Abstract
Mapping of antigenic peptide sequences from proteins of relevant pathogens recognized by T helper (Th) and by cytolytic T lymphocytes (CTL) is crucial for vaccine development. In fact, mapping of T-cell epitopes provides useful information for the design of peptide-based vaccines and of peptide libraries to monitor specific cellular immunity in protected individuals, patients and vaccinees. Nevertheless, epitope mapping is a challenging task. In fact, large panels of overlapping peptides need to be tested with lymphocytes to identify the sequences that induce a T-cell response. Since numerous peptide panels from antigenic proteins are to be screened, lymphocytes available from human subjects are a limiting factor. To overcome this limitation, high throughput (HTP) approaches based on miniaturization and automation of T-cell assays are needed. Here we consider the most recent applications of the HTP approach to T epitope mapping. The alternative or complementary use of in silico prediction and experimental epitope definition is discussed in the context of the recent literature. The currently used methods are described with special reference to the possibility of applying the HTP concept to make epitope mapping an easier procedure in terms of time, workload, reagents, cells and overall cost.
1. Introduction
Mapping of T epitopes on protein antigens derived from relevant pathogens implies the identification of amino acid sequences that are recognized by CD4 or CD8 T cells. The term “epitope” is frequently used interchangeably for “immunodominant peptide”. For the sake of precision, though, epitope should refer to the shortest sequence that maintains stimulatory capacity for T cells, while the immunodominant peptide can be of any length and its deletions allow identification of the corresponding epitope.
Epitope mapping is important in vaccine development [1, 2] for several reasons: (1) epitopes are presented in the context of one or few MHC alleles [3–7] and in some instances they are promiscuous (i.e. presented by more alleles) [8–11] or universal (i.e. presented by most of the alleles) [12–16]; (2) epitopes represent the antigenic portion of a protein and thus the nonantigenic regions can be deleted; (3) epitopes restricted to different alleles can be collected to obtain selected peptide libraries that are recognized by the majority of the immune population [17, 18], thereby providing a valuable diagnostic tool [19–27]; (4) epitopes can be selected to construct peptide-based vaccines.
Peptides derived from HIV and restricted to human DR alleles were used as pools to prime mice to induce vigorous CD4 responses [28]. Peptides as strings of beads were produced as a recombinant protein to express and immunize against P. falciparum epitopes [29]. As additional examples, peptides as strings of beads have also been used as immunogens specific for HIV [30–32], for mycobacyteria [33], for tumor-associated viral antigens [34], for LCMV [35] and for immunodominant epitopes of five different viruses [36]. In addition, selected peptide fragments can be assembled as mosaic proteins to produce polyvalent vaccines for coverage of potential T-cell epitopes in HIV variants. [37].
In fact, epitope variability in pathogens like HIV [38] is instrumental for development of escape mutants [39] and represents the greatest challenge for the immune system that must deal with these mutations.
The converse situation, defined as deimmunization, permits selective removal by protein re-engineering of immunogenic epitopes identified on proteins to be used as therapeutic agents. In this case, in fact, potential antigenicity must be avoided to improve clinical effectiveness. This innovative possibility has been described in detail [40, 41].
In the present review we mention the predictive and experimental systems that can be used for T-cell epitope mapping. Since the experimental work requires functional T lymphocytes, the number of available cells is a limiting factor with current methods [42]. Furthermore, the time to perform the assays, the cost of reagents and the workload are additional limiting factors in epitope mapping. Peptide synthesis in particular has a remarkable impact when epitope mapping is budgeted. Therefore, in addition to in silico epitope prediction, as discussed later, any miniaturization that reduces the amount of required peptides is helpful. The possibility of using a high throughput (HTP) approach based on assay miniaturization and automation to test extended peptide panels will be dealt with in more detail. The HTP approach, in fact, permits reduction of the number of cells to be tested, of the amount of tested peptides, of the cost of reagents and of the workload referred to the information that can be obtained. The HTP concept can be applied to different techniques that can be automated and miniaturized, as described later.
2. Predictive Models of T-Cell Responses
Algorithms have been developed to test the potential capacity of a given peptide to bind a predetermined MHC allele. From this information it can be assumed that a T-cell with such a specificity can be present in the T-cell repertoire. Obviously, specific T-cell are expected to be at very low frequency in the specific naïve repertoire, while their frequency should be much higher in a primed repertoire. As pointed out later, this is probably the major difference between in silico and in vitro epitope mapping: in the former case an assumption is made while in the latter case an experimental approach is taken that must take into account the naïve or memory state of the specific repertoire that is being tested. Another important point is that in silico screening is faster and cheaper and certainly permits deletion of certain peptides that have no MHC binding potential and thus can be excluded from the list of potential candidates. Therefore, a preliminary prediction may result in remarkable reduction in the number of peptides to be synthesized and screened against responding lymphocytes.
Predictive methods have been developed for human and murine MHC alleles, including class I and class II molecules [43–50]. A large scale validation study for CTL epitope prediction has been recently published by comparing five different web-based tools [51]. Prediction of binding affinity, in addition to specificity, has been recently discussed in [43]. An updated description of predictive methods can be found in the Los Alamos Database website [38]. This site has links to tools for T epitope prediction based on MHC binding and also on cleavage sites to predict antigenic peptide fragments produced upon processing by the human proteasome [52, 53]. In fact, while MHC-peptide binding predictions consider virtual peptides derived from arbitrary cleavage of the protein sequence, anticipation of peptide fragments, that can be actually produced by antigen processing and presenting cells, can be of remarkable relevance also to reduce the number of peptides to be examined as possible candidates for immunogenicity. The combined predictions for TAP binding peptides [54–56] and protein cleavage have been modeled and considered for their potential applications, in combination with MHC binding predictions. [57, 58]. The data generated by using prediction tools to define MHC or TAP binding peptides have been collected in a dedicated database [59]. This comprehensive database includes a large collection of peptide sequences whose binding affinities for MHC or TAP molecules have been assayed experimentally.
The in silico methods based on MHC-peptide binding prediction carry the obvious advantage that T epitopes can be defined independently of the primed or naïve status of a subject. Several recent reviews have reported on this subject and its integration with in vitro data [45, 60, 61]. Peptide binding to MHC class I for CD8 T-cell recognition is certainly predictable with high confidence, while MHC class II binding algorithms have been recently criticized with respect to reliability [62].
It should be stressed here that a higher integration between in silico prediction and in vitro experiments [60] should be considered, each one of the two approaches with its pros and cons.
These aspects are considered in more detail in the different chapters.
MHC binding is a prerequisite for epitope presentation to specific T cells and thus prediction of MHC binding screens for sequences that are potentially antigenic. Nevertheless, MHC binding, even if experimentally determined to appreciate actual peptide binding to solid phase MHC [63], does not grant in itself that a T-cell with such a specificity is present indeed in the T-cell repertoire. Therefore, experimental data generated by in vitro work in which T cells are challenged with peptides, are necessary to confirm the prediction.
3. Experimental Approaches for T-Cell Epitope Mapping and Data Collection
Several assays measure various features of T-cell activation that follows antigen recognition, as described below in more detail. Using different methods, immunodominant peptides have been identified on proteins of various pathogens. The best example of an available database for experimentally identified epitopes recognized by CD4 and CD8 T-cell s is represented by the Los Alamos National Laboratory database [38] that provides updated information on HIV T epitopes. Another exhaustive database is represented by the Immune Epitope Data Base (IEDB, http://tools.immuneepitope.org) [64–66], in which an accurate description of the individual contributions is given. This database is fed with experimental data obtained by literature searches or by voluntary submissions, with an accurate description of methods used for epitope identification.
4. Peptide Layout for High Throughput T Epitope Mapping
One aspect of the T epitope mapping methodologies is represented by the use of synthetic peptides. The straightforward approach is to produce a complete panel of peptides, with a given length and a predetermined overlapping sequence, that encompass the candidate protein. This comprehensive procedure, though, is limited by the cost of peptide synthesis and thus peptide panels can be restricted by deleting those peptides that have been predicted as non binders using in silico screening. A detailed discussion on synthetic peptides to be used for epitope mapping has been published [67].
An epitope with a sequence spanning two sequential peptides would be missed if it is not encompassed by the overlapping sequence. The ideal peptide scan should be with one amino acid progression (sequential peptides). Apart from increased cost, this would be possible when mapping epitopes recognized by antibodies, that are available, in practice, at unlimited amount from diluted serum and that can be tested on microarray formats. For cellular assays that detect T-cell immunity, available PBMC are an incumbent limitation. In order to induce CTL and Th responses, 15–18 mer peptides overlapping by 11 amino acid residues are commonly used. This peptide size is selected so to have potential binding capacity to MHC class I and class II. In order to avoid the need for a CD4 peptide panel and a CD8 peptide panel, a compromise can be reached as described in an extensive study [68] using peptides derived from the immunodominant pp65 protein of cytomegalovirus. Even if it was confirmed that 9 mers are optimal stimulators for CD8 responses, it was clearly shown that 15 mers can also be used to detect CTL activation. This paper also presents the conundrum of which should be the optimal overlap between contiguous peptides in order not to miss a stimulatory epitope. It is concluded that in the case of 15 mers overlapping by 11 residues, each possible 9 mer is present in two 15 mers, except for the one with a central location, that is, present only once in the middle of the 15 mer. To obviate this limit and to have all 9 mers, including the central ones, represented in at least two 15 mers, peptides should overlap by 3 residues. This implies a 25% increase in the number of peptides. Therefore, also in this case a compromise must be accepted between number/cost of synthesized peptides and confidence that no epitopes are present in suboptimal configurations (i.e. at the extreme N- or C-terminus of longer peptides). Another point to be considered is the calculated risk of missing some epitopes when overlapping instead of sequential peptides are used. It is reasonable to say that it is acceptable to miss some epitopes when dealing with large, highly antigenic proteins on which numerous epitopes are expected, as in the case of proteins derived from pathogens. On the contrary, in the case of small proteins or antigens on which a few antigenic epitopes are predicted, as in the case of tumor antigens, the search should be more thorough.
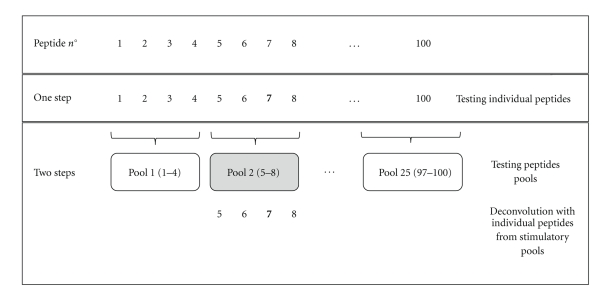
If sequential peptides from panels are individually tested (Figure 1), this results in the use of large numbers of lymphocytes for screening, independently of the method in use (see following paragraphs). Alternatively, discrete pools of peptides can be used for preliminary screening, followed by deconvolution based on testing individual peptides present in the stimulatory pools (Figure 1).
Figure 1.
Conventional layout of peptide panels for T epitope mapping. In this example of a panel of 100 peptides, peptides can be tested individually in one step or as pools in two steps. Peptide 7 in bold represents the immunodominant peptide. Response to a positive pool can be further deconvoluted by testing the individual peptides encompassed by the stimulatory pool. The one step approach is simpler, but it requires more cells. The two-step approach requires fewer cells, but cells are needed at two different times. Thus, the cases in which a small fraction of pools are positive can take advantage of this approach, while in the worst case, in which the majority of the pools are positive and need to be individually deconvoluted, the two-step approach provides no significant advantage.
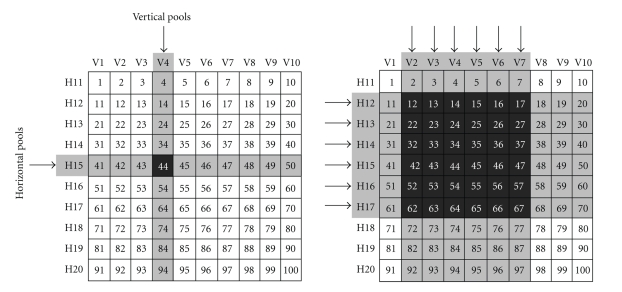
In order to save on cells, peptide matrices have been introduced [69, 70]. In this case, horizontal and vertical peptide pools are assembled as shown in Figure 2. In this example, 20 pools are screened instead of testing 100 individual peptides. The intersection resulting from response to one vertical and one horizontal pool reveal the antigenic peptide. This is a convenient approach in principle. In fact, unambiguous definition of the antigenic peptide results at the intersection (left panel). Nevertheless, in case more peptides are antigenic, the intersections can be ambiguous (right panel) and a second deconvolution step with individual candidate peptides present in the stimulatory pools is needed as detailed in the figure legend. In the layout of this type of experiments, two types of peptide containing plates should be designed: the first one containing the peptides as pools or in a matrix format, the second one containing the full peptide panel. The second deconvolution step can be run by seeding lymphocytes only in the wells that contain peptides of stimulatory pools, rather than preparing a new plate each time with the ambiguous peptides contained in the stimulatory pools. This can reduce errors and labour. Handling of large peptide panels can benefit from automated dispensers that use individual 96 or 384 channels for distribution from masterplates into secondary replica plates.
Figure 2.
Epitope mapping with peptide matrices. As an example, a panel of 100 peptides is considered. Instead of being tested individually, peptides can be arranged in a matrix of 10 vertical pools (V1-V10) and 10 horizontal pools (H11-H20). The left panel depicts an unambiguous identification. In fact, if peptide 44 only is antigenic, pools V4 and H15 are stimulatory (arrows), thereby pointing at the relevant peptide at the intersection. The right panel depicts an ambiguous identification. In fact, if V2-V7 and H12-H7 pools are stimulatory (arrows), all peptides in the shaded area can be antigenic, but the same result would be obtained if only peptides 12,23,34,45,56,67 were antigenic. Therefore, the intersections of the stimulatory pools could indicate from a minimum of 6 to a maximum of 36 antigenic peptides. This ambiguity would require an additional deconvolution step in which individual peptides identified by the intersection area are individually tested.
Conventional peptide synthesis on mg scale provides more material than actually needed. In fact, peptides are currently used at 0.1–10 ug/mL in culture. Miniaturized peptide synthesis on pins [71–73] or on paper spots [74, 75] permits a less expensive peptide synthesis. In addition, microsynthesis in 96 well plate formats permits an easier transfer of peptides to 96 well or to 384 well plates, thereby abating the risk of positional errors or cross-contaminations.
Overlapping peptides have also been applied on microarray slides for antibody epitope mapping [76], but this approach is not applicable at the present time to T epitope mapping with functional assays. In fact, there is no evidence that solid phase peptides can be captured by membrane MHC and pulse APC for presentation to specific T cells. Furthermore, at least one specific T-cell should be present in the microarea of one peptide spot and this depends on the frequency of specific T cells, as discussed in a later paragraph. Biotechnological developments in this field are desirable, even though it should be noted that the microarray slide approach has already been used with MHC-peptide complexes to retain and stimulate specific T cells, as described later.
When the immunodominant protein of a given pathogen is not known, conventional overlapping peptides panels cannot be designed. Therefore, combinatorial peptide libraries consisting of 10-mer mixtures containing every possible combination of residues with only one fixed position could reveal the unknown epitopes [77–79].
5. Current Methods for T Epitope Mapping
Once peptide panels in different formats have been produced, T cells can be tested with different methods to determine the stimulatory immunodominant peptides. Methods to detect T-cell responses can be defined as dynamic or static [42], depending on whether a T-cell function is detected or not. Most of the assays reveal a functional T-cell response as upregulation of activation markers, cytokine synthesis, proliferation, cytolytic, and helper function. Only MHC-peptide multimers (MM) can be used independently of T-cell functions. These fluorescent reagents, in fact, bind and label the specific T-cell receptors by simulating an antigen presenting cell that displays an MHC-peptide complex.
All functional assays require also an intact APC function. Therefore, optimized culture conditions are needed, in particular to provide all specific T cells a chance to interact with an APC. This means that cell density (cells versus surface) is more important than cell concentration (cells versus volume). This is crucial in order not to underestimate responses, particularly if the method reads the actual frequency of specific T cells (e.g. ELIspot, ICS).
5.1. MHC-Peptide Multimers
Multimers constructed with specific MHC class I alleles and carrying a well-defined MHC binding peptide and a fluorescent tag [80] are simple to use, require no culture steps and are revealed by flow cytometric analysis. Additional phenotypic and functional markers can be simultaneous identified on the multimer (MM) binding cells [81]. Cells are not sacrificed and thus can be further cultured or even used for in vivo reinfusion for adoptive immunoreconstitution [82]. Besides these advantages, major limitations are represented by the fact that purified MHC class I molecules are available for a limited number of alleles and by the fact that the antigenic peptide(s) need to be known in advance, as a result of preliminary epitope mapping. In order to apply MM to epitope mapping, a novel approach has been proposed [83] based on reversible binding of MHC and peptide. Here, the first peptide contains a UV cleavable site that results in its dissociation from MHC once exposed to UV light. If a second peptide with the same MHC binding capacity is present, it can replace the cleavable peptide and generate a new MM with a different specificity (i.e. recognized by another T-cell receptor differing for fine specificity but not for MHC restriction). If this process is applied in microplates in which different second peptides are distributed in wells, a whole panel of MM can be constructed for T-cell screening. This approach is appealing, but it is still limited to a few MHC class I alleles and has not been proven to function with MHC class II molecules.
Multimers with MHC class II alleles [84] are not as broadly available as MHC class I multimers and technological improvements are still under development [85].
5.2. Solid Phase MHC-Peptide Complexes
Spotting MHC class I molecules on microarray slides, followed by binding of immunodominant peptides, has been used to create solid phase MHC-peptide complexes that can be recognized by specific T cells. Lymphocytes bind and can be selectively retained [86]. In addition, if the slide is also coated with an anticytokine antibody, the cytokine secreted by specifically activated T cells is captured as a spot and can be revealed by immunofluorescence with an appropriate scanner [87]. This method could be applied for epitope mapping by deposition of different peptides on the MHC spots, but the complexity of the manipulations and of the instrumentation required make this approach not broadly applicable at the present time.
5.3. Lymphoproliferation
The 3H-thymidine uptake assay is sensitive and it has been extensively used in the past. Nevertheless, it requires a radioactive reagent, no data on cell phenotype, frequency or secreted cytokines are provided and 3–5 culture days are needed. Finally, cells are sacrificed and cannot be used any further. The classical format is the 96 well plate, with no possible extension to the 384 well format because of the lack of appropriate harvesters on the market. For these reasons, lymphoproliferation is loosing followers, even though most of the initial epitope mapping data were obtain with this technique. Lymphoproliferation resurged when the CFSE dye dilution method was established. A major advantage in this case is that the labeling dye is diluted between daughter cells at each cell division and thus fluorescence quantitation can estimate the number of duplications the cells went through. An accurate report with methodological details has been recently published [88]. A comparison of proliferation versus ICS for T epitope mapping has also been reported [89].
5.4. Activation Markers Induced on Specific T Cells
Several activation markers have been used for identification, enumeration, and selection of antigen specific CD4 and CD8 T cells. These markers for antigen activated T cells include CD137 for detection of CD8 cells [90] and CD154 for detection of CD4 cells [91]. Coexpression of CD25 and CD134 also identifies antigen specific CD4 T cells [92]. Therefore, the same markers can also be applied for epitope mapping. The use of these markers is advantageous on one side, as it combines functional activity with phenotypic parameters, but it may be cumberson for HTP epitope mapping.
5.5. ELIspot
T cells that secrete a given cytokine (IFNg most of the times) as a consequence of antigen recognition on APC can be detected by ELIspot. In this technique, originally developed to detect antibody secreting B cells [93], lymphocyte cultures are performed in 96 well plates, whose bottom is a nitrocellulose or PVDF membrane coated with a primary anticytokine antibody. The secreted cytokine is captured by the solid phase antibody in the proximity of the producing T-cell. Thus, antigen specific T cells can be revealed as a spot by a second biotinylated antibody, followed by a streptavidin-enzyme conjugate and the appropriate substrate that is enzymatically cleaved so to generate an insoluble product [94]. Spots that form are eventually enumerated by visual inspection under low magnification. This assay requires simple instrumentation and thus it is broadly used. The development of dedicated scanners for spot identification and enumeration of positive cells made it applicable to large-scale screening. A further improvement is the simultaneous analysis of two cytokines produced by a single cell using two detection reagents labeled with different fluorochromes [95]. This enables enumeration of single or double producing T cells. Two limitations of ELIspot are represented by the fact that T-cell phenotype cannot be defined, unless separated subsets are used, and that the actual number of secreting cells can be underestimated. In fact, if more than one specific T-cell interacts with the same APC, one spot may contain more than one specific cell. The 384 well format can be proposed for ELIspot thanks to the availability of appropriate scanners.
5.6. Intracytoplasmic Cytokine Staining (ICS)
In this technique T cells are stimulated with antigen and APC. Responding specific T cells synthesize cytokines that are retained in the cytoplasm because of the addition of drugs that inhibit secretion. Intracytoplasmic cytokines are stained with fluorescent antibodies after cell permeabilization with a detergent. This technique, developed in the late ‘90 s [96] proved very reliable and informative [21, 97]. The main advantage of ICS is that numerous phenotypic markers can be tested on specific T cells and different cytokines can be detected in a single cell using monoclonals labeled with different colors, the only limit being on the flow cytometer used for the analysis (and on the proficiency of the flow cytometrist). Thus, multiparametric and polychromatic flow cytometry identifies antigen specific T cells and simultaneously evaluates different effector functions [98–100]. The assay can be run in round bottom 96 well plates, in combination with automatic samplers that feed the flow cytometer. In the perspective of further extending the assay to the 384 well plate format, HTP-ICS analysis may be foreseen in the near future. Limitations of ICS are represented by the cost of reagents and instrumentation, by the fact that cells are cultured under non-physiological conditions with an inhibitor of protein secretion and by the fact that synthesized cytokines cannot be quantitated. The analysis of mean fluorescence intensity for the stained cytokines, though, may provide some indications for a relative semiquantitative evaluation.
5.7. Cytokine Secretion and Cell Surface Capture (CSC)
In this assay, T cells are activated using culture conditions in larger volumes (tubes, 24 well plates, flasks). After 18–36 hours stimulation, cytokine secreting T cells are identified with an anti-CD45 antibody conjugated with an anticytokine antibody. During a short additional incubation step, secreted cytokines are captured by the bispecific antibody that decorates all PBMC. The cytokine captured on the membrane of the producing T-cell is revealed by a second antibody specific for the same cytokine, that is, directly or indirectly fluorescent for flow cytometric analysis or sorting. Alternatively, the second antibody can be recognized by magnetic beads for preparative separation of specific T cells on magnetic columns [101]. CSC is convenient for ex vivo selection of specific T lymphocytes and it has been used for selection of virus specific T cells [102, 103] that can be subsequently reinfused for adoptive immunoreconstitution [104, 105]. Cytokine positive cells can also be stained with additional markers for extensive phenotyping, but since numerous staining steps are needed, this system is not advisable for screening of peptide panels.
5.8. Cytokine Secretion and Well Surface Capture (Cell-ELISA)
has been reported in 96 well plates [106] and recently proposed, as a variation on the theme of ELIspot and ELISA, in 384 or 1536 well plates [107, 108]. Secreted cytokines, as readout of T-cell activation, can be tested in culture supernatants by conventional ELISA. This procedure can be simplified and miniaturized using cell-ELISA, an assay in which wells are coated with the first anticytokine antibody. Medium and antigens are dispensed automatically and the plates are frozen for later use. When needed, plates are thawed and seeded with PBMC. During the 18–36 hours incubation, specific T cells responding to antigens or peptides secrete the tested cytokine that is captured by the solid phase antibody. Plates are eventually frozen before final processing. For development, plates are processed following a conventional miniaturized ELISA protocol for a quantitative evaluation of produced cytokines using standard curves. Before development, culture supernatants can be saved for cytokine profiling using different HTP approaches, like multiparametric flow cytometric assays or microarrays. Major advantages of this method are that an HTP approach can be taken for handling the multiwell plates.
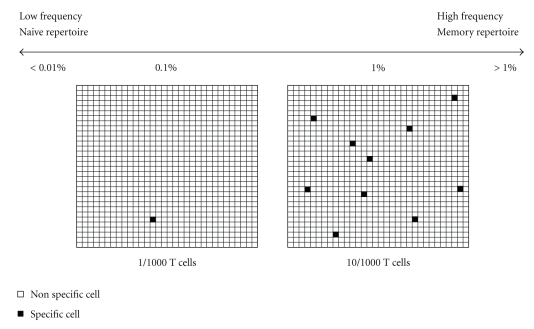
In addition, secreted cytokines are quantitated and the 1536 well plate format permits the use of as few as 104 PBMC per tested antigen. This implies some considerations regarding the frequency of antigen specific T cells. In fact, as shown in Figure 3, a reasonable frequency of memory T cells specific for a recall antigen can range between 0.1% and 1%. Lower frequencies should be expected for T cells specific for a single epitope. Therefore, each culture well should contain a sufficient number of specific cells that results in a positive signal. Sensitivity of cell-ELISA has been estimated around 0.01% specific T cells diluted into autologous non specific T cells by using established lines as standards or PBMC with known frequencies of specific cells [107]. This sensitivity range is similar, if not higher, than other conventional methods including MM, ICS, ELIspot, lymphoproliferation. Therefore, in order to be confident that a detectable number of antigen specific T cells is seeded in each well, we estimate that 104 PBMC per well is the lowest threshold. This consideration applies to scaling down of all types of lymphocyte based assays for evaluation of memory responses. An intermediate format that proved convenient was the 384 small well plate, in which each well has a culture volume of 20 uL instead of 40 uL. Therefore, 2 × 104 PBMC can be seeded instead of 4 × 104, leaving the same cell density at the bottom of the culture well.
Figure 3.
Frequency of specific T cells and culture size. The frequency of antigen specific T cells must be kept in mind when scaling down culture conditions. Reasonable frequencies for specific T cells approximately range from >1% to <0.1% for memory responses to recall protein antigens. Responses to individual epitopes may be at lower frequencies. Frequencies of specific T cells in the naïve repertoire may be <0.01%. Panels represent 1000 lymphocytes containing 1 specific cell (0.1%, left panel) or 10 specific cells (1%, right panel). In case 1000 cells containing 1 specific cell are seeded in a microwell, the specific T-cell may or may not be dispensed according to Poissonian distribution and the signal provided by one single cell may not be detected. In case 1000 cells containing 1% specific cells are seeded, 10 specific cells can provide a detectable positive signal. Therefore, it can be assumed that the smallest number of cells per well should be >103 cells. This reasoning applies to any T-cell assay adapted to 384 or 1536 well plates. Obviously, for very low frequency responses, more cells need to be seeded, but in this case the intrinsic sensitivity of the assay should be checked to establish a frequency threshold. Unfortunately, reliable cellular standards are not available, unless established antigen specific T-cell lines are used, as discussed in [108].
This obvious limitation due to reduction of the number of tested T cells applies to all assays, even if positive cells can be tested individually in principle. The issue of background noise should be considered, since it is possible to detect ex vivo activated T cells in most lymphocyte samples from healthy subjects, with remarkable variability. Therefore, HTP techniques conflict with low frequencies of responding cells. In case a naïve repertoire needs to be analyzed, one possibility can be to go through a preliminary expansion phase with a pool of peptides and then screen the expanded population at a later stage. This obviously requires a second sample of autologous APC, that can be PBMC or EBV transformed B cells for both CD4 and CD8 responses. The generation of antigen specific human T-cell lines from a naïve repertoire for epitope mapping has long been known [109, 110], but a recent appealing technique has opened new perspectives. This “artificial lymph node” technique, described as lymphoid tissue equivalent module, permits T-cell priming in vitro under optimal conditions (www.vaxdesign.com/). This is relevant in human studies, in which antigens cannot be used in vivo for priming like in animal studies. Nevertheless, studies using this approach and evaluation of the system have not yet been published.
After collection of supernatants for cytokine testing, T cells can be saved from wells in which cytokine production was detected for further expansion to generate established T-cell lines. An additional advantage of cell-ELISA is that it can be applied in large scale, collaborative studies. In fact, batches of plates containing predispensed antigen panels can be prepared in the central laboratory and distributed to peripheral laboratories. Here plates are thawed and seeded with PBMC. After incubation, the plates are frozen again and shipped back to the central laboratory for development. This permits the design of multicenter studies on epitope mapping in patients or vaccinees, for whom evaluation of specific cellular immunity would not be feasible with other methods. Simplified culture conditions including HEPES buffer, that avoids the use of a CO2 atmosphere, and a water bath or heating block in place of an incubator, in addition to the possibility of using dry plates for easier storage and shipment, make the assay appealing for use in collaborative studies in resource poor settings. A recent study validated cell-ELISA demonstrating its reproducibility [111]. The possibility of using frozen PBMC and whole blood was also demonstrated. It should be noted that the phenotype of responding cells cannot be defined, unless purified CD4 and CD8 subsets are tested [107] and that the frequency of antigen specific T cells cannot be determined by cell-ELISA. Nevertheless, the number of specific T cells can be indirectly inferred by the amount of secreted IFNg, that has been estimated to be in the range of 1–3 pg per antigen specific cell during a 24 hours culture (Li Pira et al. ms in prep).
6. Conclusions
T-cell epitope mapping allows to identify immunodominant regions on antigenic proteins. Most assays require functional T lymphocytes. Since extended peptide panels are generally tested, large amounts of PBMC are required. Therefore, efforts have been made to reduce the size of culture conditions, independently of the assay selected to define antigen specific responses. The HTP approach is based on miniaturized formats, now available as 384 and 1536 well plates, that require automation to dispense peptides and cells and for final processing. Also dedicated scanners are needed to scan these plates. Several assays described in this review can be adapted to 384 well plates, like ICS and ELIspot, but only cell-ELISA was demonstrated to be feasible in 1536 well plates. This resulted in miniaturized cultures that contained 104 PBMC per well. This figure may represent the lowest scaling down threshold that still grants the presence of a detectable number of specific T cells, considering reasonable concentrations of specific memory T cells around 0.1% or less.
Miniaturized assays described here can be considered for two main applications. The first one is HTP epitope mapping, that is, testing extended peptide panels with PBMC obtained from reasonable blood samples. The second one is analysis of specific cellular immunity, that is, testing T-cell responses towards a limited array of antigen preparations (proteins or peptide libraries) using PBMC obtained from small blood samples. It should be noted that many of the assays described here can also be scaled up for preparative purposes to purify virus specific T cells for adoptive immunoreconstitution, as recently reviewed [112]. In conclusion, T-cell epitope mapping is relevant for vaccine development, for dissection of the quality of response in vaccinees or in infected subjects and for definition of peptide libraries as screening tools of cellular immunity. Thus, we foresee continuous improvements in miniaturization and automation of assays to provide an increasing amount of peptide sequence information to be deposited in dedicated databases.
Acknowledgments
This work was supported by Grants from CIPE (Rome, 2007), Regione Liguria (Genoa, 2007); Ministry of Health, Rome (Finalized Project 2008); Ministry of Higher Education, Rome (FIRB RBNEOI-RB9B.003, FIRB RBIP064CRT-006); Italian Health Institute, Rome (AIDS 40G.33, 45G.17, 50G.25); EU, Bruxelles (LSHB-CT-2005-018680).
Abbreviations
- Th:
T helper lymphocyte;
- CTL:
cytolytic T lymphocyte;
- HTP:
high throughput;
- MM:
multimer;
- APC:
antigen presenting cell;
- MHC:
major histocompatibility complex;
- ICS:
intracytoplasmic cytokine staining;
- CSC:
cytokine secretion and capture;
- ELISA:
enzyme linked immuno-sorbent assay.
References
- 1.Sette A, Peters B. Immune epitope mapping in the post-genomic era: lessons for vaccine development. Current Opinion in Immunology. 2007;19(1):106–110. doi: 10.1016/j.coi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Salit RB, Kast WM, Velders MP. Ins and outs of clinical trials with peptide-based vaccines. Front Biosci. 2002;7:204–213. doi: 10.2741/salit. [DOI] [PubMed] [Google Scholar]
- 3.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nature Reviews Immunology. 2008;8(8):607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen PE. Recent advances in antigen processing and presentation. Nature Immunology. 2007;8(10):1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- 5.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annual Review of Immunology. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 6.Watts C, Powis S. Pathways of antigen processing and presentation. Reviews in Immunogenetics. 1999;1(1):60–74. [PubMed] [Google Scholar]
- 7.Van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annual Review of Immunology. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 8.Frahm N, Yusim K, Suscovich TJ, et al. Extensive HLA class I allele promiscuity among viral CTL epitopes. European Journal of Immunology. 2007;37(9):2419–2433. doi: 10.1002/eji.200737365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masemola AM, Mashishi TN, Khoury G, et al. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from Southern Africa. Journal of Immunology. 2004;173(7):4607–4617. doi: 10.4049/jimmunol.173.7.4607. [DOI] [PubMed] [Google Scholar]
- 10.Kaumaya PTP, Kobs-Conrad S, Seo SYH, et al. Peptide vaccines incorporating a ’promiscuous’ T-cell epitope bypass certain haplotype restricted immune responses and provide broad spectrum immunogenicity. Journal of Molecular Recognition. 1993;6(2):81–94. doi: 10.1002/jmr.300060206. [DOI] [PubMed] [Google Scholar]
- 11.Ho PC, Mutch DA, Winkel KD, et al. Identification of two promiscuous T cell epitopes from tetanus toxin. European Journal of Immunology. 1990;20(3):477–483. doi: 10.1002/eji.1830200304. [DOI] [PubMed] [Google Scholar]
- 12.Adar Y, Singer Y, Levi R, et al. A universal epitope-based influenza vaccine and its efficacy against H5N1. Vaccine. 2009;27(15):2099–2107. doi: 10.1016/j.vaccine.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Létourneau S, Im E-J, Mashishi T, et al. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS ONE. 2007;2(10, article e984) doi: 10.1371/journal.pone.0000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenstein JL, Schad VC, Goodwin WH, et al. A universal T cell epitope-containing peptide from hepatitis B surface antigen can enhance antibody specific for HIV gp120. Journal of Immunology. 1992;148(12):3970–3977. [PubMed] [Google Scholar]
- 15.Kumar A, Arora R, Kaur P, Chauhan VS, Sharma P. “Universal” T helper cell determinants enhance immunogenicity of a Plasmodium falciparum merozoite surface antigen peptide. Journal of Immunology. 1992;148(5):1499–1505. [PubMed] [Google Scholar]
- 16.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. European Journal of Immunology. 1989;19(12):2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 17.Bui H-H, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7, article 153 doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longmate J, York J, La Rosa C, et al. Population coverage by HLA class-I restricted cytotoxic T-lymphocyte epitopes. Immunogenetics. 2001;52(3-4):165–173. doi: 10.1007/s002510000271. [DOI] [PubMed] [Google Scholar]
- 19.Li Pira G, Bottone L, Ivaldi F, et al. Identification of new Th peptides from the cytomegalovirus protein pp65 to design a peptide library for generation of CD4 T cell lines for cellular immunoreconstitution. International Immunology. 2004;16(5):635–642. doi: 10.1093/intimm/dxh065. [DOI] [PubMed] [Google Scholar]
- 20.Li Pira G, Bottone L, Ivaldi F, et al. Generation of cytomegalovirus (CMV)-specific CD4 T cell lines devoid of alloreactivity, by use of a mixture of CMV-phosphoprotein 65 peptides for reconstitution of the T helper repertoire. Journal of Infectious Diseases. 2005;191(2):215–226. doi: 10.1086/427040. [DOI] [PubMed] [Google Scholar]
- 21.Kern F, Surel IP, Brock C, et al. T-cell epitope mapping by flow cytometry. Nature Medicine. 1998;4(8):975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 22.Provenzano M, Selleri S, Jin P, et al. Comprehensive epitope mapping of the Epstein-Barr virus latent membrane protein-2 in normal, non tumor-bearing individuals. Cancer Immunology, Immunotherapy. 2007;56(7):1047–1063. doi: 10.1007/s00262-006-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duraiswamy J, Burrows JM, Bharadwaj M, et al. Ex vivo analysis of T-cell responses to Epstein-Barr virus-encoded oncogene latent membrane protein 1 reveals highly conserved epitope sequences in virus isolates from diverse geographic regions. Journal of Virology. 2003;77(13):7401–7410. doi: 10.1128/JVI.77.13.7401-7410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leen AM, Christin A, Khalil M, et al. Identification of hexon-specific CD4 and CD8 T-cell epitopes for vaccine and immunotherapy. Journal of Virology. 2008;82(1):546–554. doi: 10.1128/JVI.01689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veltrop-Duits LA, Heemskerk B, Sombroek CC, et al. Human CD4+ T cells stimulated by conserved adenovirus 5 hexon peptides recognize cells infected with different species of human adenovirus. European Journal of Immunology. 2006;36(9):2410–2423. doi: 10.1002/eji.200535786. [DOI] [PubMed] [Google Scholar]
- 26.Frahm N, Korber T, Adams CM, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. Journal of Virology. 2004;78(5):2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauer GM, Ouchi K, Chung RT, et al. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. Journal of Virology. 2002;76(12):6104–6113. doi: 10.1128/JVI.76.12.6104-6113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingston B, Crimi C, Newman M, et al. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. Journal of Immunology. 2002;168(11):5499–5506. doi: 10.4049/jimmunol.168.11.5499. [DOI] [PubMed] [Google Scholar]
- 29.Joshi MB, Gam AA, Boykins RA, et al. Immunogenicity of well-characterized synthetic Plasmodium falciparum multiple antigen peptide conjugates. Infection and Immunity. 2001;69(8):4884–4890. doi: 10.1128/IAI.69.8.4884-4890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Groot AS, Marcon L, Bishop EA, et al. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005;23(17-18):2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 31.McMichael AJ, Hanke T. HIV vaccines 1983–2003. Nature Medicine. 2003;9(7):874–880. doi: 10.1038/nm0703-874. [DOI] [PubMed] [Google Scholar]
- 32.Cebere I, Dorrell L, McShane H, et al. Phase I clinical trial safety of DNA- and modified virus Ankara-vectored human immunodeficiency virus type 1 (HIV-1) vaccines administered alone and in a prime-boost regime to healthy HIV-1-uninfected volunteers. Vaccine. 2006;24(4):417–425. doi: 10.1016/j.vaccine.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 33.De Groot AS, McMurry J, Marcon L, et al. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine. 2005;23(17-18):2121–2131. doi: 10.1016/j.vaccine.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 34.Toes REM, Hoeben RC, van der Voort EIH, et al. Protective anti-tumor immunity induced by vaccination with recombinant adenoviruses encoding multiple tumor-associated cytotoxic T lymphocyte epitopes in a string-of-beads fashion. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14660–14665. doi: 10.1073/pnas.94.26.14660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitton JL, Sheng N, Oldstone MBA, McKee TA. A “string-of-beads” vaccine, comprising linked minigenes, confers protection from lethal-dose virus challenge. Journal of Virology. 1993;67(1):348–352. doi: 10.1128/jvi.67.1.348-352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An L-L, Whitton JL. A multivalent minigene vaccine, containing B-cell, cytotoxic T- lymphocyte, and Th epitopes from several microbes, induces appropriate responses in vivo and confers protection against more than one pathogen. Journal of Virology. 1997;71(3):2292–2302. doi: 10.1128/jvi.71.3.2292-2302.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer W, Perkins S, Theiler J, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nature Medicine. 2007;13(1):100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 38.Korber BTM, Brander C, Haynes BF, et al. HIV Molecular Immunology. Los Alamos, NM, USA: Los Alamos National Laboratory; 2007. [Google Scholar]
- 39.McMichael AJ, Phillips RE. Escape of human immunodeficiency virus from immune control. Annual Review of Immunology. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 40.De Groot AS, Knopf PM, Martin W. De-immunization of therapeutic proteins by T-cell epitope modification. Developments in Biologicals. 2005;122:171–194. [PubMed] [Google Scholar]
- 41.De Groot AS, Scott DW. Immunogenicity of protein therapeutics. Trends in Immunology. 2007;28(11):482–490. doi: 10.1016/j.it.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Kern F, LiPira G, Gratama JW, Manca F, Roederer M. Measuring Ag-specific immune responses: understanding immunopathogenesis and improving diagnostics in infectious disease, autoimmunity and cancer. Trends in Immunology. 2005;26(9):477–484. doi: 10.1016/j.it.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Sieker F, May A, Zacharias M. Predicting affinity and specificity of antigenic peptide binding to major histocompatibility class I molecules. Current Protein and Peptide Science. 2009;10(3):286–296. doi: 10.2174/138920309788452191. [DOI] [PubMed] [Google Scholar]
- 44.Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics. 2008;9(supplement 12, article S22) doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong JC, Tan TW, Ranganathan S. Methods and protocols for prediction of immunogenic epitopes. Briefings in Bioinformatics. 2007;8(2):96–108. doi: 10.1093/bib/bbl038. [DOI] [PubMed] [Google Scholar]
- 46.Bui H-H, Schiewe AJ, Von Grafenstein H, Haworth IS. Structural prediction of peptides binding to MHC class I molecules. Proteins. 2006;63(1):43–52. doi: 10.1002/prot.20870. [DOI] [PubMed] [Google Scholar]
- 47.Martin W, Sbai H, De Groot AS. Bioinformatics tools for identifying class I-restricted epitopes. Methods. 2003;29(3):289–298. doi: 10.1016/s1046-2023(02)00351-1. [DOI] [PubMed] [Google Scholar]
- 48.De Groot AS, Bosma A, Chinai N, et al. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine. 2001;19(31):4385–4395. doi: 10.1016/s0264-410x(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 49.Rammensee H-G, Bachmann J, Emmerich NPN, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 50.Roberts CGP, Meister GE, Jesdale BM, Lieberman J, Berzofsky JA, De Groot AS. Prediction of HIV peptide epitopes by a novel algorithm. AIDS Research and Human Retroviruses. 1996;12(7):593–610. doi: 10.1089/aid.1996.12.593. [DOI] [PubMed] [Google Scholar]
- 51.Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8, article 424 doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kloetzel P-M, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Current Opinion in Immunology. 2004;16(1):76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Advances in Immunology. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 54.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhütter H-G. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. Journal of Immunology. 2003;171(4):1741–1749. doi: 10.4049/jimmunol.171.4.1741. [DOI] [PubMed] [Google Scholar]
- 55.Tenzer S, Peters B, Bulik S, et al. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cellular and Molecular Life Sciences. 2005;62(9):1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PubMed] [Google Scholar]
- 56.Larsen MV, Lundegaard C, Lamberth K, et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. European Journal of Immunology. 2005;35(8):2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 57.Brusic V, Bajic VB, Petrovsky N. Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods. 2004;34(4):436–443. doi: 10.1016/j.ymeth.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Petrovsky N, Brusic V. Virtual models of the HLA class I antigen processing pathway. Methods. 2004;34(4):429–435. doi: 10.1016/j.ymeth.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Lata S, Bhasin M, Raghava GP. MHCBN 4.0: a database of MHC/TAP binding peptides and T-cell epitopes. BMC Research Notes. 2009;2, aricle 61 doi: 10.1186/1756-0500-2-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walshe VA, Hattotuwagama CK, Doytchinova IA, et al. Integrating in silico and in vitro analysis of peptide binding affinity to HLA-Cw∗0102: a bioinformatic approach to the prediction of new epitopes. PloS One. 2009;4(11, article e8095) doi: 10.1371/journal.pone.0008095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schirle M, Weinschenk T, Stevanović S. Combining computer algorithms with experimental approaches permits the rapid and accurate identification of T cell epitopes from defined antigens. Journal of Immunological Methods. 2001;257(1-2):1–16. doi: 10.1016/s0022-1759(01)00459-8. [DOI] [PubMed] [Google Scholar]
- 62.Gowthaman U, Agrewala JN. In silico tools for predicting peptides binding to HLA-class II molecules: more confusion than conclusion. Journal of Proteome Research. 2008;7(1):154–163. doi: 10.1021/pr070527b. [DOI] [PubMed] [Google Scholar]
- 63.Wulf M, Hoehn P, Trinder P. Identification of human MHC class I binding peptides using the iTOPIA- epitope discovery system. Methods in Molecular Biology. 2009;524:361–367. doi: 10.1007/978-1-59745-450-6_26. [DOI] [PubMed] [Google Scholar]
- 64.Vita R, Peters B, Sette A. The curation guidelines of the immune epitope database and analysis resource. Cytometry A. 2008;73(11):1066–1070. doi: 10.1002/cyto.a.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters B, Sette A. Integrating epitope data into the emerging web of biomedical knowledge resources. Nature Reviews Immunology. 2007;7(6):485–490. doi: 10.1038/nri2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vita R, Zarebski L, Greenbaum JA, et al. The immune epitope database 2.0. Nucleic Acids Research. 2009;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodda SJ. Peptide libraries for T cell epitope screening and characterization. Journal of Immunological Methods. 2002;267(1):71–77. doi: 10.1016/s0022-1759(02)00141-2. [DOI] [PubMed] [Google Scholar]
- 68.Kiecker F, Streitz M, Ay B, et al. Analysis of antigen-specific T-cell responses with synthetic peptides—what kind of peptide for which purpose? Human Immunology. 2004;65(5):523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 69.Kern F, Bunde T, Faulhaber N, et al. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. Journal of Infectious Diseases. 2002;185(12):1709–1716. doi: 10.1086/340637. [DOI] [PubMed] [Google Scholar]
- 70.Precopio ML, Butterfield TR, Casazza JP, et al. Optimizing peptide matrices for identifying T-cell antigens. Cytometry A. 2008;73(11):1071–1078. doi: 10.1002/cyto.a.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maeji NJ, Bray AM, Geysen HM. Multi-pin peptide synthesis strategy for T cell determinant analysis. Journal of Immunological Methods. 1990;134(1):23–33. doi: 10.1016/0022-1759(90)90108-8. [DOI] [PubMed] [Google Scholar]
- 72.Reece JC, McGregor DL, Geysen HM, Rodda SJ. Scanning for T helper epitopes with human PBMC using pools of short synthetic peptides. Journal of Immunological Methods. 1994;172(2):241–254. doi: 10.1016/0022-1759(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 73.Valerio RM, Bray AM, Campbell RA, et al. Multipin peptide synthesis at the micromole scale using 2-hydroxyethyl methacrylate grafted polyethylene supports. International Journal of Peptide and Protein Research. 1993;42(1):1–9. doi: 10.1111/j.1399-3011.1993.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 74.Kramer A, Reineke U, Dong L, et al. Spot synthesis: observations and optimizations. Journal of Peptide Research. 1999;54(4):319–327. doi: 10.1034/j.1399-3011.1999.00108.x. [DOI] [PubMed] [Google Scholar]
- 75.Reineke U, Volkmer-Engert R, Schneider-Mergener J. Applications of peptide arrays prepared by the spot-technology. Current Opinion in Biotechnology. 2001;12(1):59–64. doi: 10.1016/s0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 76.Nahtman T, Jernberg A, Mahdavifar S, et al. Validation of peptide epitope microarray experiments and extraction of quality data. Journal of Immunological Methods. 2007;328(1-2):1–13. doi: 10.1016/j.jim.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 77.Hemmer B, Pinilla C, Appel J, Pascal J, Houghten R, Martin R. The use of soluble synthetic peptide combinatorial libraries to determine antigen recognition of T cells. Journal of Peptide Research. 1998;52(5):338–345. doi: 10.1111/j.1399-3011.1998.tb00658.x. [DOI] [PubMed] [Google Scholar]
- 78.Sung M-H, Zhao Y, Martin R, Simon R. T-cell epitope prediction with combinatorial peptide libraries. Journal of Computational Biology. 2002;9(3):527–539. doi: 10.1089/106652702760138619. [DOI] [PubMed] [Google Scholar]
- 79.Sospedra M, Pinilla C, Martin R. Use of combinatorial peptide libraries for T-cell epitope mapping. Methods. 2003;29(3):236–247. doi: 10.1016/s1046-2023(02)00346-8. [DOI] [PubMed] [Google Scholar]
- 80.Altman JD, Moss PAH, Goulder PJR, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 81.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nature Reviews Immunology. 2004;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 82.Cobbold M, Khan N, Pourgheysari B, et al. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. Journal of Experimental Medicine. 2005;202(3):379–386. doi: 10.1084/jem.20040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Toebes M, Coccoris M, Bins A, et al. Design and use of conditional MHC class I ligands. Nature Medicine. 2006;12(2):246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 84.Kwok WW, Gebe JA, Liu A, et al. Rapid epitope identification from complex class-II-restricted T-cell antigens. Trends in Immunology. 2001;22(11):583–588. doi: 10.1016/s1471-4906(01)02038-5. [DOI] [PubMed] [Google Scholar]
- 85.Cecconi V, Moro M, Del Mare S, Dellabona P, Casorati G. Use of MHC class II tetramers to investigate CD4+ T cell responses: problems and solutions. Cytometry A. 2008;73(11):1010–1018. doi: 10.1002/cyto.a.20603. [DOI] [PubMed] [Google Scholar]
- 86.Chen DS, Soen Y, Stuge TB, et al. Marked differences in human melanoma antigen-specific T cell responsiveness after vaccination using a functional microarray. PLoS Medicine. 2005;2(10, article e265) doi: 10.1371/journal.pmed.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stone JD, Demkowicz WE, Jr., Stern LJ. HLA-restricted epitope identification and detection of functional T cell responses by using MHC-peptide and costimulatory microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3744–3749. doi: 10.1073/pnas.0407019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wallace PK, Tario JD, Jr., Fisher JL, Wallace SS, Ernstoff MS, Muirhead KA. Tracking antigen-driven responses by flow cytometry: monitoring proliferation by dye dilution. Cytometry A. 2008;73(11):1019–1034. doi: 10.1002/cyto.a.20619. [DOI] [PubMed] [Google Scholar]
- 89.Tesfa L, Volk H-D, Kern F. Comparison of proliferation and rapid cytokine induction assays for flow cytometric T-cell epitope mapping. Cytometry A. 2003;52(1):36–45. doi: 10.1002/cyto.a.10023. [DOI] [PubMed] [Google Scholar]
- 90.Wolfl M, Kuball J, Ho WY, et al. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110(1):201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frentsch M, Arbach O, Kirchhoff D, et al. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nature Medicine. 2005;11(10):1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 92.Zaunders JJ, Munier ML, Seddiki N, et al. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) Journal of Immunology. 2009;183(4):2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- 93.Czerkinsky CC, Nilsson LA, Nygren H, et al. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. Journal of Immunological Methods. 1983;65(1-2):109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 94.Czerkinsky C, Andersson G, Ekre H-P, Nilsson L-A, Klareskog L, Ouchterlony O. Reverse ELISPOT assay for clonal analysis of cytokine production. I. Enumeration of gamma-interferon-secretion cells. Journal of Immunological Methods. 1988;110(1):29–36. doi: 10.1016/0022-1759(88)90079-8. [DOI] [PubMed] [Google Scholar]
- 95.Gazagne A, Claret E, Wijdenes J, et al. A Fluorospot assay to detect single T lymphocytes simultaneously producing multiple cytokines. Journal of Immunological Methods. 2003;283(1-2):91–98. doi: 10.1016/j.jim.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 96.Waldrop SL, Pitcher CJ, Peterson DM, Maino VC, Picker LJ. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. Journal of Clinical Investigation. 1997;99(7):1739–1750. doi: 10.1172/JCI119338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sylwester AW, Mitchell BL, Edgar JB, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. Journal of Experimental Medicine. 2005;202(5):673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Rosa SC, Lu FX, Yu J, et al. Vaccination in humans generates broad T cell cytokine responses. Journal of Immunology. 2004;173(9):5372–5380. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- 99.Precopio ML, Betts MR, Parrino J, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8+ T cell responses. Journal of Experimental Medicine. 2007;204(6):1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nature Reviews Immunology. 2008;8(4):247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 101.Brosterhus H, Brings S, Leyendeckers H, et al. Enrichment and detection of live antigen-specific CD4+ and CD8+ T cells based on cytokine secretion. European Journal of Immunology. 1999;29(12):4053–4059. doi: 10.1002/(SICI)1521-4141(199912)29:12<4053::AID-IMMU4053>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 102.Rauser G, Einsele H, Sinzger C, et al. Rapid generation of combined CMV-specific CD4+ and CD8+ T-cell lines for adoptive transfer into recipients of allogeneic stem cell transplants. Blood. 2004;103(9):3565–3572. doi: 10.1182/blood-2003-09-3056. [DOI] [PubMed] [Google Scholar]
- 103.Li Pira G, Ivaldi F, Tripodi G, Martinengo M, Manca F. Positive selection and expansion of cytomegalovirus-specific CD4 and CD8 T cells in sealed systems: potential applications for adoptive cellular immunoreconstitution. Journal of Immunotherapy. 2008;31(8):762–770. doi: 10.1097/CJI.0b013e3181826232. [DOI] [PubMed] [Google Scholar]
- 104.Feuchtinger T, Matthes-Martin S, Richard C, et al. Safe adoptive transfer of virus-specific T-cell immunity for the treatment of systemic adenovirus infection after allogeneic stem cell transplantation. British Journal of Haematology. 2006;134(1):64–76. doi: 10.1111/j.1365-2141.2006.06108.x. [DOI] [PubMed] [Google Scholar]
- 105.Mackinnon S, Thomson K, Verfuerth S, Peggs K, Lowdell M. Adoptive cellular therapy for cytomegalovirus infection following allogeneic stem cell transplantation using virus-specific T cells. Blood Cells, Molecules, and Diseases. 2008;40(1):63–67. doi: 10.1016/j.bcmd.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 106.McKinney DM, Skvoretz R, Qin M, et al. Characterization of an in situ IFN-gamma ELISA assay which is able to detect specific peptide responses from freshly isolated splenocytes induced by DNA minigene immunization. Journal of Immunological Methods. 2000;237(1-2):105–117. doi: 10.1016/s0022-1759(00)00138-1. [DOI] [PubMed] [Google Scholar]
- 107.Li Pira G, Ivaldi F, Bottone L, Manca F. High throughput functional microdissection of pathogen-specific T-cell immunity using antigen and lymphocyte arrays. Journal of Immunological Methods. 2007;326(1-2):22–32. doi: 10.1016/j.jim.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 108.Li Pira G, Ivaldi F, Dentone C, et al. Evaluation of antigen-specific T-cell responses with a miniaturized and automated method. Clinical and Vaccine Immunology. 2008;15(12):1811–1818. doi: 10.1128/CVI.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Manca F, Habeshaw J, Dalgleish A. The naive repertoire of human T helper cells specific for gp120, the envelope glycoprotein of HIV. Journal of Immunology. 1991;146(6):1964–1971. [PubMed] [Google Scholar]
- 110.Manca F, Fenoglio D, Valle MT, et al. Human CD4+ T cells can discriminate the molecular and structural context of T epitopes of HIV gp120 and HIV p66. Journal of Acquired Immune Deficiency Syndromes and Human Retrovirology. 1995;9(3):227–237. [PubMed] [Google Scholar]
- 111.Li Pira G, Ivaldi F, Moretti P, Risso M, Tripodi G, Manca F. Validation of a miniaturized assay based on IFNg secretion for assessment of specific T cell immunity. Journal of Immunological Methods. 2010;355(1-2):68–75. doi: 10.1016/j.jim.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 112.Li Pira G, Kapp M, Manca F, Einsele H. Pathogen specific T-lymphocytes for the reconstitution of the immunocompromised host. Current Opinion in Immunology. 2009;21(5):549–556. doi: 10.1016/j.coi.2009.08.006. [DOI] [PubMed] [Google Scholar]