Abstract
The phosphoinositide 3-kinase (PI3K) pathway is a major target for cancer drug development. PI-103 is an isoform-selective class I PI3K and mammalian target of rapamycin inhibitor. The aims of this work were as follows: first, to use magnetic resonance spectroscopy (MRS) to identify and develop a robust pharmacodynamic (PD) biomarker for target inhibition and potentially tumor response following PI3K inhibition; second, to evaluate mechanisms underlying the MRS-detected changes. Treatment of human PTEN null PC3 prostate and PIK3CA mutant HCT116 colon carcinoma cells with PI-103 resulted in a concentration- and time-dependent decrease in phosphocholine (PC) and total choline (tCho) levels (P < 0.05) detected by phosphorus (31P)- and proton (1H)-MRS. In contrast, the cytotoxic microtubule inhibitor docetaxel increased glycerophosphocholine and tCho levels in PC3 cells. PI-103–induced MRS changes were associated with alterations in the protein expression levels of regulatory enzymes involved in lipid metabolism, including choline kinase α (ChoKα), fatty acid synthase (FAS), and phosphorylated ATP-citrate lyase (pACL). However, a strong correlation (r2 = 0.9, P = 0.009) was found only between PC concentrations and ChoKα expression but not with FAS or pACL. This study identified inhibition of ChoKα as a major cause of the observed change in PC levels following PI-103 treatment. We also showed the capacity of 1H-MRS, a clinically well-established technique with higher sensitivity and wider applicability compared with 31P-MRS, to assess response to PI-103. Our results show that monitoring the effects of PI3K inhibitors by MRS may provide a noninvasive PD biomarker for PI3K inhibition and potentially of tumor response during early-stage clinical trials with PI3K inhibitors.
Keywords: PI3K, Choline Kinase, MRS, phosphocholine, biomarkers
Introduction
Magnetic resonance spectroscopy (MRS) is increasingly used for the noninvasive assessment of cellular metabolism, both in vitro (tissue cultures, body fluids, tissue extracts, and isolated tissues) and in vivo (small animals and humans; ref. 1). Phosphorous (31P)-MRS measures signals from endogenous metabolites such as ATP, indicative of cellular energy status, as well as phosphocholine (PC), phosphoethanolamine (PE), glycerophosphocholine (GPC), and glycerophosphoethanolamine (GPE) arising from the synthetic and degradative pathways of major phospholipids (1). Proton (1H)-MRS can measure the total choline (tCho) peak consisting of PC, GPC, free choline, and corresponding ethanolamine metabolites (1). The levels of lactate, creatine, glutamine/glutamate, and other amino acids can also be obtained from the 1H-MR spectra (1). In the field of cancer, MRS has emerged as a promising tool for characterizing disease and assessing response to therapy (2,3). Higher levels of phosphomonoesters comprising PC and PE were observed in tumors compared with the corresponding normal tissues (4,5). Other studies showed that progression from the normal to the malignant phenotype is associated with an increase in PC and tCho (6). Recently, an increase in the MRS-detected PC/GPC ratio was reported in a mouse model of early gastrointestinal tumorigenesis, highlighting its use as a potential biomarker for monitoring disease progression (7). Similar findings have also been reported in human colorectal cancer (8).
With the emergence of promising new molecularly targeted agents for the treatment of cancer (9), new technologies for the screening and early detection of response to therapy are required. The development of noninvasive endpoints such as MRS is desirable because it may avoid the need for tumor biopsy (10,11).
One oncogenic pathway that is highly and frequently activated in a wide range of tumor types is the production of the phospholipid, phosphatidylinositol-3,4,5-triphosphate by phosphoinositide 3-kinases (PI3K), triggering cell growth, proliferation, survival, motility, invasion, and angiogenesis (12). The use of the PI3K inhibitors wortmannin and LY294002, despite their lack of specificity, has provided initial proof of concept for the anticancer activity of PI3K inhibitors; more recently, compounds that inhibit class I PI3K with increased specificity have been discovered (13-17), including the pyridofuropyrimidine lead compound PI-103 that we identified (14). Detailed characterization was carried out on this compound (17-20). PI-103 showed antiproliferative activity against a range of human cancer cell lines in vitro as well as significant antitumor activity in human tumor xenografts in athymic mice (19,20). PI-103 is now used widely as a chemical tool compound (17) and has been recommended (21) as a replacement for the less potent and selective LY294002 when used alongside wortmannin to show the involvement of PI3K in cellular processes.
Using MRS, we have previously reported that blockade of PI3K with LY294002 and wortmannin in human breast cancer cells was associated with a decrease in PC levels as well as (LY294002 only) an elevation in GPC content (22). With the emergence of more selective and drug-like PI3K inhibitors, we have used MRS to test whether the novel class I PI3K and mammalian target of rapamycin (mTOR) inhibitor PI-103 would result in similar metabolic alterations in human carcinoma cell lines with activating genetic abnormalities in the PI3K pathway. MRS-detected changes were compared with the effects of the cytotoxic microtubule inhibitor docetaxel that is widely used in prostate cancer, with the objective of ruling out nonspecific antiproliferative effects associated with cytotoxicity. We have also investigated potential mechanisms underlying the observed metabolic changes. We report a concentration- and time-dependent decrease in PC and tCho levels following PI3K pathway inhibition with PI-103. Furthermore, we discovered that PC concentrations correlated with choline kinase α (ChoKα) protein expression levels, indicating that ChoKα deregulation following PI3K inhibition is one of the main mechanisms underlying the reduction in PC detected by MRS.
Materials and Methods
Cell culture and treatment
The human PTEN null PC3 prostate adenocarcinoma and PIK3CA mutant HCT116 colorectal carcinoma cell lines (American Type Culture Collection) were cultured in DMEM (Life Technologies) supplemented with 10% FCS (PAA Labs Ltd), 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies) at 37°C in 5% CO2. Cells were cultured according to the supplier's instructions and used at passages 6 to 12. Cell viability was routinely >90%, as judged by trypan blue exclusion. Both cell lines routinely tested negative for Mycoplasma by PCR. PTEN status was verified by protein expression using Western blotting in house, and PIK3CA status was determined or verified experimentally by sequencing. In the case of the HCT116 cell line, single nucleotide polymorphism profiling was also used to confirm that the genotypes matched those provided in the Cancer Genome Project Cosmic database.
Both cell lines were treated with PI-103. PC3 cells were also treated with docetaxel (Sigma). GI50 values (concentrations causing 50% inhibition of proliferation of tumor cells) were determined using the sulforhodamine B assay following 96 hours continuous exposure to compounds (20). Cell culture conditions were selected to ensure similar availability of choline per cell in control and treated cells over the time course of treatment. At the required time points, cells underwent trypsinization and trypan blue exclusion assay (23). The effect of treatment on cell number was monitored by counting the number of viable attached cells in a treated flask and comparing that number with the number of attached cells in a control flask.
Flow cytometry
Cell cycle analysis was performed as previously described (24). Cell volume was estimated using the Time of Flight parameter on the Elite ESP Beckman Coulter (High Wycombe) cell sorter. Pulse width is measured as the particle (cell) moves through the laser beam and the width of the laser beam is subtracted, through machine hardware, from the measurement. The calibration of the machine for diameter measurements was performed using beads of known diameters: 5, 10, and 15 μm. Assuming a spherical particle, a quantity proportional to volume was calculated by measuring cell diameter from a pulse width signal, using the formula (4/3 × π × r3).
Immunoblotting
Western blotting was performed as previously described (22). Western blots were probed for pAKT (Ser473), total AKT, pS6RP (Ser240/244), total S6RP (Cell Signaling), and glyceraldehyde-3-phosphate dehydrogenase (Chemicon). Protein expression levels of ChoKα were detected using the human ChoKα monoclonal antibody (25). The blots were also probed for fatty acid synthase (FAS; Cell Signaling) as well as phosphorylated (Ser454) and total ATP-citrate lyase (ACL, Cell Signaling). Western blot bands were quantified by densitometry using Image Quant version 5 (GE Healthcare).
In vitro 1H- and 31P-MRS of cell extracts
To obtain an MR spectrum, an average of 4 × 107 cells in logarithmic phase were extracted from cell culture using the dual phase extraction method, as previously described (24,26). Before acquisition of the MRS spectra, the water-soluble metabolites were resuspended in deuterium oxide for 1H-MRS or deuterium oxide with 10 mmol/L EDTA (pH 8.2) for 31P-MRS. 1H- and 1H-decoupled 31P-MR spectra were acquired and metabolites were quantified as previously reported (24).
Statistical analysis
Data are presented as the mean ± SD (n ≥ 3). Statistical significance of differences was determined by unpaired two-tailed Student's standard t tests with a P value of ≤0.05 considered to be statistically significant. Pearson correlation coefficients were calculated using SPSS version 17.
Results
Concentration-response as well as time-response experiments were performed using the PTEN null PC3 human prostate cell line. MR-detected changes were compared with effects on cell number as well as downstream markers of PI3K pathway inhibition. We have examined consistency using the PIK3CA mutant HCT116 human colorectal carcinoma cell line. Results in PC3 cells were also compared with the action of the cytotoxic microtubule inhibitor docetaxel, widely used for the treatment of prostate cancer, to rule out nonspecific effects associated with cytotoxicity.
1H- and 31P-MRS of cell extracts
MRS of aqueous extracts from cells treated in vitro with the class I PI3K and mTOR inhibitor PI-103 was used to identify potential biomarkers of PI3K pathway inhibition.
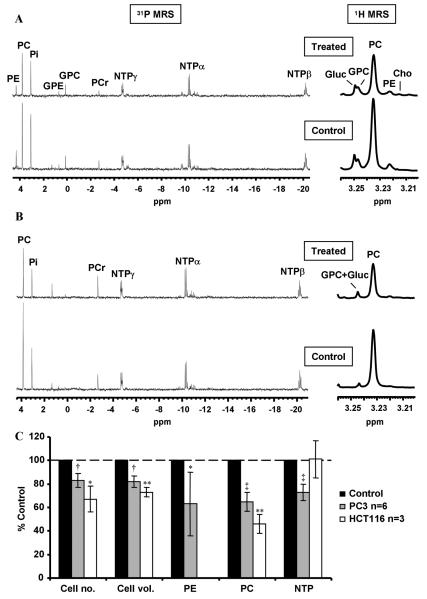
The PTEN null human prostate cell line PC3 was treated with PI-103 for 24 hours at pharmacologically active concentrations corresponding to 1×, 2.5×, 5×, and 10× GI50 (GI50 = 100 nmol/L). Examples of the 31P-MR spectra of control and PI-103–treated PC3 cells are illustrated in Fig. 1A. As summarized in Table 1, 31P-MRS showed that PI-103 treatment for 24 hours led to a significant concentration-dependent reduction in PC, PE, and nucleotide triphosphate (NTP) levels. Levels of PC were reduced from 15.6 ± 1.4 fmol/cell in DMSO-treated control cells to 12.0 ± 0.7 fmol/cell, P < 0.003 using 1× GI50, and to 9.0 ± 0.2 fmol/cell, P < 0.0001 using PI-103 at 10× GI50 (Supplementary Table S1). PI-103 at 5× GI50 caused a time-dependent decrease in PC starting from 6 hours following treatment. For example, at 8 hours, PI-103–treated cells contained 13.2 ± 2.0 fmol/ cell compared with 19.3 ± 3.5 fmol/cell in DMSO-treated control cells (P < 0.02). As shown in Table 2A, the decrease in PC levels in PI-103–treated cells was maintained for up to 24 hours. PI-103 at 5× GI50 also caused a significant time-dependent decrease in PE and NTP levels, but GPC or GPE were not affected (Table 2A).
Figure 1.
Metabolic changes caused by PI-103 in human PTEN null PC3 prostate and PIK3CA mutant HCT116 colon carcinoma cell lines. A, representative in vitro 31P-MRS and expansion of the choline-containing metabolites region of 1H-MR spectra of PC3 aqueous cell extracts following treatment with PI-103 (5× GI50, 500 nmol/L) compared with vehicle-treated (DMSO) control; Gluc, glucose. B, representative in vitro 31P-MRS and expansion of the choline-containing metabolites region of 1H-MR spectra of HCT116 aqueous cell extracts following treatment with PI-103 (5× GI50, 5 μmol/L) compared with vehicle-treated (DMSO) control. C, comparison of cellular effects and 31P-MRS–detected metabolic changes caused by PI-103 (5× GI50) in PC3 prostate cancer and HCT116 colon cancer cells at 24 h. Results are expressed as % treated versus control (% T/C); columns, mean of at least three separate experiments; bars, SD. Statistically significantly different from the control; *, P ≤ 0.05; **, P < 0.01; †, P ≤ 0.005; and ‡, P < 0.0005; two-tailed unpaired t test was used for all comparisons.
Table 1.
Concentration-response analysis of inhibition with PI-103 (24 h): effects on cell number, cell volume and MRS-detected metabolite levels in PTEN null human prostate cancer cells PC3.
| PI- 103/nM |
Cell no. |
P | Cell vol. |
P | PE (31P) |
P | PC (31P) |
P | GPC (31P) |
P | NTP (31P) |
P | PC (1H) |
P | GPC (1H) |
P | tCho (1H) |
P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 96(±8) | ns | 91(±3) | ns | 51(±14) | 0.04 | 80(±1) | 0.001 | 135(±29) | ns | 76(±4) | 0.01 | 78(±5) | 0.02 | 94(±34) | ns | 78(±6) | 0.03 |
| 250 | 91(±6) | ns | 81(±14) | ns | 36(±6) | 0.0003 | 65(±7) | 0.003 | 121(±39) | ns | 77(±8) | 0.02 | 62(±6) | 0.002 | 180(±55) | ns | 76(±8) | 0.01 |
| 500 | 79(±8) | 0.05 | 70(±9) | 0.04 | 38(±8) | 0.01 | 58(±13) | 0.04 | 124(±35) | ns | 65(±8) | 0.03 | 65(±11) | 0.04 | 121(±9) | ns | 77(±3) | 0.006 |
| 1000 | 79(±5) | 0.03 | 72(±12) | ns | 43(±5) | 0.003 | 56(±5) | 0.007 | 147(±63) | ns | 75(±7) | 0.03 | 59(±4) | 0.004 | 122(±33) | ns | 74(±4) | 0.01 |
NOTE:
-Data are expressed as %T/C and presented as the mean ± SD, n ≥ 3.
-Two-tailed unpaired t test was used to compare results in treated cells to controls.
-GPE in PC3 cells was not consistently detectable due to the low levels of this metabolite and was not affected by treatment with PI-103.
-Experiments were not designed to measure levels of other metabolites present in 1H-MR spectra such as lactate, alanine, glutamine/glutamate or acetate.
Table 2.
Time-response analysis of inhibition with PI-103 (5×GI50): effects on cell number, cell volume and metabolite levels in PTEN null human prostate cancer cells PC3:
| A. 31P-MRS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time/h | Cell no. | P | Cell Vol. |
P | PE | P | PC | P | GPC | P | NTP | P |
| 2 | 104(±4) | ns | 98(±3) | ns | 77(±16) | ns | 93 (±6) | ns | 111(±21) | ns | 91(±5) | ns |
| 4 | 103(±10) | ns | 97(±14) | ns | 78(±20) | ns | 87(±9) | ns | 70(±16) | ns | 84(±6) | 0.02 |
| 6 | 96(±10) | ns | 94(±9) | ns | 87(±20) | ns | 79(±10) | 0.006 | 97(±17) | ns | 81(±13) | 0.02 |
| 8 | 108(±9) | ns | 91(±12) | 0.006 | 64(±19) | 0.02 | 69(±8) | 0.002 | 102(±41) | ns | 78(±6) | 0.002 |
| 12 | 101(±10) | ns | 84(±14) | 0.003 | 61(±19) | 0.01 | 57(±10) | 0.001 | 85(±13) | ns | 73(±14) | 0.02 |
| 16 | 88(±4) | 0.01 | 81(±9) | 0.005 | 62(±17) | 0.03 | 56(±8) | 0.002 | 120(±66) | ns | 81(±10) | 0.05 |
| 24 | 83(±6) | 0.001 | 82(±12) | 0.002 | 63(±27) | 0.03 | 65(±8) | 0.0002 | 88(±30) | ns | 73(±7) | 0.0003 |
NOTE:
-Data are expressed as %T/C and presented as the mean ± SD, n ≥ 3.
-Two-tailed unpaired t test was used to compare results in treated cells to controls within the same time-point.
-GPE in PC3 cells was not consistently detectable due to the low levels of this metabolite and was not affected by treatment with PI-103.
1H-MR spectra of extracts of control and PI-103–treated PC3 cells were also investigated (Fig. 1A). Concentration-response analysis showed a statistically significant concentration-dependent decrease in PC levels in PI-103–treated PC3 cells compared with controls. As shown in Table 1, changes in PC observed by 1H-MRS were similar to those detected by 31P-MRS. Furthermore, a concentration-dependent decrease in the tCho (PC+ GPC+ choline) levels was also detected (Table 1). Table 2B shows the time-dependent changes in PC and tCho levels following treatment with PI-103 detected by 1H-MRS. In PC3 cells, tCho levels are dominated by the PC signal; hence, changes in tCho reflected those seen in PC levels over the time course of treatment.
Table 2.
| B. 1H-MRS | ||||||
|---|---|---|---|---|---|---|
| Time/h | PC | P | GPC | P | tCho | P |
| 4 | 99(±8) | ns | 107(±23) | ns | 106(±8) | ns |
| 6 | 80(±9) | 0.01 | 102(±17) | ns | 86(±9) | 0.03 |
| 8 | 71(±8) | 0.002 | 98(±17) | ns | 78(±10) | 0.01 |
| 12 | 62(±7) | 0.002 | 96(±18) | ns | 67(±5) | 0.002 |
| 16 | 58(±3) | 0.0001 | 134(±50) | ns | 61(±4) | 0.0006 |
| 24 | 61(±5) | 0.00002 | 106(±21) | ns | 69(±8) | 0.0003 |
NOTE: See (A)
The PIK3CA mutant human colorectal cell line HCT116 was treated with PI-103 for 24 hours at a pharmacologically active concentration corresponding to 5× GI50 (GI50 = 1 μmol/L). Analysis of 31P-MR spectra of control and PI-103–treated HCT116 cells (Fig. 1B) showed that treatment for 24 hours led to a 54 ± 8% (P = 0.01) decrease in PC levels relative to control, changing from 14.7 ± 1.2 fmol/cell in DMSO-treated control cells to 6.7 ± 0.7 fmol/cell (P = 0.004) following treatment. Levels of NTP were not altered by PI-103, and GPC, GPE, and PE signals were small and not accurately measurable. 1H-MRS (Fig. 1B) showed a significant decrease in the levels of PC (down to 52 ± 1%, P = 0.0005) as well as tCho (down to 62 ± 4%, P = 0.006). As shown in Fig. 1C, PI-103 caused similar effects on PC levels in HCT116 to those seen in PC3 cells.
Relationship of MRS changes with markers of PI3K pathway inhibition
Similar to our previous results (20), 24-hour treatment with PI-103 (5× GI50) resulted in a decrease in the number of treated cells per flask compared with controls both in PC3 cells (down to 83 ± 6%, P = 0.001) and HCT116 cells (down to 69 ± 11%, P = 0.05), consistent with decreased proliferation. In PC3 cells, PI-103 caused both concentration and time-dependent changes in cell number (Tables 1 and 2A). The decrease in PC levels was detectable using PI-103 for 24 hours at 1× GI50 (Table 1). Treatment at 5× GI50 did not show reduced cell number for up to 12 hours, whereas a significant decrease in PC levels was detected as early as 6 hours after PI-103 treatment (Table 2A and B).
Western blotting analysis of downstream components of the PI3K pathway was performed. Using PC3 cells, PI-103 blocked PI3K signaling as indicated by decreases in pAKT (Ser473) and pS6RP (Ser240/244) in treated compared with control cells. This effect was observed at a PI-103 concentration of 1× GI50 (Supplementary Table S2A; Fig. 2A). At 5× GI50, PI-103 decreased pAKT (Ser473) and pS6RP (Ser240/244), which was detectable at 2 hours following treatment and was sustained over the time course of treatment (Supplementary Table S2B; Fig. 2B). Overall, no effect on total AKT or S6RP was detected, indicating that the effects seen were on the phosphorylation of these proteins. Treatment of HCT116 cells with PI-103 for 24 hours at 5× GI50 reduced levels of pAKT (Ser473) and pS6RP (Ser240/244; Fig. 2C), confirming PI3K pathway inhibition in these cells. These results showed that inhibition of the PI3K pathway was occurring at PI-103 concentrations and time points that resulted in the MRS changes reported above.
Figure 2.
Molecular effects of inhibition with PI-103 on the PI3K signaling pathway in PTEN null PC3 prostate and PIK3CA mutant HCT116 colon carcinoma cell lines. A, representative Western blots showing decreases in molecular biomarkers in the PI3K signaling pathway in PC3 prostate cancer cells following treatment with PI-103 at 1×, 2.5×, and 5× GI50, 24 h posttreatment. B, representative Western blots showing decreases in molecular biomarkers in the PI3K signaling pathway in PC3 prostate cancer cells at selected time points posttreatment with PI-103 (5× GI50). C, representative Western blots showing inhibition of molecular biomarkers in the PI3K signaling pathway and changes in protein expression levels of enzymes involved in lipid metabolism in HCT116 colon cancer cells following treatment with PI-103 (5× GI50).
Cell cycle analysis by flow cytometry showed that 24-hour treatment with PI-103 at 5× GI50 caused an increase in the G1 cell population and a decrease in S phase fraction in both cell lines (Fig. 3A–C). In PC3 cells, cell cycle effects of PI-103 were both concentration- and time-dependent (Fig. 3A and B). These data show that our MRS-detected PC changes occurred at low concentrations and preceded PI-103–mediated cell cycle effects.
Figure 3.
Effects of inhibition with PI-103 on cell cycle distribution in PTEN null PC3 prostate and PIK3CA mutant HCT116 colon carcinoma cell lines. A, representative flow cytometry analysis histograms showing the cell cycle distribution of PC3 cells with vehicle treatment (DMSO, control, solid) or following treatment with PI-103 at 1×, 2.5×, and 5× GI50, 24 h posttreatment (empty). B, representative flow cytometry analysis histograms showing the cell cycle distribution of PC3 cells with vehicle treatment (DMSO, control, solid) or following treatment with PI-103 (5× GI50, empty) at selected time points posttreatment. C, representative flow cytometry analysis histograms showing the cell cycle distribution of HCT116 cells with vehicle treatment (DMSO, control, black) or following treatment with PI-103 (grey).
Mechanism for PC depletion
PI3K signaling is well known to regulate cell size (27). This will have an effect on cellular metabolism and hence necessitates normalizing metabolite concentrations to cell volume. We carried out cell volume measurements using flow cytometry on cell samples used for MRS analysis, to determine whether cell volume changes caused by PI-103 contribute to our MRS-detected changes. In PC3 cells, PI-103 at 5× GI50 caused a decrease in cell volume compared with controls starting from 8 hours following treatment (Table 2A). After normalization for cell volume, there is no significant effect of PI-103 on PE or NTP (P > 0.1), but a significant average decrease of 30% (P < 0.05) in PC levels is still detected relative to controls starting from 8 hours following treatment (Supplementary Table S3). Treatment of HCT116 cells with PI-103 reduced cell volume to 73 ± 4% (P = 0.01), relative to controls. Normalizing to cell volume, PC levels in treated cells were still decreased compared with control treatment (down to 63 ± 9%, P = 0.03).
To rule out nonspecific antiproliferative effects associated with cytotoxicity, PC3 cells were also treated with the antimitotic agent docetaxel. Docetaxel was selected as this is used clinically in prostate cancer patients (28). Treatment with docetaxel at 5× GI50 (GI50 = 4.25 nmol/L) for 24 hours decreased cell number to 51 ± 6% (P = 0.001). In direct contrast to the decrease observed with PI-103, 31P-MRS showed that docetaxel increased concentrations of PC (up to 132 ± 12%, P = 0.02), PE (up to 173 ± 23%, P = 0.01), GPC (up to 667 ± 98%, P = 0.002) and NTP (up to 175 ± 19%, P = 0.006). Docetaxel acts through the inhibition of microtubular depolymerization leading to the inhibition of mitosis and induction of apoptosis. In line with this, docetaxel-treated cells were arrested in G2-M phase (from 19 ± 2% to 67 ± 4%, P = 0.0001), resulting in an increase in average cell volume to 138 ± 11% (P = 0.01). Normalizing to cell volume, no change in PC concentrations was detected compared with control, but increases in PE (up to 126 ± 12%, P = 0.04), GPC (up to 487 ± 76%, P = 0.003), and NTP (up to 127 ± 10%, P = 0.02) were still evident. An increase in choline-containing metabolites was also detected by 1H-MRS following treatment with docetaxel, contrasting with the decrease caused by PI-103 (Fig. 4A).
Figure 4.
Investigation of mechanisms underlying MRS-detected changes in PC levels following treatment of PTEN null human prostate cancer cells PC3 with PI-103. A, comparison of cellular effects and 1H-MRS–detected metabolic changes caused by PI-103 and docetaxel (DTX) in PC3 cells at 5× GI50, 24 h. B, representative Western blots showing changes in protein expression levels of FAS, pACL (Ser454), and ChoKα at selected time points posttreatment with PI-103 (5× GI50). C, changes in PC (31P-MRS) normalized to cell volume and ChoKα protein expression levels (densitometry) in PC3 cells over the time course of treatment with PI-103 (5× GI50). Results are expressed as % T/C; points, mean of at least three separate experiments; bars, SD. Statistically significantly different from the control; *, P ≤ 0.05; **, P < 0.01; †, P ≤ 0.005; ‡, P < 0.0005; two-tailed unpaired t test was used for all comparisons.
Several reports have identified a link between PI3K signaling pathway and enzymes involved in choline and lipid metabolism including ChoK (29), FAS (30) and ACL (31). Hence, the effect of PI-103 on the expression of these enzymes was determined to investigate potential mechanisms underlying MRS-detected PC depletion. Western blotting of cell extracts obtained from cells used for MRS analysis showed that PI-103 caused a small decrease in the ACL phosphorylation (Ser454) in PC3 cells (Fig. 4B). The decrease in pACL (Ser454) was maintained up to 24 hours posttreatment. Quantification using densitometry showed a significant decrease in pACL (Ser454) levels reaching a minimum of 67 ± 14% (P = 0.03) in 6 hour–treated cells compared with control cells (Supplementary Table S4). However, no correlation (r2 = 0.3, P = 0.2) was found between changes in the levels of PC (measured by MRS) and pACL (Ser454; quantified by densitometry). A small statistically significant decrease (down to 84 ± 4%, P = 0.04) in FAS levels was seen only at 4 hours after PI-103 treatment (Supplementary Table S4; Fig. 4B). Treatment of HCT116 cells with PI-103 at 5× GI50 for 24 hours also resulted in a decrease in pACL (Ser454; Fig. 2C) and FAS expression compared with controls. However, no correlation was found between PC and pACL (r2 = 0.8, P = 0.1) or FAS (r2 = 0.2, P = 0.5).
Levels of ChoKα were also analyzed by Western blotting, using an antibody against human ChoKα (25). A decrease in ChoKα expression was detectable from 4 hours following treatment of PC3 cells with PI-103 at 5× GI50 (Fig. 4B). ChoKα levels, quantified by densitometry, decreased over the time course of treatment reaching a minimum of 59 ± 13% (P = 0.01) in 16 hour–treated cells relative to control (Supplementary Table S4; Fig. 4C). From 6 hours onwards, a strong correlation was found between PC levels (measured by MRS) and ChoKα levels (r2 = 0.9, P = 0.009; Fig. 4C). ChoKα levels also decreased in HCT116 cells (Fig. 2C) and correlated with the decrease in PC concentrations (r2 = 1, P = 0.02) following PI-103 treatment.
Discussion
PI-103 is a potent inhibitor of class I PI3K and mTOR that was synthesized following a high-throughput screen to identify inhibitors of the enzymatic activity of recombinant PI3K p110α (13-17). Detailed pharmacologic characterization has been carried out on PI-103 (17-20). PI-103 is widely used as a chemical probe for the PI3K/mTOR pathway (17,21) and has inhibitory properties similar to several clinical candidates (32). Identification of noninvasive biomarkers of target inhibition and potentially of tumor response to this novel treatment would be of value in the clinical development of PI3K inhibitors. In this study, we have used MRS to search for potential noninvasive biomarkers of the effects of PI-103 in two human cancer cell lines, PTEN null PC3 prostate and PIK3CA mutant HCT116 colon, in vitro. Using 31P-MRS, a significant decrease in PC concentrations was detected in both tumor lines following PI-103 treatment. A significant reduction in PE and NTP levels was also detected in the human prostate cancer cell line PC3, but no effect on GPC or GPE was observed. In the HCT116 colon cancer cells, NTP levels were not affected by PI-103 treatment while other phosphometabolites including PE, GPE and GPC were below the detection limit of our MR spectrometer.
We have previously reported a similar decrease in PC concentrations following inhibition of PI3K signaling with LY294002 and wortmannin (22). This indicates that the decrease in PC levels is likely to be related to the action of these inhibitors and PI-103 on their common target PI3K, while the increase in GPC observed following treatment with LY294002 (22) may be related to other off-target effects of this weakly potent and nonselective compound. It is important to note, however, that a decrease in PC concentrations was also observed following inhibition of other signaling molecules, for example: mitogen-activated protein kinase (33), FAS (34), HIF-1α (35) and phospholipase Cγ1 (36). Although this might suggest that these changes may not be specific to the PI3K pathway, we have shown that they are different from those seen with for example, inhibitors of HSP90 (23) and HDAC (37,38). Furthermore, assessing the effect of the antimitotic agent docetaxel, we detected a decrease in cell number but an increase in PE and GPC levels. This further supports the view that our MRS-detected changes are a consequence of the inhibition of cell signaling and not simply a nonspecific antiproliferative effects associated with cytotoxicity.
Although changes in PC and/or tCho may not be specific to drugs acting on a particular molecular target or pathway, they can nevertheless provide valuable biomarkers of response (11). Many biomarkers are not specific to a given pathway, but nevertheless have considerable clinical value (e.g., [18F]-2-fluoro-deoxy-d-glucose). Significant PC changes have been observed at 1x GI50 concentrations of PI-103, which are achieved in animal studies (20). Furthermore, preliminary evidence suggests that PI3K inhibitors with improved pharmaceutical properties compared with PI-103 also show similar effects (data not shown). We have demonstrated that we can measure choline-containing metabolites in spectra from tumors from patients in the clinic (2,39). Based on our observation that changes in these metabolites parallel target inhibition in cells, it seems reasonable to hypothesize that our findings reported herein may have the potential to translate to animal and human studies.
Using 1H-MRS, we were able to detect a similar decrease in PC levels to that measured by 31P-MRS in both PC3 and HCT116 cells following treatment with PI-103. More importantly, the composite tCho peak decreased significantly, indicating that the effect of PI-103 can be followed in vivo despite the overlap of the choline-containing metabolite peaks in 1H-MR spectra. This increases the potential for the translation of our work into the clinic as 1H-MRS can be performed on many current clinical MRS systems, in contrast to 31P-MRS, which requires specialized hardware. 1H-MRS also has the advantage of higher sensitivity compared to 31P-MRS, thus reducing measurement times.
The antiproliferative effects of PI-103 were previously investigated using a panel of human cancer cell lines with an activated PI3K pathway caused by different genetic abnormalities (20). Using the PTEN null PC3 cells, PI-103 was found to be ~50-fold more potent than the commonly used PI3K inhibitor LY294002 (20). Similarly, our results show that a low concentration of PI-103 equal to 1x GI50 was sufficient to significantly decrease PC levels, whereas LY294002 required a ~50-fold higher concentration to cause a similar decrease in PC levels in the human breast cancer cell line MDA-MB-231 (22). PI-103 showed a concentration-dependent effect on cell proliferation and G1 cell cycle arrest as well as the decrease in PC levels. PI-103 treatment for 24 h at 1x GI50 resulted in a significant reduction in PC detected by MRS, which was associated with pAKT depletion. Higher concentrations of PI-103 were required to affect cell number and cell cycle. Furthermore, a time-dependent decrease in PC levels starting from 6 hours following treatment with PI-103 (5x GI50) was observed. This was associated with a reduction in pAKT but preceded G1 cell cycle arrest and reduction of cell number. These results further confirm that the decrease in PC levels is related to the inhibition of PI3K signaling, rather than being a consequence of the effect of PI-103 on cell growth. Thus, MRS-detected PC may represent a sensitive and early pharmacodynamic biomarker for response to PI3K inhibition.
PC plays important roles in lipid biosynthesis and cell signaling (40). The concentration of PC in cells is, therefore, likely to result from an interplay between both metabolic and signaling networks. Aberrant choline metabolism has been demonstrated in cancer cells in culture in which elevated choline levels may be indicative of membrane turnover (40), increased malignant potential, or upregulation of signaling processes due to oncogenic activation events (41,42).
In the present study, we have investigated possible mechanisms underlying MR-detectable metabolic changes with the focus on the decrease in PC levels. The aim was to establish the link between PI3K signaling and phospholipid metabolism and, in turn, to help optimize the use of MRS-detectable changes as potential biomarkers for inhibitors of this pathway.
First, we have investigated the decrease in cell size following PI3K/mTOR inhibition as a possible cause for the reduction in MRS-detectable lipid biosynthesis. An enhanced lipid synthesis following AKT activation was previously reported, attributed to the acceleration of the de novo membrane synthesis required to allow the increased cell volume (31,43). In line with this report, inhibition of the PI3K pathway using PI-103 resulted in a significant decrease in treated PC3 and HCT116 cells volume compared to controls. However, this reduction in cell volume was not enough to explain the decrease in PC levels detected by MRS in our system.
Second, we ruled out the possibility that the decrease in PC levels was due to nonspecific effects associated with cytotoxicity by comparing MRS-detected changes in PC3 cells with the effects of the clinically relevant microtubule inhibitor docetaxel. Treatment with this antimitotic agent caused a decrease in cell number but had no significant effect on PC levels. We have previously reported similar results using doxorubicin (23). Further work could be performed with other cytotoxics in the future.
Finally, the effects of PI-103 on enzymes involved in lipid metabolism were explored. As discussed in our previous report (22) and references therein, oncogene- and/or mitogen-signal transduction effectors are known to modulate many phosphatidylcholine-specific enzymes. More specifically, the PI3K pathway has been shown to regulate ChoK, the enzyme responsible for phosphorylation of choline into PC (29). Furthermore, activation of AKT has been shown to induce expression of FAS and ACL, enzymes catalyzing the de novo synthesis of long-chain fatty acids, thus increasing membrane lipid biosynthesis (31,43,44). Hence, we hypothesized that modulation of PI3K pathway using PI-103 would affect these enzymes and thus modulate choline metabolism leading to the changes we detected by MRS. Our results showed a consistent decrease in ChoKα protein expression following treatment with PI-103 in both cell lines. Furthermore, a strong correlation between the decrease in PC levels detected by MRS and ChoKα expression was observed in the human prostate carcinoma cell line PC3 following treatment with PI-103. In contrast, our results showed no obvious relationship between the reduction in PC levels and FAS or pACL protein expression. This indicates that the decrease in ChoKα protein expression induced by PI-103 plays a major role in the MRS-detected PC depletion that we detected in our study.
It is important to note that the involvement of other enzymes related to choline metabolism, such as phospholipases D, C and A2 (45,46) as well as choline transporters (47) cannot be excluded. Further work is required to identify whether these enzymes are also involved in MRS changes detected following PI3K inhibition. We have previously published a genome-wide cDNA microarray profiling study of mRNAs that show altered expression in response to PI-103 in the PTEN null human glioblastoma cells (Supplementary data published therein; ref. 19). Inspection of the data shows changes in the expression of genes involved in glucose and cholesterol biosynthesis. Thus further studies of pathways involved in tumor metabolism should be carried out in the future.
Taken together, our results strongly support a link between ChoKα and its product PC and the PI3K signaling pathway. Because ChoKα protein levels are decreased, one possibility is that ChoKα could be regulated at downstream of mTOR (48). However, GSK-3 could also play a role in lipid metabolism control through ACL (49). Inhibitors that act at the different key components in the PI3K-AKT-mTOR network (32) could be used to further clarify the association between MR-detectable metabolic changes and the PI3K signaling pathway.
In conclusion, our study has shown that MRS can be used to detect a decrease in PC and tCho levels that is associated with the inhibition of PI3K by the class I PI3K and mTOR inhibitor PI-103 in human prostate and colon carcinoma cell lines. We have also identified inhibition of ChoKα as a factor involved in the observed decrease in PC levels following treatment with PI-103. Monitoring the changes in PC and tCho levels by MRS (using either 1H or 31P) may provide noninvasive pharmacodynamic biomarkers of PI3K pathway inhibition and potentially of tumor response in solid tumors during clinical trials with novel PI3K inhibitors.
Supplementary Material
Acknowledgements
We thank J. Titley for the help with flow cytometry analyses.
Financial Support: Association for International Cancer Research grant 03-304 (N.M.S. Al-Saffar), Cancer Research UK and Engineering and Physical Sciences Research Council Cancer Imaging Centre in association with the Medical Research Council and Department of Health (United Kingdom) grant C1060/ A10334 and C1060/6916 (M.O. Leach and L.E. Jackson), Cancer Research UK grant C309/A2187 and C309/A8274 (P. Workman, F.I. Raynaud, and P.A. Clarke), and Comunidad de Madrid (S-BIO/0280/2006), EU grant (LSHG-CT-2006-037278), Ministerio de Sanidad (RD06/0020/0016), and MICIN (SAF2008-03750; J.C. Lacal). P. Workman is a Cancer Research UK Life Fellow. The Institute of Cancer Research coauthors acknowledge National Health Service funding to the Biomedical Research Centre.
References
- 1.Gadian DG. The information available from NMR. NMR and its applications to living systems. 2 ed. Oxford University Press Inc; New York: 1995. pp. 29–64. [Google Scholar]
- 2.Payne GS, Leach MO. Applications of magnetic resonance spectroscopy in radiotherapy treatment planning. (Spec No 1).Br J Radiol. 2006;79:S16–S26. doi: 10.1259/bjr/84072695. [DOI] [PubMed] [Google Scholar]
- 3.Shah N, Sattar A, Benanti M, Hollander S, Cheuck L. Magnetic resonance spectroscopy as an imaging tool for cancer: a review of the literature. J Am Osteopath Assoc. 2006;106:23–7. [PubMed] [Google Scholar]
- 4.Negendank W. Studies of human tumours by MRS: a review. NMR Biomed. 1992;5:303–24. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 5.Smith TAD, Bush C, Jameson C, et al. Phospholipid metabolites, prognosis and proliferation in human breast carcinoma. NMR Biomed. 1993;6:318–23. doi: 10.1002/nbm.1940060506. [DOI] [PubMed] [Google Scholar]
- 6.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–4. [PubMed] [Google Scholar]
- 7.Backshall A, Alferez D, Teichert F, et al. Detection of metabolic alterations in non-tumor gastrointestinal tissue of the Apc(Min/+) mouse by (1)H MAS NMR spectroscopy. J Proteome Res. 2009;8:1423–30. doi: 10.1021/pr800793w. [DOI] [PubMed] [Google Scholar]
- 8.Chan EC, Koh PK, Mal M, et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) J Proteome Res. 2009;8:352–61. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 9.Collins I, Workman P. New approaches to molecular cancer therapeutics. Nat Chem Biol. 2006;2:689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- 10.Beloueche-Babari M, Chung YL, Al-Saffar NM, Falck-Miniotis M, Leach MO. Metabolic assessment of the action of targeted cancer therapeutics using magnetic resonance spectroscopy. Br J Cancer. 2010;102:1–7. doi: 10.1038/sj.bjc.6605457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Workman P, Aboagye EO, Chung YL, et al. Minimally invasive pharmacokinetic and pharmacodynamic technologies in hypothesis-testing clinical trials of innovative therapies. J Natl Cancer Inst. 2006;98:580–98. doi: 10.1093/jnci/djj162. [DOI] [PubMed] [Google Scholar]
- 12.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–19. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa M, Kaizawa H, Moritomo H, et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase P110α inhibitors. Bioorg Med Chem. 2006;14:6847–58. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa M, Kaizawa H, Moritomo H, et al. Synthesis and biological evaluation of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem Lett. 2007;17:2438–42. doi: 10.1016/j.bmcl.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 15.Hayakawa M, Kaizawa H, Kawaguchi K, et al. Synthesis and biological evaluation of imidazo[1,2-a]pyridine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2007;15:403–12. doi: 10.1016/j.bmc.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa M, Kawaguchi K, Kaizawa H, et al. Synthesis and biological evaluation of sulfonylhydrazone-substituted imidazo[1,2-a]pyridines as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2007;15:5837–44. doi: 10.1016/j.bmc.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 17.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 Kinome: From Chemical Tools to Drugs in the Clinic. Cancer Res. 2010;70:2146–57. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan QW, Knight ZA, Goldenberg DD, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–9. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillard S, Clarke PA, Te-Poele R, et al. Molecular pharmacology of phosphatidylinositol 3-kinase inhibition in human glioma. Cell Cycle. 2009;8:443–53. doi: 10.4161/cc.8.3.7643. [DOI] [PubMed] [Google Scholar]
- 20.Raynaud FI, Eccles S, Clarke PA, et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–50. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 21.Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beloueche-Babari M, Jackson LE, Al Saffar NMS, et al. Identification of magnetic resonance detectable metabolic changes associated with inhibition of phosphoinositide 3-kinase signaling in human breast cancer cells. Mol Cancer Therap. 2006;5:187–96. doi: 10.1158/1535-7163.MCT-03-0220. [DOI] [PubMed] [Google Scholar]
- 23.Chung YL, Troy H, Banerji U, et al. Magnetic resonance spectroscopic pharmacodynamic markers of the heat shock protein 90 inhibitor 17-allylamino,17- demethoxygeldanamycin (17AAG) in human colon cancer models. J Natl Cancer Inst. 2003;95:1624–33. doi: 10.1093/jnci/djg084. [DOI] [PubMed] [Google Scholar]
- 24.Al-Saffar NS, Troy H, Ramĺrez de Molina A, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of the choline kinase inhibitor MN58b in Human Carcinoma models. Cancer Res. 2006;66:427–34. doi: 10.1158/0008-5472.CAN-05-1338. [DOI] [PubMed] [Google Scholar]
- 25.Gallego-Ortega D, Ramĺrez de Molina A, Gutierrez R, et al. Generation and characterization of monoclonal antibodies against choline kinase alpha and their potential use as diagnostic tools in cancer. Int J Oncol. 2006;29:335–40. [PubMed] [Google Scholar]
- 26.Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: Evaluation by NMR spectroscopy. Mag Res Med. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- 27.Kozma SC, Thomas G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays. 2002;24:65–71. doi: 10.1002/bies.10031. [DOI] [PubMed] [Google Scholar]
- 28.Mackler NJ, Pienta KJ. Drug insight: Use of docetaxel in prostate and urothelial cancers. Nat Clin Pract Urol. 2005;2:92–100. doi: 10.1038/ncpuro0099. [DOI] [PubMed] [Google Scholar]
- 29.Ramĺrez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and P13K. Oncogene. 2002;21:937–46. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- 30.Wang HQ, Altomare DA, Skele KL, et al. Positive feedback regulation between AKT activation and fatty acid synthase expression in ovarian carcinoma cells. Oncogene. 2005;24:3574–82. doi: 10.1038/sj.onc.1208463. [DOI] [PubMed] [Google Scholar]
- 31.Porstmann T, Griffiths B, Chung YL, et al. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–81. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 32.Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls, and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Beloueche-Babari M, Jackson LE, Al Saffar NMS, Workman P, Leach MO, Ronen SM. Magnetic resonance spectroscopy monitoring of mitogen-activated protein kinase signaling inhibition. Cancer Res. 2005;65:3356–63. doi: 10.1158/10.1158/0008-5472.CAN-03-2981. [DOI] [PubMed] [Google Scholar]
- 34.Ross J, Najjar AM, Sankaranarayanapillai M, Tong WP, Kaluarachchi K, Ronen SM. Fatty acid synthase inhibition results in a magnetic resonance-detectable drop in phosphocholine. Mol Cancer Ther. 2008;7:2556–65. doi: 10.1158/1535-7163.MCT-08-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan BF, Black K, Robey IF, Runquist M, Powis G, Gillies RJ. Metabolite changes in HT-29 xenograft tumors following HIF-1α inhibition with PX-478 as studied by MR spectroscopy in vivo and ex vivo. NMR Biomed. 2005;18:430–9. doi: 10.1002/nbm.977. [DOI] [PubMed] [Google Scholar]
- 36.Beloueche-Babari M, Peak JC, Jackson LE, Tiet MY, Leach MO, Eccles SA. Changes in choline metabolism as potential biomarkers of phospholipase C{γ}1 inhibition in human prostate cancer cells. Mol Cancer Ther. 2009;8:1305–11. doi: 10.1158/1535-7163.MCT-09-0039. [DOI] [PubMed] [Google Scholar]
- 37.Chung YL, Troy H, Kristeleit R, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts. Neoplasia. 2008;10:303–13. doi: 10.1593/neo.07834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sankaranarayanapillai M, Tong WP, Yuan Q, et al. Monitoring histone deacetylase inhibition in vivo: noninvasive magnetic resonance spectroscopy method. Mol Imaging. 2008;7:92–100. [PubMed] [Google Scholar]
- 39.Murphy PS, Viviers L, Abson C, et al. Monitoring Temozolomide treatment of low-grade glioma with proton magnetic resonance spectroscopy. Br J Cancer. 2004;90:781–6. doi: 10.1038/sj.bjc.6601593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–39. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 41.Lacal JC, Moscat J, Aaronson SA. Novel source of 1,2-diacylglycerol elevated in cells transformed by Ha-ras oncogene. Nature. 1987;330:269–72. doi: 10.1038/330269a0. [DOI] [PubMed] [Google Scholar]
- 42.Ronen SM, Jackson LE, Beloueche M, Leach MO. Magnetic resonance detects changes in phosphocholine associated with Ras activation and inhibition in NIH 3T3 cells. Br J Cancer. 2002;84:691–6. doi: 10.1054/bjoc.2000.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porstmann T, Santos CR, Griffiths B, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porstmann T, Santos CR, Lewis C, Griffiths B, Schulze A. A new player in the orchestra of cell growth: SREBP activity is regulated by mTORC1 and contributes to the regulation of cell and organ size. Biochem Soc Trans. 2009;37:278–83. doi: 10.1042/BST0370278. [DOI] [PubMed] [Google Scholar]
- 45.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–6. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 46.Spadaro F, Ramoni C, Mezzanzanica D, et al. Phosphatidylcholine-specific phospholipase C activation in epithelial ovarian cancer cells. Cancer Res. 2008;68:6541–9. doi: 10.1158/0008-5472.CAN-07-6763. [DOI] [PubMed] [Google Scholar]
- 47.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–30. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 48.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–81. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 49.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.