37°C.
Taken together, our temperature-dependent experiments indicate that stabilization of the tertiary structure is very sensitive to changes in Mg2+ concentration for both RNA constructs. However, the Mg2+-induced tertiary structure stabilization is more pronounced for Gsw than for Gswloop, an effect that might be attributed to the reduced number of tertiary contacts due to the mutations in Gswloop.
DISCUSSION
Riboswitches constitute evolutionary optimized RNA elements that sense the concentration of small ligands and thereby function as very sensitive regulators of gene expression. However, not only nucleotides in the sensor region were found to be essential for ligand-binding efficiency, but also organization of global RNA architecture involving structured regions remote from the ligand-binding core plays an important role for regulatory function (17,18). With the intricate network of obligatory inter- and intramolecular interactions that sensitively defines their function, riboswitch modules might constitute interesting targets for RNA-targeted drug design (39). A pre-requisite to design molecules to target riboswitches is a detailed understanding of the RNA architecture, the RNA functional stability and the role of RNA–Mg2+ interactions as well as the correlation of these properties with cellular function.
The long-range tertiary loop–loop interaction in RNA–ligand complexes of the guanine-sensing riboswitch aptamer domain is mainly stabilized by two inter-helical base-quadruples and could already be observed in the ligand-free state of the RNA (24,25,29). Within the mutant riboswitch (G37A/C61U), the interactions between the two loop regions can form. The double mutation, however, induces dramatic changes in the conformational ensemble characteristics of the free state of the RNA and, in turn, influences its ligand-binding capability. We observed that the mutations induce a drastic Mg2+ dependence of RNA structural characteristics that strongly differs from those of the wild-type Gsw. The residue-specific resolution in the NMR spectra and the fact that various imino proton reporter signals participate in the formation of the different characteristic inter- and intramolecular RNA tertiary interactions allows the dissection of individual structural contributions to the RNA characteristics.
Mg2+ ions induce different structural effects in the ligand-free RNA conformation depending on the [RNA]:[Mg2+] ratio: we find three different regimes that in turn determine the capability of Gswloop for ligand binding.
The free state of the riboswitch variant Gswloop undergoes considerable dynamics. NMR spectra in the absence of Mg2+ indicate the formation of the predicted secondary structure, but there is no evidence of any tertiary interaction. Neither imino proton signals indicative for the long-range loop–loop interaction nor for nucleotides in the ligand-binding pocket could be detected. The observation that the mutant RNA cannot bind the ligand in the absence of Mg2+ confirms the biological relevance of the long-range tertiary loop–loop interaction for ligand binding in the guanine-sensing riboswitch (22,24).
The presence of Mg2+ ions, however, restores the capability of ligand binding for the mutant Gswloop. This observation raises the question to what extent the destabilized structural pre-formation is compensated by RNA–Mg2+ interactions to obtain a binding-competent RNA conformation in turn.
Even before a stable long-range tertiary interaction is formed, the guanine-sensing riboswitch is ligand-binding competent as evidenced by the NMR-spectroscopic characterization in [Gswloop]:[Mg2+] ratios up to ∼1:18. Here, coupled binding of ligand and Mg2+ induces two structural effects, the formation of the long-range tertiary element and organization of the characteristic ligand-binding pocket. For this [Gswloop]:[Mg2+] concentration regime, restriction of conformational dynamics due to the addition of Mg2+ might induce an RNA conformational ensemble for which loop–loop interactions become dynamically accessible and thus enable ligand binding.
Following a further increase in Mg2+ concentration ([RNA]:[Mg2+] ratios >1:18) the appearance of characteristic imino proton reporter signals in the NMR spectra indicate the stabilization of the tertiary long-range interaction already in the ligand-free state of the RNA. Under these conditions, the dynamic ensemble of RNA conformations is now skewed towards conformations with potential loop–loop interactions. Thus, high concentrations of Mg2+ are able to compensate for the RNA structural defects caused by the mutations. In addition, significant chemical shift changes compared to the previously reported concentration regimes are detected for signals in the helices P2 and P3, indicating packing of these helices. This observation might be associated with the combined effects of Mg2+ binding and global RNA folding. Besides formation of the ligand-binding pocket, addition of ligand to Gswloop at high Mg2+ concentrations causes chemical shift changes for residues located in the loop regions. Since the long-range tertiary loop–loop interaction is already pre-formed in the absence of ligand at high Mg2+ concentrations, these findings indicate that upon addition of hypoxanthine not only ligand binding but also a conformational rearrangement takes place. A similar structural rearrangement in regions remote from the core region of the RNA upon ligand binding can also be detected for the wild-type Gsw despite the fact that here, the tertiary interaction is already pre-formed in the free RNA and in the absence of Mg2+ (29,31).
Earlier, we showed that hypoxanthine forms a low-affinity initial encounter complex with the guanine-sensing riboswitch aptamer domain. By analyzing the mutant RNA, for which we can fine-tune the degree of formation of the tertiary interactions by variation of the Mg2+ concentration, we could decipher pre-requisites for formation of such an initial encounter complex. For Gswloop in the presence of high concentrations of Mg2+, both, specific binding of the cognate ligand hypoxanthine and low-affinity ligand binding of the non-cognate ligand adenine was detected. For this Mg2+ concentration regime, not only structural characteristics in the ligand-free state, namely pre-formation of the long-range tertiary structural element but also the ligand-binding characteristics resemble the situation observed for the wild-type Gsw (31). These observations suggest that an RNA conformational state capable of forming such an initial encounter complex depends on the presence of a partially organized ligand-binding core region. Such partial pre-organization of the core region might be induced by restriction of RNA conformational dynamics through stabilization of remote regions. This restriction can be mediated either by the stable tertiary loop–loop interaction in the wild-type Gsw or by Mg2+-stabilized tertiary loop–loop interactions in the mutant Gswloop. When no stable long-range loop–loop interaction is present, as e.g. for Gswloop in the absence of Mg2+, a low-affinity complex formation cannot be observed, suggesting that the core region is not pre-organized to the same extent. Based on fluorescence data, structural pre-organization in the ligand-binding core region upon addition of Mg2+ was proposed also for an adenine-sensing riboswitch aptamer domain (26).
Our NMR-spectroscopic results indicate that for Gswloop, restriction of conformational ensemble dynamics can be compensated through RNA-Mg2+ interactions but is essential for ligand-binding activity. However, the Mg2+-induced compaction of the RNA seems to be specific, resulting in very similar intramolecular long-range RNA tertiary interactions as observed in the NMR spectra of the wild-type Gsw. Additionally, no misfolded intermediates or non-native conformations could be detected. The possibility for inducing different structural events, namely the formation of the long-range tertiary loop–loop interaction and the ligand-binding pocket independently through variation of the Mg2+ concentration might thus further enable the dissection of the conformational contributions to the kinetics of ligand binding and RNA folding.
The stability of the long-range tertiary structural element is linked to ligand-binding capability for the guanine-sensing riboswitch. Functional stability is significantly affected by RNA–Mg2+ interactions not only for the mutant Gswloop but also for the wild-type Gsw as evidenced by our thermal unfolding experiments. Mg2+ induces the appearance of an additional unfolding transition for Gswloop ([RNA]:[Mg2+] ratio >1:18) that is attributed to the thus-stabilized tertiary structure and pronounced shifts of the melting transition related to the long-range tertiary interactions. For the wild-type Gsw, our data support previous chemical-probing studies that observed a temperature-dependent increase in reactivity for nucleotides in the loop region elements, which was found to be dependent on the presence of Mg2+ (40). Our temperature-dependent results suggest that in the case of a pre-formed loop–loop interaction, the ensemble of RNA conformations becomes further restricted by association of Mg2+ ions that might increase the formation of definite native contacts. When the specific RNA–RNA interactions are absent, the degree of compaction of the RNA ensemble in the ligand-free state is decreased and the free energy barrier might be determined by additional entropy changes.
Our studies strongly support the notion that even nucleotides remote from the ligand-binding region can be crucial for the regulatory capacity as also for the guanine-sensing riboswitch element (22,24). A destabilization in the long-range tertiary element dramatically alters structural and functional characteristics of the RNA conformational ensemble. The topology of the long-range tertiary loop–loop interaction and its conformational dynamics constitute crucial factors in the folding of the riboswitch element. Our results indicate that a stable long-range tertiary interaction reduces the RNA conformational dynamics which might also be accomplished by Mg2+ ions. The possible compensation of stable intramolecular RNA interactions by Mg2+ ions illustrates the significance of RNA–Mg2+ interactions for structure and function of the riboswitch aptamer domains as also observed for other RNAs (2,36,41). For the mutant RNA our results show that the dynamics of the conformational ensemble strongly affect the ligand binding capability of the guanine-sensing riboswitch aptamer domain. We propose that reduction of the conformational ensemble dynamics in the ligand-free RNA that is accomplished by conformational constraining of the helical elements P2 and P3 is the minimal pre-requisite to partially pre-organize and thus optimize the core region of the RNA for ligand binding. Residues involved in the tertiary long-range interaction may have been evolutionarily optimized to increase the functionality of this regulatory element to function even independently of other cofactors abundant in the cell, as for example Mg2+ ions. Our data indicate that the presence of Mg2+ not only compensates for a mutationally destabilized long-range interaction but also for temperature-induced increase of conformational dynamics for the guanine-sensing riboswitch aptamer domain. Thus, an RNA structural stabilization induced by Mg2+ ions seems to be essentially required for riboswitch function in temperature ranges of cellular processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (SSP: ‘Sensory and regulatory RNAs in prokaryotes’) and the Aventis foundation; H.S. is member of the DFG-supported ‘Cluster of Excellence: Macromolecular Complexes’. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Elke Stirnal and Christian Richter for excellent technical assistance.
REFERENCES
- 1.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 3.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 4.Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. Riboswitches: ancient and promising genetic regulators. Chembiochem. 2009;10:400–416. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 5.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 6.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulshina N, Edwards TE, Ferré-D'Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 12.McDaniel BA, Grundy FJ, Kurlekar VP, Tomsic J, Henkin TM. Identification of a mutation in the Bacillus subtilis S-adenosylmethionine synthetase gene that results in derepression of S-box gene expression. J. Bacteriol. 2006;188:3674–3681. doi: 10.1128/JB.188.10.3674-3681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 15.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA. 2008;14:822–828. doi: 10.1261/rna.988608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serganov A. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Blouin S, Lafontaine DA. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA. 2007;13:1256–1267. doi: 10.1261/rna.560307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heppell B, Lafontaine DA. Folding of the SAM aptamer is determined by the formation of a K-turn-dependent pseudoknot. Biochemistry. 2008;47:1490–1499. doi: 10.1021/bi701164y. [DOI] [PubMed] [Google Scholar]
- 21.Baird NJ, Ferre-D'Amare AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 23.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 24.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 25.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 27.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wöhnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Noeske J, Buck J, Fürtig B, Nasiri HR, Schwalbe H, Wöhnert J. Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noeske J, Schwalbe H, Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck J, Fürtig B, Noeske J, Wöhnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl Acad. Sci. USA. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodenhausen G, Ruben DJ. Natural abundance Nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 33.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 34.Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 35.Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 36.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowan JA. Metallobiochemistry of RNA Co(NH3)6(3+) as a probe for Mg(2+)(aq) binding sites. J. Inorg. Biochem. 1993;49:171–175. doi: 10.1016/0162-0134(93)80002-q. [DOI] [PubMed] [Google Scholar]
- 38.Sosnick TR, Fang X, Shelton VM. Application of circular dichroism to study RNA folding transitions. Meth. Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 39.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 40.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyle AM. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.