Abstract
Purpose
As genome-scale technologies begin to unravel the complexity of the equivalent tumours in adults, detailed characterisation of high grade gliomas in children have until recently been lacking. We have sought to validate and extend investigations of the differences between paediatric and adult tumours.
Experimental Design
We carried out copy number profiling by array CGH using a 32K BAC platform on 63 formalin-fixed paraffin-embedded (FFPE) cases of high grade glioma arising in children and young people (<23 yrs).
Results
The genomic profiles of these tumours could be subclassified into four categories – those with stable genomes, which was associated with a better prognosis; those with aneuploid or highly rearranged genomes; and those with an amplifier genotype, which had a significantly worse clinical outcome. Independent of this was a clear segregation of cases with 1q gain (more common in children) from those with concurrent 7 gain / 10q loss (a defining feature of adults). Detailed mapping of all the amplification and deletion events revealed numerous low frequency amplifications including IGF1R, PDGFRB, PIK3CA, CDK6, CCND1, CCNE1; and novel homozygous deletions encompassing unknown genes including those at 5q35, 10q25, and 22q13. Despite this, aberrations targeting the “core signalling pathways” in adult glioblastomas are significantly under-represented in the paediatric setting.
Conclusions
These data highlight that whilst there are overlaps in the genomic events driving gliomagenesis of all ages, the paediatric disease harbours a distinct spectrum of copy number aberrations compared to adults.
Keywords: paediatric, glioblastoma, PDGFRA, amplification, deletion, array CGH
INTRODUCTION
The use of genome-scale profiling techniques to identify the key genetic aberrations underlying various tumour types has lead to fundamental discoveries about the drivers of oncogenesis, as well as providing the rationale for specific targeted therapies in these lesions. Until recently, the application of such studies to the fields of high grade glioma specifically, and childhood tumours in general, have lagged behind the adult epithelial cancers. This is now rapidly changing, with large screens of adult glioblastoma through collaborative networks (1) or single institutions (2) joining an increasing number of smaller independent studies (3-7) in comprehensively mapping the glioblastoma genome.
There are also beginning to emerge genomic studies specifically addressing childhood cancers, and there is mounting evidence that the paediatric high grade glioma genome has certain key differences with that of histologically similar adult tumours. An early study using metaphase comparative genomic hybridisation (CGH) (8) highlighted distinct chromosomal changes in 23 childhood cases, a result borne out in a later 10K SNP array study of a further 14 high grade tumours (9), and more recent studies of 18 paediatric glioblastoma on Illumina 100K arrays (10), and 20 high grade tumours using molecular inversion probes (11). Most recently, we have participated in a collaborative effort to carry out molecular profiling of 78 paediatric high grade gliomas by Affymetrix 500K SNP and U133 Plus2.0 expression arrays (12). From all these studies, it seems clear that whilst there are many large-scale chromosomal and specific genetic amplification/deletion events common to tumours from patients of all ages, there are certain events found at significantly different frequencies in paediatric versus adult lesions.
One of the most immediately apparent differences was the high frequency of chromosome 1q gains and 16q losses, and lower frequency of (often concurrent) gain of chromosome 7 and loss of 10q in childhood cases compared to adults. Although there were numerous low frequency amplifications and deletions such as MYC/MYCN, CCND2, KRAS, and CDKN2C which appeared to show the paediatric high grade glioma genome to be similar to clinical secondary adult glioblastomas (13, 14), a lack of IDH1 mutations in the childhood setting demonstrated the distinct biological pathways active during pathogenesis (12).
The most common amplification in the paediatric cases was at 4q12, with shortest region of overlap and expression analyses identifying the amplicon driver to be PDGFRA (12). This was present in up to 17% of primary paediatric glioblastoma, and 29% of diffuse intrinsic pontine glioma (DIPG), and was also found in 50% of cases of high grade glioma arising as a secondary malignancy after cranio-spinal radiation (post-IR). Many cases without PDGFRA amplification were still found to show overexpression of a specific PDGFRA associated gene signature, which was itself distinct from that observed in adult cases with the 4q12 amplification. Taken together, PDGF-driven signalling appears to be preferentially activated in the majority of paediatric tumours, in contrast to adults, where EGFR is implicated as the predominant target (12).
Although these studies are beginning to unravel the key features of the paediatric high grade glioma genome, the total number of cases studied remains considerably smaller than for adult tumours. This is of particular importance given the lower frequency of the majority of genetic aberrations detected in childhood cases. Validating these low-frequency events in independent cohorts as being recurrent abnormalities, as well as the likely identification of novel isolated copy number changes will aid our understanding of the key pathways underlying the diversity of high grade gliomas in children. To this end we have carried out an array CGH study of 63 cases of paediatric high grade glioma from formalin-fixed, paraffin-embedded archival pathology specimens on a 32K tiling-path BAC platform.
MATERIALS AND METHODS
Samples and DNA extraction
High-grade glioma samples from 63 patients (< 23 years old) treated at the Royal Marsden Hospital (RMH), Sutton, and Newcastle Royal Infirmary, UK, were obtained after approval by Local and Multicenter Ethical Review Committees. The collection consisted of 37 glioblastoma multiforme (GBM), 14 anaplastic astrocytomas (AA), 4 anaplastic oligodendrogliomas (AOG), 4 diffuse intrinsic (brain stem) gliomas (DIPG) and (two astroblastoma, one oligoastrocytoma and one gliosarcoma). All cases were archival formalin-fixed paraffin-embedded (FFPE) tissues. The presence of tumour tissue in these samples and the tumour type was verified on a haematoxylin and eosin (H&E) stained section independently by two neuropathologists (DWE and SA-S). Nine of the cases were previously profiled from a frozen tumour specimen in the collaborative SNP study (12). DNA was extracted using the DNeasy Tissue Kit (Qiagen, Crawley, UK) according to the manufacturer’s protocol and quantitated on a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Array CGH
All raw and processed data have been deposited in Array Express1 (E-TABM-857). The array CGH platform used in this study was constructed at the Breakthrough Breast Cancer Research Centre and comprises 31,619 overlapping bacterial artificial chromosome (BAC) probes covering the human genome at an approximate resolution of 50kb (A-MEXP-1734). Hybridisations were carried out as previously described (15) and slides scanned using an Axon 4000B scanner (Axon Instruments, Burlingame, CA, USA) with images analysed using Genepix Pro 4.1 software (Axon Instruments). The median localised background slide signal for each clone was subtracted and each clone Cy5/Cy3 ratio normalised by local regression (loess) against fluorescence intensity and spatial location. Clones overlapping known copy number variants were removed for statistical and visualisation purposes, but not for mapping of specific amplifications and deletions, which was done according to the March 2006 build of the human genome sequence (hg18).
Data analysis
All data transformation and statistical analysis were carried out in R 2.9.02 and BioConductor 2.43, making extensive use of modified versions of the package aCGH in particular (15). For identification of DNA copy number alterations, data were smoothed using a local polynomial adaptive weights procedure for regression problems with additive errors, with thresholds for assigning ‘gain’ and ‘loss’ set at 0.1 (3 × standard deviation of control hybridisations). For visualisation purposes, the processed log2 ratios were coloured green (gain) or red (loss) after segmentation and copy number determination.
In order to assess the significance of the genomic alterations, we applied an algorithm similar to those previously described (Genomic Identification of Significant Targets in Cancer, GISTIC (13) and Genome Topography Scanning, GTS (16)), taking into account the frequency, amplitude and focality of the observed amplifications (log2 ratio > 1.0) and deletions (log2 ratio < −0.75). This was calculated as the product of the absolute log2 ratio, the number of clones in each segment and the frequency within the entire cohort, scaled to the absolute maximum for amplifications/deletions separately, and overplotted on the frequency histogram for gains and losses described above.
Fluorescent in situ hybridisation (FISH)
FISH analysis was carried out on FFPE sections as previously described (17). Probes directed against MYCN (pool of clones RP11-1183P10, RP11-674F13 and RP11-754G14), PIK3CA (RP11-4B14, RP11-642A13, RP11-379M20), PDGFRA (RP11-819D11, RP11-58C6), SKP2 (RP11-749P08, CTD-2010F22), PDGFRB (RP11-211F05, RP11-21I20), MYC (RP11-440N18, RP11-237F24, CTD-2034C18), CDK4 (RP11-66N19, RP11-277A02, RP11-672O16), MDM2 (RP11-611O02, RP13-618A08, CTD-2067J14), and IGF1R (CTD-2015I17, RP11-203H14, RP11-189B22) were labelled with Cy5 (GE Healthcare, Amersham, UK), while chromosome-specific control probes at loci of no copy number change were labelled with fluorescein (GE Healthcare). Hybridized preparations were counterstained with DAPI in antifade (Vector Laboratories Inc., Burlingame, CA, USA). Images were captured using a cooled charge-coupled device camera (Photometrics, Tuscon, AZ, USA).
Statistics
All statistical tests were performed in R2.9.0. Correlations between categorical values were performed using the Chi-square and Fisher’s exact tests. Correlations between continuous variables were performed using Student’s t test or the Mann-Whitney U test. Cumulative survival probabilities were calculated using the Kaplan-Meier method on uniformly treated patients within our cohort from the same institution (RMH), with differences between survival rates analysed with the log-rank test. Important prognostic information (including extent of resection, Karnofsky performance score) was not available for all cases in this retrospective study, so multivariate analysis was not able to be performed. All tests were two-tailed, with a confidence interval of 95%. P values of less than 0.05 were considered statistically significant.
RESULTS
Distinct patterns of copy number change in the paediatric high grade glioma genome
Previously we have utilised whole genome amplification strategies for array CGH studies of tumours extracted from formalin-fixed paraffin-embedded specimens (18). However, in this study we were able to utilise a cohort of samples for which sufficient material was available in order to avoid this. We were able to generate high quality copy number profiles from an unselected series of 63 paediatric high grade gliomas using 32K tiling-path BAC arrays from which the tumour cell purity could be verified as >90% without the need for additional steps.
We observed a mean number of large-scale (whole chromosome or chromosomal arms) gains and losses of 5.8 per sample (median 4, range 0-22), with more losses (mean 3.5, median 3, range 0-14) than gains (mean 2.3, median 2, range 0-11). There were a further mean of 1.8 focal amplifications/deletions per sample (median 1, range 0-11), again with a slightly increased number of deletions (mean 1.0, median 0, range 0-8) compared to amplifications (mean 0.8, median 0, range 0-4). The list of observed alterations are given for the full dataset in Supplementary Table S1.
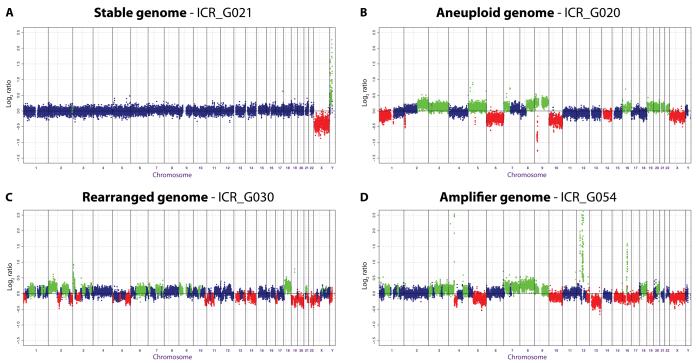
We were able to subtype the samples into four groups based upon the pattern of their genomic profiles. Firstly was a group of tumours that had a very stable genome, with few (<3), low-level, focal changes. This subtype comprised 13/63 (20.6%) cases, and included eight tumours (12.7%) that harboured no detectable copy number alterations on our 32K BAC platform (Figure 1A). The second type contained only large, single copy alterations involving whole chromosomes or chromosomal arms, resulting in aneuploidy in the absence of any high-level amplifications in 22/63 (34.9%) cases, the largest subgroup we observed (Figure 1B). The third type harboured numerous, low-level, intra-chromosomal breaks resulting in multiple gains and losses and a highly rearranged genome. This group was also defined for this purpose by exclusion of cases with bona-fide amplicons, and comprised 11/63 (17.5%) of the cohort (Figure 1C). Finally we considered those tumours with single or multiple high-level (log2 ratio > 1.0) amplifications, regardless of the genomic background, as belonging to the fourth, ‘amplifier’ subtype. This group consisted of 17/63 (27.0%) of cases (Figure 1D).
Figure 1. Paediatric high grade gliomas are comprised of different subtypes of copy number profiles.
Sample genome plots are given for (A) “Stable” (B) “Aneuploid” (C) “Rearranged” and (D) “Amplifier” genomes within our sample cohort. Log2 ratios for each clone (y axis) are plotted according to chromosomal location (x axis). The centromeres are represented by vertical lines. Points are coloured green and red to represent gains and losses, respectively.
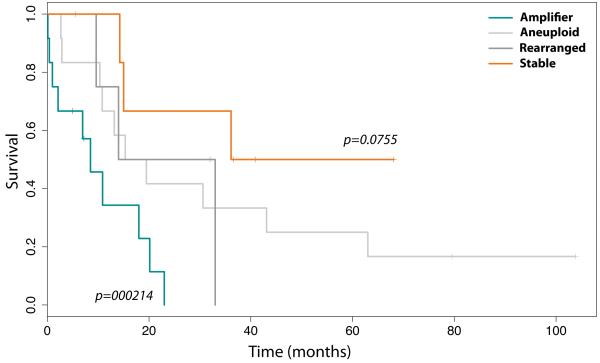
There were no significant correlations between genomic subtype and WHO grade or histology (p > 0.05, Fishers exact test), with glioblastomas, anaplastic astrocytomas and anaplastic oligodendrogliomas spread across all subtypes. Of note, there were no “stable” genomic cases amongst the series of five patients that were treated for a previous malignancy by cranio-spinal radiation (post-IR) (Supplementary Table S1). There was also no association of copy number profiles with age at diagnosis (p > 0.05, Mann-Whitney U test), although the “amplifier’ group did not include any infant tumours (<3 years). However, when we investigated the overall survival of the patients treated at a single institution (RMH), we detected significant differences byretrospective univariate analysis in the clinical outcome of cases according to the genomic profile of the tumour. The “stable” genome cases showed a trend towards better prognosis when compared with all other cases (p = 0.0755, log-rank test), whilst the samples with an “amplifier” genome had a significantly shorter time to death (p=0.00214, log-rank test) (Figure 2). The “aneuploid” and “rearranged” cases fell in between, and were representative of the survival characteristics of the cohort as a whole, suggesting that they may need to be considered together as falling between the extremes of the other two groups.
Figure 2. Genomic subtypes of paediatric high grade glioma have prognostic relevance.
Kaplan-Meier plot for overall survival of paediatric high grade gliomas treated at a single institution stratified according to genomic subtype. The “stable” genome cases showed a trend towards better prognosis when compared with all other cases (p = 0.0755, log-rank test), whilst the samples with an “amplifier” genome had a significantly shorter time to death (p=0.00214, log-rank test)
One of the defining features of paediatric high grade glioma is the frequent gain of chromosome 1q (12/63, 19.0% vs 17/189 9.0% adult cases (1), p= 0.039, Fishers exact test) and loss of 16q (11/63, 17.5% vs 14/189, 7.4%, p= 0.028, Fishers exact test); in contrast to adult glioblastoma cases, where gains of chromosome 7 (12/63, 19.0% vs 140/189, 74.1%, p<0.0001, Fishers exact test), and losses of 10q (10/63, 15.9% vs 152/189, 80.4%, p<0.0001, Fishers exact test) predominate. In our FFPE cohort, we noticed a clear distinction of 1q gain cases from those with concurrent 7 gain / 10q loss (7+/10q−, 8/63, 12.7%), with only a single case harbouring both abnormalities. Neither event was significantly associated with any clinicopathological parameters, although there was a trend towards shorter survival in the 1q+ cases (p=0.0865, log-rank test). Neither abnormality was seen in any infant cases.
Mapping of focal amplifications and deletions to known oncogenes and novel loci
As well as large scale alterations, we observed numerous focal amplifications and deletions. In summary, we identified 47 unique amplification and 32 unique deletions. All these events are detailed in full in Supplementary Table S2 (amplifications) and Supplementary Table S3 (deletions).
The most common amplicon was at 4q12 (10/63, 15.9%), and deletion at 9p21 (10/63, 15.9% - eight homozygous, two hemizygous). Mapping the shortest region of overlap (SRO) in these cases narrowed these regions specifically to PDGFRA and CDKN2A, respectively, confirming the initial observations that these are by far the most common amplifications/deletions in paediatric high grade glioma (12). Other common events included amplification of MYCN at 2p24 (3/63, 4.7%) or MYC at 8q24 (2/63, 3.2%), together giving a frequency of 7.9% (5/63) of cases with genomic MYC family dysregulation; and 3/63 (4.7%) EGFR amplification at 7p12 – a lower frequency than observed in our recent chromogenic in situ hybridisation study of a larger cohort of which this series is a subset, reflecting the focal nature of the amplification event in a small number of tumours identified by molecular pathology (19).
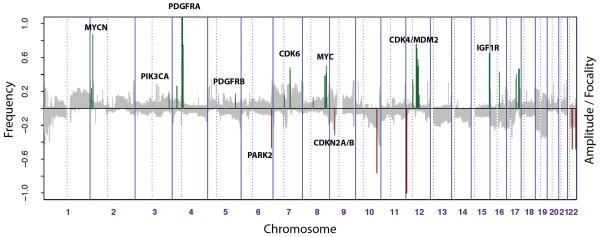
For the remaining aberrations, we highlighted the SROs where they were found to be recurrent. However as most were present only in a single case, and we were unable to narrow down gained/lost regions, with the result that we identified a total of 1026 amplified, and 1243 deleted genes across our series. In order to facilitate the identification of key oncogenic events in paediatric high grade glioma, we sought to assign significance to the genomic aberrations we observed. Inspired by algorithms such as GISTIC (13) and GTS (16), we developed a simple measure based upon three key features of our data for each clone on the array – (a) frequency of high level amplification/homozygous deletion, (b) absolute magnitude of the change, and (c) focality of the segmented copy number change. This amplitude/focality measure was then scaled to the maximum and minimum for amplifications/deletions respectively, and plotted over the frequency of low-level gains and losses on the same histogram (Figure 3).
Figure 3. Summary and significance of genomic aberrations in paediatric high grade glioma.
The proportion of tumours in which each clone is gained or lost is plotted in grey (y axis) for each BAC clone according to genomic location (x axis). A measure of the frequency, amplitude and focality of high level events was calculated for each affected clone and was overplotted for amplifications (green) and deletions (red), scaled to the absolute maximum for each.
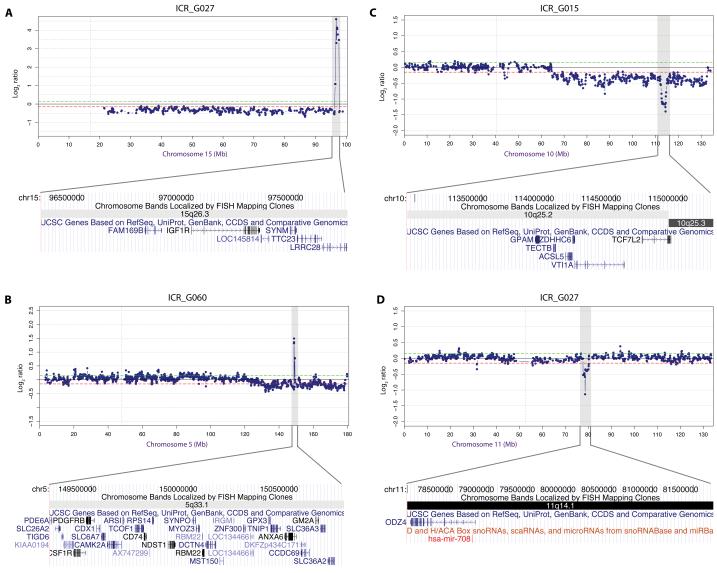
As well as PDGFRA (the highest scoring gene) and CDKN2A, this analysis highlighted the importance of several known oncogenes, amplified at low frequency in our series, but at high magnitude, and in a focally restricted manner. These included PIK3CA (3q26), CDK6 (7q21) and CDK4 (12q14), the first two previously reported in adult glioblastoma, but not in paediatric cases, and present here in a single case. We also identified amplifications of two additional receptor tyrosine kinases – IGF1R at 15q26 (Figure 4A) and PDGFRB at 5q33 (Figure 4B). Such an approach further highlighted the potential significance of known deletions targeting PARK2 at 6q6 and MGMT, PTPRE and others at 10q26; as well as unique events for which the candidate gene is unknown at 10q25 (Figure 4C) and 11q14 (Figure 4D).
Figure 4. Novel low-frequency amplifications and fine-mapping focal deletions in paediatric high grade glioma.
Chromosome plots for (A) chromosome 15, targeting IGF1R; (B) chromosome 5, targeting PDGFRB/CSF1R; (C) chromosome 10, mapping a deletion at 10q25.2-q25.3; and (D) chromosome 11, resolving a deletion at 11q14 to ODZ4 and hsa-mir-708. Log2 ratios for each clone are plotted (y axis) for each BAC clone according to location (x axis) along the length of the chromosome, with genes and microRNA within the minimal regions plotted underneath according to positional information from the UCSC Genome Browser (hg18).
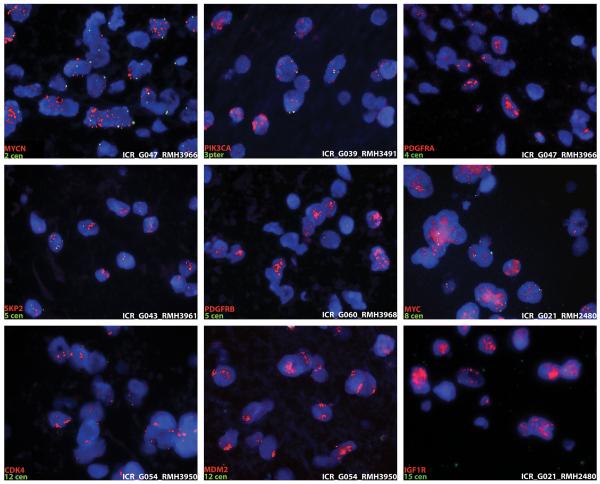
We were able to validate a total of 9 of these lower frequency amplification events by carrying out fluorescent in situ hybridisation on our FFPE sections using specific probes against MYCN, PIK3CA, PDGFRA, SKP2, PDGFRB, MYC, CDK4, MDM2 and IGF1R (Figure 5).
Figure 5.
FISH validation of low frequency amplifications in paediatric high grade glioma. Specific probes for MYCN, PIK3CA, PDGFRA, SKP2, PDGFRB, MYC, CDK4, MDM2, and IGF1R were labelled with Cy5 (red) and co-hybridised to interphase nuclei on FFPE specimens with chromosome-specific control probes labelled with fluoroscein.
Glioblastoma core signalling pathways are not commonly activated by copy number changes in paediatric patients
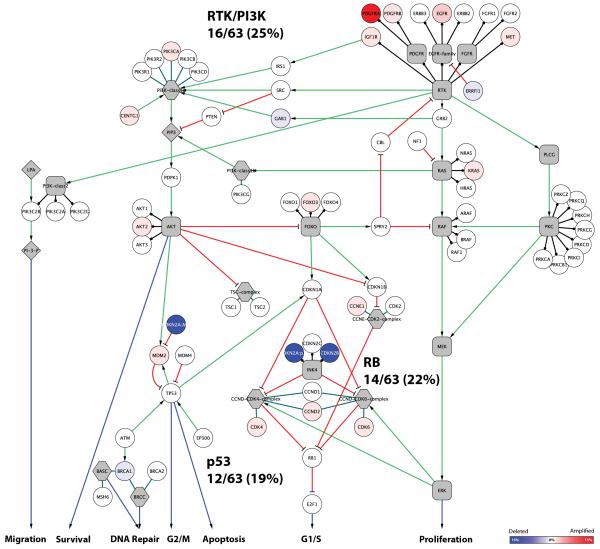
One of the most important findings from recent large-scale genomic profiling studies of adult glioblastoma was the identification of three ‘core signalling pathways’ which were abrogated by amplification, deletion and/or mutations of key genes in the vast majority of cases. Considering only the copy number data from these studies, 59%, 70% and 66% of cases were found to have at least one genetic event targeting the receptor tyrosine kinase/PI3 kinase (RTK/PI3K), p53, or RB pathways respectively (1, 2).
We mapped the copy number changes in our paediatric cases to the same pathways, which included many of the genes described above, as well as others described in adult glioblastoma including MET, KRAS, and AKT2 (RTK/PI3K), MDM2 (p53) and CCND2 (RB). Despite this, we observed a significantly lower frequency of pathway dysregulation compared to that reported in adults – 16/63 (25%) RTK/PI3K, 12/63 (19%) p53 and 14/63 (22%) RB (all p < 0.0001, Fishers exact test) (Figure 6). Even after removing the “stable genome” subtype from this analysis, it is apparent that paediatric tumours demonstrate targeting of these core pathways by copy number alterations in less than half as many instances than in adults.
Figure 6. Glioblastoma “core signalling pathways” are dysregulated by copy number changes less frequently in paediatric than adult tumours.
Signalling pathway heatmap of interactions defined by the TCGA (1). Genes with amplifications are shown in red, genes with focal deletion are shown in blue. The overall frequency of copy number alteration in paediatric high grade glioma for each pathway is listed, and is significantly lower than in adults (25% RTK/PI3K, 19% p53 and 22% RB versus 59%, 70% and 66% for adult glioblastoma respectively).
To explore whether other canonical pathways may be activated by this mechanism preferentially in childhood tumours, we mapped amplified/deleted genes in those tumours without “core” pathway targeting via GenMAPP. Although there were isolated cases with clear genomic events linked to activation of the Sonic Hedgehog (GLI2 amplification, HHIP deletion), and Notch (DLL3 amplification, DLK1 deletion) pathway activation, there was no consistently targeted pathway in these cases, nor specific enrichment of any additional pathway across the entire cohort.
DISCUSSION
We have previously been part of a collaborative study setting out to comprehensively map the copy number alterations present in the paediatric high grade glioma genome, in which we used Affymetrix 500K SNP arrays on a series of 78 cases available as frozen tumour samples (12). Those data revealed an overlapping, but distinct, underlying molecular genetics of the childhood disease when compared with recent large-scale genomic analyses of adult high grade glioma (1, 2). As well as the common amplification/deletion targets of PDGFRA and CDKN2A/B, there were numerous low frequency events targeting both well-recognised oncogenes and novel loci. The purposes of the present study were three-fold: (i) to validate the high frequency events in an independent set of samples, analysed on an independent microarray platform; (ii) to extend the sample set to provide evidence of ‘recurrence’ for the low frequency events previously reported; and (iii) to identify novel low frequency events, which by their nature may have been missed in the earlier study.
The most frequent focal events were PDGFRA amplification and CDKN2A/B deletion, and the most common large scale gains and losses included chromosomes 1q and 16q respectively. The PDGFRAamp, 1q+, 16q− events were significantly more common in the childhood setting (10, 11, 20), although it is important to note that they are present in a proportion of adult tumours. Similarly, we observed a group of tumours in our cohort containing aberrations more commonly associated with the adult disease, namely EGFRamp, 7+, 10q−, albeit at significantly reduced frequencies. That they tended towards exclusivity suggests they represent archetypes for different ends of the spectrum of the disease.
One of the most intriguing differences observed in the paediatric setting was the presence of a proportion of cases of high grade tumours with very few, or even no detectable copy number alterations. This was true on both BAC (approx 32,000 probes, 100kb resolution) and SNP (approx 500,000 probes, 6kb resolution) platforms (12), and is in direct contrast to data from adult tumours (1, 2). This “stable” genomic profile is independent of histological grade or type, and appears to convey an improved survival in patients with high grade glioma, in contrast to those patients with an “amplifier” genomic profile, who do significantly worse.
Another of the defining features of the paediatric high grade glioma genome is the numerous low frequency amplifications and deletions present only in isolated cases in any given study. By nearly doubling the number of these rare tumours for which we have genomic data, we have been able to certain aberrations as recurrent across 132 cases. These include amplifications of known oncogenes within the “core signalling pathways” described in adult glioblastoma, such as CDK6 (10), MET and CCND2, as well as novel targets. These include ID2 at 2p25, previously found in association with the MYCN amplicon at 2p24, possibly part of a single event, identified here as an independent target in its own right. ID2 is a helix-loop-helix transcription factor which has previously been shown to be widely expressed in astrocytic tumours (21, 22), and may play a role in negatively regulating cell differentiation and promoting cell survival (23, 24). Another amplicon at 17q22 was also confirmed in the FFPE series, with an SRO analysis identifying RNF43 as the most likely target. RNF43 is ubiquitin ligase that promotes cell growth and is upregulated in colon cancer (25, 26), but has not previously been implicated in gliomagenesis.
Homozygous deletions now apparent as recurrent lesions include those at 14q32, encompassing a large number of microRNAs, as well as the gene DLK1. DLK1 is a delta-like homolog which acts to inhibit Notch signalling through specific binding interactions with the receptor (27), and may play diverse roles in cellular transformation and differentiation (28). Although we have now observed two cases of homozygous deletion, other mechanisms of down-regulation may be active, as DLK1 is present at an imprinted locus, with increased methylation upstream of GTL2 leading to reduced expression in other tumour types (29). Other deletions may have a more complicated role in gliomagenesis such as those on chromosome 16q. The SNP study identified a large deletion in a single tumour which is present as two separate events at 16q12 and 16q21 in two independent cases here, targeting numerous candidates including clusters of Iroquois homeobox genes, metallothioneins and coiled-coil domain containing genes. By contrast, a homozygous deletion observed in the present study overlaps two independent loci previously reported at 11q14 to target a single microRNA, hsa-mir-708, and a single gene, ODZ4. Although little appears known about mir-708, the odd Oz/ten-m homolog 4 is expressed in the developing and adult central nervous system, and appears to act as an important transcriptional regulator associated with neurodevelopment (30, 31).
Finally, we were also able to identify several novel amplifications and deletions, the significance of many of which is not yet clear. There were some genes identified that were also present in adult glioblastoma studies which had not previously been reported in paediatric high grade glioma, such as AKT2, CCNE1, GLI2, MDM2, PARK2, and PIK3CA. There were other previously unreported genes which may be associated with specific glioblastoma-related signalling pathways such as AKTIP (16q12), an Akt-interacting protein which acts as an activator of the PI3K pathway (32); and PIK3C3 (18q12), also known as Vps34, a member of the PI3K family associated with autophagy (33). There were numerous others with potential functional relevance unknown.
We also noted rare amplifications at receptor tyrosine kinases considered less likely to be driven by copy number gain. Firstly, was a very high level gain of IGF1R at 15q26 (11). Insulin-like growth factor signalling has previously been implicated in gliomagenesis, primarily on the basis of high levels of the ligand IGF2 in glioblastoma specimens (34). The growth-promoting effects of IGF2 that were demonstrated were mediated via IGF1R and the PI3K regulatory subunit PIK3R3. Of particular relevance to the childhood setting was the observation of a mutual exclusivity between IGF2 associated tumours and EGFR-driven cases, suggesting that the IGF pathway may play a prominent role in paediatric tumours, possibly in concert with PDGFR-related signalling.
Secondly was an amplicon at 5q33 which included PDGFRB (and another receptor tyrosine kinase CSF1R). Given the clear importance of PDGFR signalling on paediatric high grade gliomas, it is perhaps unsurprising that there may be multiple mechanisms active in driving tumorigenesis through a common pathway. To this end, we also observed recurrent amplification of the ligand PDGFB (22q13) in the previous SNP study (12), and here further observed focal copy number gain at 7p22 encompassing PDGFA. That these unique genomic events have thus far been found to be restricted to paediatric tumours adds further evidence to a distinct underlying genetics driving archetypal high grade gliomas in children, one that is largely PDGF-driven, and forms a discrete pole within the diversity of glioma biology. Understanding the most appropriate ways of efficaciously targeting these pathways in the most appropriate patient populations will hopefully overcome the disappointing early phase clinical trials observed thus far with PDGFR inhibitors.
STATEMENT OF TRANSLATIONAL RELEVANCE.
Paediatric high grade gliomas represent clinically devastating and biologically understudied tumours of the central nervous system. Little is known about the key genomic alterations which arise in childhood cases, nor the specific differences with the adult disease. We present the copy number profiling of a large series of these rare tumours, and identify numerous low frequency events previously unreported in paediatric high grade glioma, including the potential therapeutic target IGF1R. Tumours could be classified into distinct genomic subtypes, with marked differences in clinical outcome, and an idealised PDGFRAamp, 1q+, 16q− genotype was considerably enriched in paediatric cases, in contrast to EGFRamp, 7+, 10q− cases more commonly associated with adults. We further highlight the importance of PDGF signalling in this context, through the most commonly observed genomic amplification of PDGFRA, as well as a unique amplification of PDGFRB, providing strong rationale for clinically targeting this pathway in children with this disease.
Supplementary Material
Supplementary Table S1 – Summary data of all cases and genomic data. All copy number changes as detected by array CGH are given for each case as well assource, histopathological diagnosis, WHO grade, overall survival, genomic subtype, and “core signalling pathway” activation. Also provided are information regarding whether the patient had previously received craniospinal radiation as treatment for an earlier malignancy, and whether the sample was included in our earlier SNP array study (nine cases) (12).
Supplementary Table S2 – Summary of all focal amplifications. Each individual focal amplification Is mapped for each case in which it is found, with shortest region of overlap reported where present in more than one case. In each instance, genes within the region are listed along with microRNAs. Frequency is given for the present data as well as our earlier SNP study (12). Overall frequency takes into account the nine cases that are present within both.
Supplementary Table S3 – Summary of all focal deletions. Each individual focal deletion Is mapped for each case in which it is found, with shortest region of overlap reported where present in more than one case. In each instance, genes within the region are listed along with microRNAs. Frequency is given for the present data as well as our earlier SNP study (12). Overall frequency takes into account the nine cases that are present within both.
ACKNOWLEDGEMENTS
We acknowledge NHS funding to the NIHR Biomedical Research Centre. This work was supported by The Royal Marsden Children’s Department Fund, Fundação para a Ciência e Tecnologia, Portugal, and Breakthrough Breast Cancer.
Footnotes
REFERENCES
- 1.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2009 doi: 10.1007/s11060-009-9959-4. [DOI] [PubMed] [Google Scholar]
- 4.Gardina PJ, Lo KC, Lee W, Cowell JK, Turpaz Y. Ploidy status and copy number aberrations in primary glioblastomas defined by integrated analysis of allelic ratios, signal ratios and loss of heterozygosity using 500K SNP Mapping Arrays. BMC Genomics. 2008;9:489. doi: 10.1186/1471-2164-9-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Tayrac M, Etcheverry A, Aubry M, et al. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer. 2009;48:55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 6.Ruano Y, Mollejo M, Ribalta T, et al. Identification of novel candidate target genes in amplicons of Glioblastoma multiforme tumors detected by expression and CGH microarray profiling. Mol Cancer. 2006;5:39. doi: 10.1186/1476-4598-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korshunov A, Sycheva R, Golanov A. Genetically distinct and clinically relevant subtypes of glioblastoma defined by array-based comparative genomic hybridization (array-CGH) Acta Neuropathol. 2006;111:465–74. doi: 10.1007/s00401-006-0057-9. [DOI] [PubMed] [Google Scholar]
- 8.Rickert CH, Strater R, Kaatsch P, et al. Pediatric high-grade astrocytomas show chromosomal imbalances distinct from adult cases. Am J Pathol. 2001;158:1525–32. doi: 10.1016/S0002-9440(10)64103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong KK, Tsang YT, Chang YM, et al. Genome-wide allelic imbalance analysis of pediatric gliomas by single nucleotide polymorphic allele array. Cancer Res. 2006;66:11172–8. doi: 10.1158/0008-5472.CAN-06-2438. [DOI] [PubMed] [Google Scholar]
- 10.Qu HQ, Jacob K, Fatet S, et al. Genome-wide profiling using single-nucleotide polymorphism arrays identifies novel chromosomal imbalances in pediatric glioblastomas. Neuro Oncol. 2010;12:153–63. doi: 10.1093/neuonc/nop001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiffman JD, Hodgson JG, VandenBerg SR, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–9. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paugh BS, Qu C, Jones C, et al. Integrated molecular profiling of pediatric high grade gliomas reveals key differences with the adult disease. J Clin Oncol. doi: 10.1200/JCO.2009.26.7252. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beroukhim R, Getz G, Nghiemphu L, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maher EA, Brennan C, Wen PY, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–13. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 15.Natrajan R, Williams RD, Hing SN, et al. Array CGH profiling of favourable histology Wilms tumours reveals novel gains and losses associated with relapse. J Pathol. 2006;210:49–58. doi: 10.1002/path.2021. [DOI] [PubMed] [Google Scholar]
- 16.Wiedemeyer R, Brennan C, Heffernan TP, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–64. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambros MB, Simpson PT, Jones C, et al. Unlocking pathology archives for molecular genetic studies: a reliable method to generate probes for chromogenic and fluorescent in situ hybridization. Lab Invest. 2006;86:398–408. doi: 10.1038/labinvest.3700390. [DOI] [PubMed] [Google Scholar]
- 18.Vuononvirta R, Sebire NJ, Dallosso AR, et al. Perilobar nephrogenic rests are nonobligate molecular genetic precursor lesions of insulin-like growth factor-II-associated Wilms tumors. Clin Cancer Res. 2008;14:7635–44. doi: 10.1158/1078-0432.CCR-08-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bax DA, Gaspar N, Little SE, et al. EGFRvIII deletion mutations in pediatric high-grade glioma and response to targeted therapy in pediatric glioma cell lines. Clin Cancer Res. 2009;15:5753–61. doi: 10.1158/1078-0432.CCR-08-3210. [DOI] [PubMed] [Google Scholar]
- 20.Zarghooni M, Bartels U, Lee E, et al. Whole-Genome Profiling of Pediatric Diffuse Intrinsic Pontine Gliomas Highlights Platelet-Derived Growth Factor Receptor {alpha} and Poly (ADP-ribose) Polymerase As Potential Therapeutic Targets. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 21.Mikami S, Hirose Y, Yoshida K, et al. Predominant expression of OLIG2 over ID2 in oligodendroglial tumors. Virchows Arch. 2007;450:575–84. doi: 10.1007/s00428-007-0394-7. [DOI] [PubMed] [Google Scholar]
- 22.Vandeputte DA, Troost D, Leenstra S, et al. Expression and distribution of id helix-loop-helix proteins in human astrocytic tumors. Glia. 2002;38:329–38. doi: 10.1002/glia.10076. [DOI] [PubMed] [Google Scholar]
- 23.Gray MJ, Dallas NA, Van Buren G, et al. Therapeutic targeting of Id2 reduces growth of human colorectal carcinoma in the murine liver. Oncogene. 2008;27:7192–200. doi: 10.1038/onc.2008.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obayashi S, Tabunoki H, Kim SU, Satoh J. Gene expression profiling of human neural progenitor cells following the serum-induced astrocyte differentiation. Cell Mol Neurobiol. 2009;29:423–38. doi: 10.1007/s10571-008-9338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiura T, Yamaguchi A, Miyamoto K. A cancer-associated RING finger protein, RNF43, is a ubiquitin ligase that interacts with a nuclear protein, HAP95. Exp Cell Res. 2008;314:1519–28. doi: 10.1016/j.yexcr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Yagyu R, Furukawa Y, Lin YM, Shimokawa T, Yamamura T, Nakamura Y. A novel oncoprotein RNF43 functions in an autocrine manner in colorectal cancer. Int J Oncol. 2004;25:1343–8. [PubMed] [Google Scholar]
- 27.Nueda ML, Baladron V, Sanchez-Solana B, Ballesteros MA, Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol. 2007;367:1281–93. doi: 10.1016/j.jmb.2006.10.043. [DOI] [PubMed] [Google Scholar]
- 28.Espina AG, Mendez-Vidal C, Moreno-Mateos MA, et al. Induction of Dlk1 by PTTG1 inhibits adipocyte differentiation and correlates with malignant transformation. Mol Biol Cell. 2009;20:3353–62. doi: 10.1091/mbc.E08-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawakami T, Chano T, Minami K, Okabe H, Okada Y, Okamoto K. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum Mol Genet. 2006;15:821–30. doi: 10.1093/hmg/ddl001. [DOI] [PubMed] [Google Scholar]
- 30.Tucker RP, Chiquet-Ehrismann R. Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev Biol. 2006;290:237–45. doi: 10.1016/j.ydbio.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 31.Zhou XH, Brandau O, Feng K, et al. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3:397–405. doi: 10.1016/s1567-133x(03)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Remy I, Michnick SW. Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Mol Cell Biol. 2004;24:1493–504. doi: 10.1128/MCB.24.4.1493-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soroceanu L, Kharbanda S, Chen R, et al. Identification of IGF2 signaling through phosphoinositide-3-kinase regulatory subunit 3 as a growth-promoting axis in glioblastoma. Proc Natl Acad Sci U S A. 2007;104:3466–71. doi: 10.1073/pnas.0611271104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1 – Summary data of all cases and genomic data. All copy number changes as detected by array CGH are given for each case as well assource, histopathological diagnosis, WHO grade, overall survival, genomic subtype, and “core signalling pathway” activation. Also provided are information regarding whether the patient had previously received craniospinal radiation as treatment for an earlier malignancy, and whether the sample was included in our earlier SNP array study (nine cases) (12).
Supplementary Table S2 – Summary of all focal amplifications. Each individual focal amplification Is mapped for each case in which it is found, with shortest region of overlap reported where present in more than one case. In each instance, genes within the region are listed along with microRNAs. Frequency is given for the present data as well as our earlier SNP study (12). Overall frequency takes into account the nine cases that are present within both.
Supplementary Table S3 – Summary of all focal deletions. Each individual focal deletion Is mapped for each case in which it is found, with shortest region of overlap reported where present in more than one case. In each instance, genes within the region are listed along with microRNAs. Frequency is given for the present data as well as our earlier SNP study (12). Overall frequency takes into account the nine cases that are present within both.