Abstract
For the past 50 years, quality control and safety tests have been used to evaluate vaccine safety. However, conventional animal safety tests need to be improved in several aspects. For example, the number of test animals used needs to be reduced and the test period shortened. It is, therefore, necessary to develop a new vaccine evaluation system. In this review, we show that gene expression patterns are well correlated to biological responses in vaccinated rats. Our findings and methods using experimental biology and genome science provide an important means of assessment for vaccine toxicity.
1. Introduction
Vaccination effectively enables the control of many infectious diseases. However, we cannot always avoid the problem of adverse reactions accompanied by vaccination. While most adverse reactions are mild and local, some vaccines have been associated with very rare but severe systemic reactions. Therefore, all vaccines for public use are made in compliance with Good Manufacturing Practices (GMP) to prevent safety problems. Furthermore, manufacturers must submit samples and results of their in-house tests for each vaccine batch to the national control authorities before vaccines are released into the market. Among many quality control tests, conventional animal safety tests are performed to detect vaccine toxicity because residual vaccine toxicity has the potential to cause adverse reactions. For example, the animal body weight change test is the most commonly used test to evaluate the toxicity of vaccines [1]. Although a good correlation of the body weight loss with a vaccine's toxicity has been shown [2, 3], a greater understanding of the molecular mechanisms involved in the reaction to a vaccines' toxicity is needed. We, therefore, attempted to measure animals' responses to vaccines by determining changes in gene expression profiles.
Gene expression profiling is a unique way to characterize how cells or tissues are affected by abnormal conditions. The measurement of gene expression levels upon exposure to toxicants can be used to identify toxic products, and to provide information about the mechanism of toxicity [4]. DNA microarray technology has opened the way for the parallel detection and analysis of expression patterns of thousands of genes in a single experiment. Furthermore, the development of high-quality gene arrays has allowed DNA microarray technology to become a standard tool in molecular toxicology. Recently, the field of toxicogenomics has validated the concept of gene expression profiles as “signatures” of toxicant classes [5–7]. These signatures have effectively directed the analytical search for predictive toxicant biomarkers and they have contributed to the understanding of the dynamic responses of molecular mechanisms associated with toxic responses. In fact, many studies of gene-expression profiles have now been reported in the toxicology field. For example, Hamadeh et al. reported patterns of gene expression in liver tissue taken from rats exposed to different chemicals [8]. DNA microarray assays have also been applied to the analysis of the side effects of medicines [9]. Recently, the United States Food and Drug Administration (FDA) and the European Medicines Agency (EMEA) have, either individually or together, started to review submissions for the qualification of biomarkers for medical products for specific purposes proposed by industry [10]. The introduction of pharmacogenomics, or pharmacogenetics, to the evaluation of medicines is a global trend.
For a better understanding of the molecular toxicology regarding vaccines, DNA microarray analysis promises to be an ideal method, as has been the case for pharmaceuticals. The FDA now encourages the voluntary submission of genomic data to the FDA outside of the regular review process [11]. However, no studies similar to those described above for pharmaceuticals have yet been conducted in the field of vaccines. At the beginning of this review, we summarized the current efforts used for the control of vaccine safety using conventional animal tests. We then referred to our recent efforts using DNA microarray analysis to identify “genetic signatures” for the toxicants remaining in vaccines. Since pertussis and influenza vaccines are among the most commonly used vaccines, we tried to develop a system to evaluate the “genetic signatures” of the toxicity of these vaccines.
2. Current Vaccine Safety Test
2.1. Body Weight Change in Vaccinated Animals
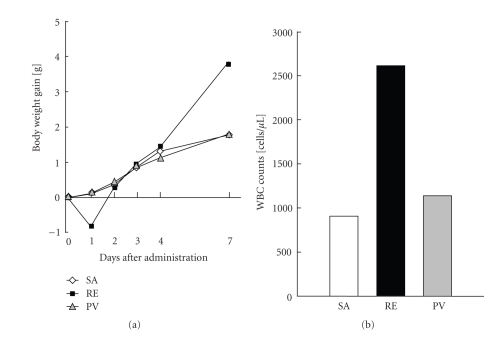
To screen for general toxicity of vaccines, the body weight of vaccine-treated animals can be analyzed as the general safety test [12]. Five mL of the vaccine are injected into the peritoneum of guinea pigs weighing 300–400 g, and the weight loss experienced by the animals is analyzed at days 1, 2, 3, 4, and 7 after administration. None of the animals should show any abnormal signs; no statistically significant (P = .01) difference in weight loss should be observed between the treated animals and the control group on any observation day. This test has been applied to a wide variety of vaccines in a unified way, and plays an important role in ensuring the safety and consistency of vaccine batches [12]. For pertussis vaccine (inactivated whole cell formulation), the effects of vaccine treatment were also measured using test for toxicity to mouse weight gain, in addition to the general safety test. All mice were weighed on days 0, 1, 2, 3, 4, and 7 after vaccine administration. The criterion for judgment is that mean body weight 3 days after injection should be no less than that at the time of injection upon statistical analysis, and no mice showed any abnormal sign during the observation periods [12]. When the reference vaccine (RE: the inactivated whole cell pertussis vaccine) was administrated, weight loss was observed on day 1 after administration (Figure 1(a)).
Figure 1.
Safety control tests for pertussis vaccines. (a) Test for toxicity to mouse weight gain. Physiological saline (SA), an inactivated whole-cell pertussis vaccine (RE), or an acellular pertussis vaccine (PV)-administered mice were weighed on 0, 1, 2, 3, 4, and 7 days postadministration. Ten mice in each group were used, and the mean changes in body weight are indicated. (b) Leukocytosis promoting activity of various pertussis vaccines. White blood cell (WBC) counts in peripheral blood were measured 3 days after vaccine administration. Ten mice in each group were used and the mean WBC counts are indicated.
2.2. Leukocytosis-Promoting Toxicity in Vaccinated Animals
To detect the toxin present in pertussis vaccines, the number of peripheral leukocytes can also be analyzed. Pertussis vaccine is injected into the peritoneum of mice at a dose of 0.5 mL. Leukocytes present in peripheral blood are then counted 3 days after injection [12]. The white blood cell (WBC) counts in peripheral blood of reference vaccine-treated mice reach approximately 2,500 cells/μL (Figure 1(b)). The standard criterion of safety for pertussis vaccine (inactivated whole cell formulation) is that the mean count of leukocytes in peripheral blood, 3 days after injection, should not exceed 10 times that before injection [12].
2.3. Leukopenic Toxicity Test in Vaccinated Animals
Quality control of influenza vaccines is performed using the general safety test and the leukopenic toxicity test (LTT), which is based on peripheral WBC counts in mice 12–18 hours after intraperitoneal injection of a vaccine. The criterion for judgment is that the leukopenic toxicity of the test sample relative to that of the toxicity reference sample should be no higher than the value corresponding to 80% of the leukocyte count of the control relative to that of the toxicity reference sample [12–14].
3. DNA Microarray-Based Safety Test
The currently used quality control and safety tests, such as the LTT and the general safety test, have been used to evaluate vaccine safety for over 50 years [3]. We are now developing a new quality control method for vaccines using DNA microarray analysis as a substitute for the conventional animal tests [15–17]. The principle of this method is to translate vaccine quality, immunogenicity, and reactogenicity, into gene expression profile data. This method is expected to be informative, rapid, and highly sensitive.
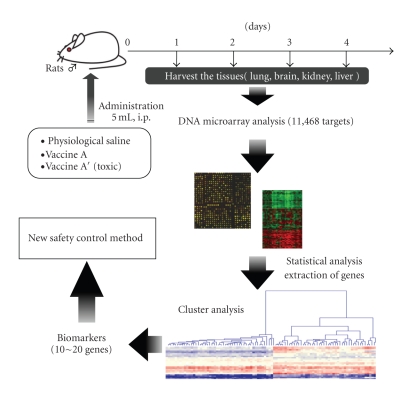
For DNA microarray analysis using vaccines, 8 week-old male rats, weighing 180–220 g, were intraperitoneally administered with 5 mL of vaccine or physiological saline (SA). Three to 6 rats were used for each group. Vaccinated rats were sacrificed to obtain whole lung, kidney, brain, and the lateral left lobe of the liver on day 1, 2, 3, and 4 postadministration (Figure 2). Tissues were immediately frozen in liquid nitrogen for storage. Thawed tissue was homogenized and poly(A)+ RNA was purified from the lysate. Cyanine 5-labeled poly(A)+ RNA was subjected to DNA microarray analysis. Blood was also collected, however, this could not be analyzed due to the low quality of purified RNA.
Figure 2.
The gene expression analysis procedure. The detail of the procedure is described in the text.
For DNA microarray analysis, a set of synthetic polynucleotides (80-mers) representing 11,468 rat transcripts and including most of the RefSeq genes deposited in the NCBI database (MicroDiagnostic, Tokyo, Japan) was arrayed on aminosilane-coated glass slides [18, 19]. Cyanine 5-labeled poly(A)+ RNA was competitively hybridized on the slide with cyanine 3-labeled common reference RNA. Hybridization signals were measured, processed into primary expression ratios ([Cyanine 5-intensity obtained from each sample]/[Cyanine 3-intensity obtained from common reference RNA]), and then normalized by multiplying normalization factors calculated for each microarray feature.
For data processing and hierarchical cluster analysis, the primary expression ratios were converted into log 2 ratios (log 2 Cyanine-5 intensity/Cyanine-3 intensity). The genes with log 2 ratios over 1 or under −1 in at least one sample were extracted from the primary data matrix, then subjected to two-dimensional hierarchical cluster analysis for samples and genes.
For the identification of biomarker genes for pertussis vaccines, we extracted differentially expressed genes from physiological saline and pertussis toxin-treated lung samples using the t-test (P < .01). Among the extracted genes, we further selected genes that exhibited mean average log 2 ratio differences greater than 0.75 between the two sample groups [17]. For influenza vaccines, we extracted differentially expressed genes from physiological saline and inactivated whole-virion vaccine-treated lung samples using the t-test (P < .005) [16].
4. Pertussis Vaccines
Pertussis, or whooping cough, is an infectious respiratory disease caused by a Gram-negative bacillus, Bordetella pertussis. Bordetella pertussis possesses several pathogenic components, including pertussis toxin (PT) [20]. PT is known as a leukocytosis promoting factor, a major contributor to the pathogenesis of pertussis, and an antigen in immunity to pertussis [21]. At present, whole-cell pertussis vaccines and acellular pertussis vaccines containing inactivated PT are in commercial use [20].
Although pertussis vaccines are effective in the prevention of whooping cough, they have occasionally caused local reactions such as redness, swelling, and pain at the injection site. However, little is known about the overall responses to these vaccines. To address this problem, we applied DNA microarray analysis and quantification of specific genes to analyze the toxicants in pertussis vaccines [15, 17]. Three preparations, an acellular vaccine containing inactivated pertussis toxin (PV), an inactivated whole-cell vaccine (RE), and a purified pertussis toxin (PT) were prepared. RE is a reference vaccine for National Quality Control Tests of pertussis vaccines in Japan and is made from formaldehyde-inactivated Bordetella pertussis preparations. Physiological saline (SA) was used as a control. For comprehensive gene expression analysis, 5 mL of SA, PV, PT, and RE were each injected into 3 rats and the vaccinated tissues, lung, brain, kidney, and liver, were harvested at 1, 2, 3, and 4 days after vaccine administration. The experiments were performed twice and purified poly(A)+ RNA from a total of 384 samples was subjected to DNA microarray analysis.
Of the 4 organs tested, the lung expressed genes that were extracted by DNA microarray analysis were classified sharply into clusters depending on sample treatment. From the DNA microarray analysis of vaccinated rat lungs at day 1, 13 genes for which expression levels were dynamically changed in response to PT treatment were identified [17] (accession numbers were updated in Table 1). Interestingly, the DNA microarray-based gene expression data correlated well with the body weight change of vaccine-treated mice (Figure 1(a)) and rats [17]. The real-time PCR quantification results of the expression levels of the 13 genes were comparable to the relative expression ratios from the DNA microarray analysis. Furthermore, cluster analysis using the 13 genes could distinguish SA- and PV-treated groups from PT- and RE-treated groups. These 13 genes are likely to be closely involved in the toxicity of pertussis vaccines. To quantify these genes in a convenient way, the QuantiGene Plex assay was applied. The QuantiGene Plex assay enabled the simultaneous analysis of the 13 genes. We evaluated the expression levels of the 13 genes in the lungs of rats vaccinated with various doses of RE. Nine genes, S100A9, S100A8, IRF7, MX2, IFI27L, BEST5, MMP9, MMP8, and CYP2E1 (indicated in bold letters in Table 1) showed dose-dependent up-or down-regulation in response to the various doses of RE treatment. RE vaccine toxicity could be measured by the expression level in lung lysate of these 9 genes. The quantification of these 9 genes using the QuantiGene Plex assay is, we believe, a promising candidate for a new control test for pertussis vaccines.
Table 1.
Biomarkers for pertussis vaccine toxicity.
| Category | Accession no. | Symbol | Brief description |
|---|---|---|---|
| Inflammation | NM_053587 | S100A9 | A calcium binding protein that may be associated with acute inflammatory processes, coupled with S100a8 |
| NM_053822 | S100A8 | May play a role in inflammatory responses such as cell motility, coupled with S100a9 | |
| NM_019323 | MCPT9 | A serine protease expressed in mast cells, but the precise function has not yet been determined | |
| NM_031530 | CCL2 | A ligand for CCR2 that acts as a chemoattractant of monocytes | |
| IFN inducible, immune response | NM_001033691 | IRF7 | Unknown |
| NM_134350 | MX2 | Involved in inhibiting vesicular stomatitis virus | |
| NM_203410 | IFI27 | Induced by steroid hormone, IFN, and LPS in endometrium at implantation, dendric cells, and macrophases | |
| NM_001007694 | IFIT3 | May induced by IFN or virus infection | |
| Y07704 | BEST5 | Induced by IFN and involved in bone formation | |
| Peptidoglycan metabolism | NM_031055 | MMP9 | Metalloproteinase involved in extracellular matrix remodeling, bone resorption, and immune responses |
| NM_022221 | MMP8 | May play a role in appositional bone formation and regulation of the extracellular matrix | |
| Xenobiotic metabolism | J02627 | CYP2E1 | Protects hepatocytes from stress-induced cell death |
| Others | NM_001106862 | NGP | Unknown |
5. Influenza Vaccines
Influenza virus triggers a highly contagious acute respiratory disease and has caused epidemics and global pandemics, partly because it possesses the capacity for gradual antigenic change in two surface antigens, hemagglutinin (HA) and neuraminidase (NA) [22]. To combat influenza, split vaccines consisting of subvirion preparations and whole-virus vaccines are manufactured using strains recommended annually by the WHO, based on the antigenic characteristics of HAs and NAs. Furthermore, the recent circulation of the highly pathogenic avian influenza A (H5N1) virus has raised concerns about the preparations for a coming influenza pandemic [23]. Many efforts are underway to develop vaccines against influenza A (H5N1).
To identify biomarkers for influenza vaccine toxicity, 3 vaccines were used: trivalent influenza HA vaccine (HAv, a split vaccine), trivalent influenza vaccine (WPv, an inactivated whole-virion vaccine), and prepandemic influenza vaccine (PDv, inactivated whole-virion (A/H5N1) absorbed onto an aluminum salt). All were produced by Kaketsuken, The Chemo-Sero-Therapeutic Research Institute, Japan. Physiological saline (SA) was used as a control. For comprehensive gene expression analysis, SA, HAv, WPv, and PDv were each injected into 5 rats, and the vaccinated tissues, lung, liver, brain, and peripheral blood, were harvested at 1, 2, 3, and 4 days after vaccine administration. Purified poly(A)+ RNA from a total of 320 samples was subjected to DNA microarray analysis [16]. Based on the analysis of pertussis vaccines, described above, the gene expression profiles from lung samples were subjected to two-dimensional hierarchical cluster analysis. PDv- and WPv-treated samples at day 1 formed an independent cluster from other samples, indicating distinct profiles in gene expression of these groups. As was the case with pertussis vaccines, we tried to identify several biomarkers from the analysis of lung gene expression. The analysis of lungs from vaccinated rats at day 1 resulted in the extraction of 76 genes, whose expression levels were statistically different between SA- and WPv-treated samples (P < .005) [16]. The cluster analysis using these 76 genes successfully distinguished WPv- and PDv-treated groups at day 1 from other groups, indicating the suitability of the 76 genes as biomarkers for influenza vaccines.
The extracted 76 genes were categorized according to function, such as interferon-inducible, chemokine and cytokine function, immune response, transcriptional activity, and so on. Among the 76 genes, 17 genes met the requirement for high expression levels and were chosen as representatives for each functional category (Table 2). Among the 17 genes, IRF7 and MX2 were also nominated for biomarkers of pertussis vaccine toxicity. Real-time PCR quantification results of the expression levels of the 17 genes were comparable to the relative expression ratios determined by DNA microarray analysis. We are now working to establish a rapid quantification system for these 17 biomarkers using the QuantiGene Plex assay.
Table 2.
Biomarkers for influenza vaccine toxicity.
| Category | Accession No. | Symbol | Brief description |
|---|---|---|---|
| IFN inducible gene | NM_172019 | IFI47 | Mouse homolog may be a guanine nucleotide-binding protein induced by IFN-gamma |
| AF329825 | TRAFD1 | Putative TRAF-interacting zinc finger protein | |
| NM_019242 | IFRD1 | May be involved in proliferation of neuronal and glial precursors | |
| IFN inducible, immune response | NM_001033691 | IRF7 | Unknown |
| NM_134350 | MX2 | Involved in inhibiting vesicular stomatitis virus | |
| Immune response | NM_172222 | C2 | Likely component of the classical pathway of the complement cascade |
| NM_012708 | PSMB9 | Subunit of the proteasome complex, which may play a role in protein catabolism | |
| NM_032056 | TAP2 | Transports peptides into the ER lumen for binding with MHC class I molecules; plays a role in antigen processing and presentation | |
| NM_033098 | TAPBP | Facilitates the binding of MHC class I molecules to the transporter associated with antigen processing (TAP) in MHC class I assembly | |
| NM_017264 | PSME1 | May play a role in proteasome activation | |
| Chemokine and Cytokine function | AF065438 | LGALS3BP | Displays differential expression in a fibroblast cell line transformed by human T-cell leukemia virus type 1 Tax protein |
| NM_012977 | LGALS9 | A highly selective urate transporter/channel | |
| NM_053819 | TIMP1 | Acts as an inhibitor of metalloprotease activity; may play a role in vascular tissue remodeling | |
| NM_023981 | CSF1 | Plays a role in macrophage formation | |
| NM_145672 | CXCL9 | Chemokine which plays a role in the recruitment of mononuclear cells and in allograft rejection | |
| XM_223236 | CXCL11 | Mouse homolog is a chemokine and is involved in the immune response | |
| Transcription activity | AJ302054 | ZBP1 | DNA binding protein; thought to bind Z-DNA, which is largely controlled by the amount of supercoiling |
6. Japanese Encephalitis Vaccines
Japanese encephalitis (JE) is a seasonal and sporadic encephalitis in East Asia caused by the JE virus. Vaccination is very important to prevent JE infection, because palliative care is the only treatment available for JE patients. Recently, a Vero cell-derived JE vaccine had been licensed in Japan as an alternative to the long-used mouse brain-derived JE vaccines. The newly developed Vero cell-derived vaccine should be at least equivalent to the mouse brain-derived vaccines, because the mouse brain-derived vaccines were considered generally safe and succeeded in the near elimination of JE in certain endemic regions. In this context, we performed DNA microarray analysis of tissues from rats administered with mouse brain-derived or Vero cell-derived JE vaccine and compared the gene expression profiles. As expected, the gene expression patterns in brain and liver were comparable between mouse brain-derived and Vero cell-derived vaccines, indicating that both vaccines possessed equivalent reactivity characteristics in rats [24].
7. Conclusions
Over recent decades, the safety control of vaccines has been assessed using several animal tests, including the body weight change test and white blood cell counts. However, conventional animal safety tests need to be improved in many aspects. For example, the number of test animals used needs to be reduced and the test period needs to be shortened. This requires the development of a new vaccine evaluation system. In this review, we showed that gene expression patterns were well correlated to the biological responsiveness of vaccinated animals. From the DNA microarray analysis of lungs from vaccinated rats, we identified 13 and 17 biomarkers to detect the toxicity of pertussis and influenza vaccines, respectively.
Furthermore, the QuantiGene Plex assay for gene expression analysis is being introduced. The QuantiGene Plex assay was revealed to be as accurate as real-time PCR and has the great benefit of being able to evaluate all biomarkers simultaneously. Using the QuantiGene Plex assay, we could rapidly and sensitively detect the gene expression changes that accompany biological reactivity in vaccinated rats.
Thus, it may be concluded that DNA microarray technology is an informative, rapid, and highly sensitive method with which to evaluate vaccine quality. Our data suggest that this new method has the potential to shorten the time for safety tests and can reduce the number of animals used. In addition, our test may contribute to the development of urgently required vaccines. Further analyses are required to confirm that gene expression changes correlate with vaccine quality.
In this review, we referred to our recent efforts of exploring new safety control methods using gene expression pattern indexes, focusing on pertussis and influenza vaccines. In the future, for the evaluation of all kinds of vaccines, microarray analysis is expected to play an important role in the new safety control test, especially for checking toxin-reactive transcripts.
Acknowledgment
This work was supported by Grants-in-Aid from the Ministry of Health, Labour and Welfare, Japan.
References
- 1.Horiuchi Y, Takahashi M, Konda T, et al. Quality control of diphtheria tetanus acellular pertussis combined (DTaP) vaccines in Japan. Japanese Journal of Infectious Diseases. 2001;54(5):167–180. [PubMed] [Google Scholar]
- 2.Kurokawa M. Toxicity and toxicity testing of pertussis vaccine. Japanese Journal of Medical Science and Biology. 1984;37(2):41–81. doi: 10.7883/yoken1952.37.41. [DOI] [PubMed] [Google Scholar]
- 3.Mizukami T, Masumi A, Momose H, et al. An improved abnormal toxicity test by using reference vaccine-specific body weight curves and histopathological data for monitoring vaccine quality and safety in Japan. Biologicals. 2009;37(1):8–17. doi: 10.1016/j.biologicals.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Lettieri T. Recent applications of DNA microarray technology to toxicology and ecotoxicology. Environmental Health Perspectives. 2006;114(1):4–9. doi: 10.1289/ehp.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobenhofer EK, Bushel PR, Afshari CA, Hamadeh HK. Progress in the application of DNA microarrays. Environmental Health Perspectives. 2001;109(9):881–891. doi: 10.1289/ehp.01109881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennie W, Pettit SD, Lord PG. Toxicogenomics in risk assessment: an overview of an HESI collaborative research program. Environmental Health Perspectives. 2004;112(4):417–419. doi: 10.1289/ehp.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrill AH, Rusyn I. Systems biology and functional genomics approaches for the identification of cellular responses to drug toxicity. Expert Opinion on Drug Metabolism & Toxicology. 2008;4(11):1379–1389. doi: 10.1517/17425255.4.11.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamadeh HK, Bushel PR, Jayadev S, et al. Gene expression analysis reveals chemical-specific profiles. Toxicological Sciences. 2002;67(2):219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- 9.Ejiri N, Katayama K-I, Kiyosawa N, Baba Y, Doi K. Microarray analysis on phase II drug metabolizing enzymes expression in pregnant rats after treatment with pregnenolone-16α-carbonitrile or phenobarbital. Experimental and Molecular Pathology. 2005;79(3):272–277. doi: 10.1016/j.yexmp.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 10.European Medicines Agency Concept Paper. Pharmacogenomics (PG) biomarker qualification: format and data standards. In: Proceedings of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use; June 2008; EMEA/CHMP/190395/2008, http://www.emea.europa.eu/pdfs/human/pharmacogenetics/19039508en.pdf. [Google Scholar]
- 11.Frueh FW. Impact of microarray data quality on genomic data submissions to the FDA. Nature Biotechnology. 2006;24(9):1105–1107. doi: 10.1038/nbt0906-1105. [DOI] [PubMed] [Google Scholar]
- 12.Minimum Requirements for Biological Products. Tokyo, Japan: National Institute of Infectious Diseases; 2006. http://www.nih.go.jp/niid/MRBP/files/seibutsuki_english.pdf. [Google Scholar]
- 13.Kurokawa M, Ishida S, Asakawa S, Iwasa S, Goto N. Toxicities of influenza vaccine: peripheral leukocytic response to live and inactivated influenza viruses in mice. Japanese Journal of Medical Science and Biology. 1975;28(1):37–52. doi: 10.7883/yoken1952.28.37. [DOI] [PubMed] [Google Scholar]
- 14.Chino F. The views and policy of the Japanese control authorities on the three Rs. Developments in Biological Standardization. 1996;86:53–62. [PubMed] [Google Scholar]
- 15.Hamaguchi I, Imai J-I, Momose H, et al. Two vaccine toxicity-related genes Agp and Hpx could prove useful for pertussis vaccine safety control. Vaccine. 2007;25(17):3355–3364. doi: 10.1016/j.vaccine.2006.12.059. [DOI] [PubMed] [Google Scholar]
- 16.Mizukami T, Imai J-I, Hamaguchi I, et al. Application of DNA microarray technology to influenza A/Vietnam/1194/2004 (H5N1) vaccine safety evaluation. Vaccine. 2008;26(18):2270–2283. doi: 10.1016/j.vaccine.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 17.Hamaguchi I, Imai J-I, Momose H, et al. Application of quantitative gene expression analysis for pertussis vaccine safety control. Vaccine. 2008;26(36):4686–4696. doi: 10.1016/j.vaccine.2008.06.086. [DOI] [PubMed] [Google Scholar]
- 18.Ito E, Honma R, Imai J-I, et al. A tetraspanin-family protein, T-cell acute lymphoblastic leukemia-associated antigen 1, is induced by the Ewing’s sarcoma-Wilms’ tumor 1 fusion protein of desmoplastic small round-cell tumor. American Journal of Pathology. 2003;163(6):2165–2172. doi: 10.1016/s0002-9440(10)63573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi S, Ito E, Honma R, et al. Dynamic regulation of gene expression by the Flt-1 kinase and Matrigel in endothelial tubulogenesis. Genomics. 2004;84(1):185–192. doi: 10.1016/j.ygeno.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Edwards KM, Decker MD. Pertussis vaccines. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 5th edition. New York, NY, USA: Elsevier; 2008. pp. 467–517. [Google Scholar]
- 21.Sato H, Sato Y. Bordetella pertussis infection in mice: correlation of specific antibodies against two antigens, pertussis toxin, and filamentous hemagglutinin with mouse protectivity in an intracerebral or aerosol challenge system. Infection and Immunity. 1984;46(2):415–421. doi: 10.1128/iai.46.2.415-421.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bridges CB, Katz JM, Levandowski RA, Cox NJ. Inactivated influenza vaccines. In: Plotkin SA, Orenstein WA, Offit PA, editors. Vaccines. 5th edition. New York, NY, USA: Elsevier; 2008. pp. 259–290. [Google Scholar]
- 23.Ungchusak K, Auewarakul P, Dowell SF, et al. Probable person-to-person transmission of avian influenza A (H5N1) The New England Journal of Medicine. 2005;352(4):333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 24.Momose H, Imai J-I, Hamaguchi I, et al. Induction of indistinguishable gene expression patterns in rats by Vero cell-derived and mouse brain-derived Japanese encephalitis vaccines. Japanese Journal of Infectious Diseases. 2010;63(1):25–30. [PubMed] [Google Scholar]