Abstract
The human red blood cell (RBC) has long been used for modeling of complex biological networks, for elucidation of a wide variety of dynamic phenomena, and for understanding the fundamental topology of metabolic pathways. Here, we introduce our recent work on an RBC metabolic model using the E-Cell Simulation Environment. The model is sufficiently detailed to predict the temporal hypoxic response of each metabolite and, at the same time, successfully integrates modulation of metabolism and of the oxygen transporting capacity of hemoglobin. The model includes the mechanisms of RBC maintenance as a single cell system and the functioning of RBCs as components of a higher order system. Modeling of RBC metabolism is now approaching a fully mature stage of realistic predictions at the molecular level and will be useful for predicting conditions in biotechnological applications such as long-term cold storage of RBCs.
1. Introduction
Systems level behaviors occur as a consequence of synergistic interactions between individual components that can function as single systems, but can also affect the dynamic state of other components. To understand how components interact in biological systems, which exhibit nonlinear behaviors, not only the network structure of the system, but also all relevant components of the system need to be integrated in a quantitative manner. Systems biology has emerged to address these problems, using a combination of computational and experimental analyses [1, 2].
Kinetic and dynamic modeling is one of the most useful approaches in systems biology [3]. The E-Cell project was launched in 1996, early in the systems biology and -omics technology era, with the aim of developing whole-cell-scale mathematical models [4, 5]. Although a number of bio-simulation-specific platforms have been released recently [6], the approach of E-Cell in a fully object-oriented fashion is unique and allows for multitimescale/multialgorithm simulations [7, 8].
When a dynamic mathematical model is obtained, multiple analyses can be conducted to elucidate the fundamental design principles of a biological system [9, 10]. Such models enable researchers to examine experimentally intractable systems; for example, maintenance of homeostasis in organisms in vivo can be mimicked in silico, with large approximations, but not in vitro. Additionally, experimentally costly analyses such as comprehensive sensitivity analysis of an enzyme activity in response to perturbation of all components of the systems' networks can be modeled. Furthermore, a successful model that can represent the dynamic behavior of the target system in the intended environment can be used to predict how the system responds to an external physiological stimulus or to modification of each component in the system, which would be extremely useful for development of biotechnological applications.
The metabolic network in human red blood cells (RBCs) has been the subject of mathematical modeling for the past three to four decades. As discussed below, a number of detailed models of RBC metabolism have been developed with various levels of abstraction, and many studies analyzing these models have produced a deeper understanding of the regulatory properties of metabolic pathways and provided novel insights for implementation of both mathematical analysis and metabolic engineering strategies, which can then be applied to many other cell types.
Our model of RBC metabolism using the E-Cell Simulation Environment focuses on the interrelationship between metabolism and the functional aspects of RBC physiology, including factors affecting allosteric transitions of hemoglobin and recent findings regarding the assembly of intracellular proteins at the plasma membrane. Our model successfully reproduced the temporal response to hypoxia, previously measured by metabolome analysis [11]. The model was then used to predict the metabolic status of RBCs in long-term cold storage, with the goal of optimizing the storage conditions [12].
2. The E-Cell System as a Cell Simulation Environment
Modeling of biological systems requires suitable abstraction of the system considering the amount, size, and speed of reactions, as well as many other factors. This process includes making decisions as to whether continuous processes should be broken down into discrete steps or, if treated as continuous processes, using deterministic or stochastic rules for modeling the process. The E-Cell System provides the simulation platform for use of these calculations to model separately, and/or in combination, multialgorithm/multi-timescale simulations [7].
E-Cell models have three fundamental object classes: “variable”, with the option of either molar concentration units or value units, “process”, for writing operations on the variables, and “system” for identifying logical and/or physical compartments with/without volume that contain the variables and processes. This object-oriented approach enables intuitive description and prevents mistakes in modeling because of the one-to-one correspondence between each chemical process and reaction process in the E-Cell model. At the same time, once the reaction-module (“process” in the E-Cell System) has been created and defined, the module is easily reusable not only in the same model but also in another models. Similarly, the calculation algorithm itself can be modified or extended as a module and switched easily by rewriting one line of the model file, for example, from a deterministic to a stochastic model.
The system is written in C++ to maximize calculation speed and the frontends are easily scriptable and extensible using the Python language. Using Python scripts, users can program the rules for simulation sessions as well as the simulation conditions themselves in a given session; for example, initial parameter settings, time-dependent or concentration-dependent perturbations, or the output form of the simulation results. Some widely used methods of ODE model-based mathematical analysis have been already provided in the E-Cell Simulation Environment. These include sensitivity analysis, bifurcation analysis, Metabolic Control Analysis (MCA), and real genetic algorithms for parameter optimization.
In the case of large-scale pathways such as whole RBC metabolism, object-oriented modeling is necessary to ensure accuracy. In addition, its extensibility is also very helpful for parametric tuning of the model.
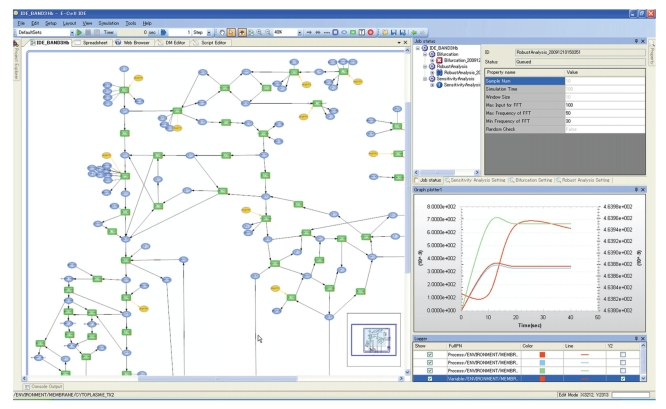
However, due to the lack of a user-friendly Graphical User Interface (GUI) for the display of modeling, in silico experimentation, and simulation results, the E-Cell System has been difficult to use, especially for biologists. Recently, the E-Cell IDE (Integrated Development Environment), a GUI -based simulation toolkit for Windows, was developed in order to allow non-expert users to edit, run, and analyze the E-Cell model (Figure 1). The pathway editor allows users to generate new pathways or customize existing pathway maps, and to set values/concentrations, reactions, and parameters directly using the GUI toolkit. This system can also read and write models in SBML (Systems Biology Markup Language) format [13], which is the most common markup language for making the models compatible with other cell simulators, for example, Copasi [14], DBSolve [15], Virtual Cell [16], Systems Biology Workbench (SBW [17]), and XPPAUT (http://www.math.pitt.edu/~bard/xpp/xpp.html). The E-Cell IDE provides a visual representation of the dynamics of the simulation on the pathway map by varying the size and width of each node or edge, respectively. The GUI tools enable the user to conduct classical mathematical analyses, such as parameter estimation, using a real number genetic algorithm and metabolic control analysis. Users can also perform CUI (Command-line User Interface)-based simulations and analyses for more complex or large-scale operations, such as situations in which the automation of many simulations or parametric computing is needed.
Figure 1.
View of the on-screen E-Cell GUI (E-Cell IDE). The E-Cell IDE has a user-friendly pathway editor (left) and a GUI-based mathematical analysis tool kit (right top). The simulated dynamics is visualized in the tracer of the time histories of variables/processes (bottom right), as well as in the pathway map (left).
Development of the next-generation E-Cell System (E-Cell Version 4) has started by aiming to simulate biological events at the molecular level, so that we can assess “cellular space” issues such as molecular fluctuation, localization, and crowding, while keeping complete compatibility and combinability with the current version (E-Cell Version 3). A detailed description, the current status, and the future vision of the E-Cell Simulation Environment are provided in recent works [7] and the project web page [8].
3. Human RBC Metabolism: A Long History of Dynamic Simulation
Human RBCs lose their mitochondria and are entirely dependent on glycolysis to produce ATP. The glucose transporter GLUT-1, expressed in RBCs, has a high affinity for extracellular glucose and remains saturated under physiological concentrations of glucose. Hexokinase (HK), which catalyzes the initial step of glycolysis in RBCs, also has a low Km (high affinity) for intracellular glucose. Thus, the rate of initiation of glycolysis is ensured even when plasma glucose concentration is low. ATP is required by ion pumps to prevent cell swelling and is also used in many other endergonic enzymatic processes. ATP is a necessary substrate for two of the initial rate-limiting steps in glycolysis (HK, PFK), but excess ATP can downregulate glycolysis through the inhibition of PFK. Most of the glucose entering a cell is converted to lactate through glycolysis (in our model, 87.5% under normoxic steady-state conditions), with the remainder entering pentose-phosphate pathways (in our model, 12.5%). This route provides reduced nicotinamide adenine dinucleotide phosphate (NADPH), which prevents oxidation of the cell directly and indirectly through the conversion of glutathione from its oxidative form back to its active reduced form.
One of the most characteristic pathways in RBCs is the Rapoport–Luebering cycle, which generates a high concentration of 2,3-diphosphoglycerate(2,3-BPG), a diversion of glycolysis from 1,3-disphosphoglycerate(1,3-BPG) that prevents excess ATP production by bypassing the process catalyzed by phosphoglycerate kinase (PGK). The increase in 2,3-BPG facilitates the release of oxygen from hemoglobin to tissues. In this manner, although the binding and release of gas molecules (oxygen, carbon dioxide, and so on) through hemoglobin requires no energy, the regulation of glycolytic flux, which is important for maintaining oxygen transport capacity through the maintenance of adequate levels of ATP and 2,3-BPG.
A single RBC can be assumed to be a closed system enclosed by a plasma membrane. Human RBCs circulate for as long as 120 days [18], and, thus, the metabolism in the cell should be robust in physiological situations. This simplicity and robustness, as well as the abundance of accumulated knowledge regarding metabolic enzymes, have made RBC metabolism a suitable subject for mathematical modeling and system level analysis of the metabolic/biological pathways. There is a long history of construction of metabolic models of human RBCs, in which RBC metabolism is described with simultaneous ordinary differential equations with different levels of detail, depending on the focus of the model. These models have very different system level properties (for comparative analysis of several RBC glycolysis models, see [19]).
The first challenge in modeling human RBC metabolism was constructing a linear glycolytic model (by Rapoport and Heinrich [20]), which was intended to test whether a linear theory suffices for a description of the steady state under several experimental conditions, and to better understand the crossover structure. The model also contributed to the discovery of the framework in “flux control theorem” [21]. Ataullakhanov et al. expanded the glycolytic model to include the pentose phosphate pathway, in which ATP and ADP are treated as parameters, to predict the dependence of glycolytic flux on ATP content [22].
An extension of the glycolytic model to include the reactions of synthesis and degradation of adenine nucleotides was provided by Schauer et al., and suggested that adenylate metabolism, the functional role of which remained poorly understood at the time, may serve to improve the stabilization of the energy charge [23].
Brumen and Heinrich were the first to attempt combining the models of energy metabolism with the models of volume regulation [24], and presented a metabolic osmotic model of RBCs [25]. Reactions synthesizing the adenylate pool, osmoregulation, electroneutrality, and ion transport were later incorporated and proved that changes in RBC volume are associated with glycolysis.
Holzhutter et al. used their mathematical model, which included glycolysis, the pentose phosphate pathway, and the 2,3-BPG shunt, for analysis of the pathology associated with pyruvate kinase deficiency [26]. They showed the effects of severely lowering the activities of several enzymes in the model and compared their results with experimental data from patients with deficiencies in these enzymes.
Joshi and Palsson constructed a model to provide a framework for the integration and interpretation of the extensive biochemical data on enzymes and metabolites by means of a comprehensive mathematical model of RBC metabolism [27, 28]. The model examines three different interdependent characteristics of RBC: the properties of the red cell membrane, the kinetics of transmembrane fluxes of chemical constituents of RBCs and plasma, and the thermodynamic formulation of the osmotic states. The model was extended to include the pH dependence of enzyme activities and mechanisms of volume regulation, namely electroneutrality and osmotic balances, accounting for the cell's interaction with the environment as well as cell metabolism [29]. They used their comprehensive model to propose emerging mathematical frameworks for metabolic analysis such as top-down analysis for revealing metabolic pools [30–32].
Mulquiney and Kuchel developed a precise model for the Rapoport–Lubering (2,3-BPG) shunt, as well as glycolysis and the pentose phosphate pathway, based on enzyme kinetics derived from their NMR assays [33, 34]. Their model also includes a detailed description of magnesium equilibrium and binding of metabolites to oxyhemoglobin (oxyHb) [35].
The first E-Cell RBC model that was published presented an analysis of the pathology associated with hereditary G6PD deficiency [36]. This model was subsequently expanded by incorporating the Joshi and Palsson model and also by the inclusion of the GSH synthesis pathway and GSSG transport system, and suggested that normally inactive pathways may have an essential role in abnormal conditions such as enzyme deficiencies.
Using E-Cell, we also constructed a model of the two metHb-reduction pathways in RBCs by expanding the Mulquiney and Kuchel model [37]. The model assessed the mechanism that switches between NADPH dependent and NADH dependent pathways of metHb reduction. The former pathway has a high response rate to hemoglobin oxidation with a limited reducing flux, and the latter has a low response rate with a high-capacity flux, correlated to the supply of NADH and NADPH from central energy metabolism.
Recently, we published a model that contains not only enzymatic or ion binding reactions combined with existing models, but also includes allosteric transitions in hemoglobin in response to the partial pressure of oxygen and the binding of plasma membrane proteins to glycolytic enzymes and hemoglobin ([11], described below).
Based on the same assumptions, Hald et al. focused on the alteration of RBC metabolism caused by changes in oxygen and carbon dioxide partial pressures during circulation, and showed that changes in these gases resulted in glycolytic flux oscillations, with consequent overshoots in levels of central metabolites [38].
4. Metabolic Model of Human RBCs Using the E-Cell System
4.1. Model Construction
4.1.1. Metabolic Reactions
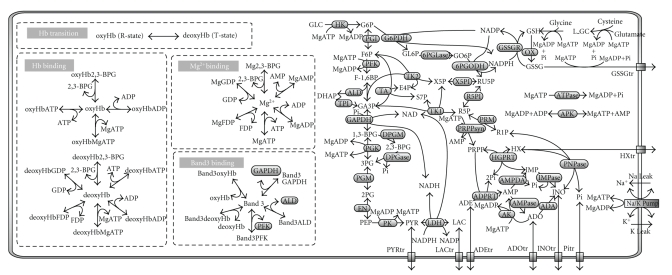
The metabolic model constructed with the E-Cell system is shown schematically in Figure 2, which includes enzyme reactions, transporter functions, binding interactions, and a process for determining hemoglobin allosteric transitions. For detailed descriptions of all equations, see the supporting material from our previous reports [11, 12].
Figure 2.
A Metabolic pathway map constructed using the E-Cell RBC model. The complete reaction network of the metabolism in the E-Cell RBC model. MgX/oxyHbX/deoxyHbX/band3X denotes the complex formed by compound “X” and Mg2+, oxyhemoglobin, deoxyhemoglobin, or band 3, respectively. Abbreviations used in this figure: GLC, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6-BP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GA3P, glyceraldehyde-3-phosphate; 1,3-BPG, 1,3-bisphosphoglycerate; 2,3-BPG, 2,3-bisphosphoglycerate; 3PG, 3-phosphoglycerate; 2PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; LAC, lactate; GL6P, gluconolactone-6-phosphate; GO6P, gluconate-6-phosphate; RU5P, ribulose-5-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; PRPP, 5-phosphoribosyl-1-phosphate; ADE, adenine; IMP, inosine monophosphate; R1P, ribose-1-phosphate; INO, inosine; ADO, adenosine; HX, hypoxanthine; AMP, adenosine monophosphate; ADP, adenosine diphosphate; ATP, adenosine triphosphate; NADP, nicotinamide adenine dinucleotide phosphate; NADPH, nicotinamide adenine dinucleotide phosphate (reduced); NAD, nicotinamide adenine dinucleotide; NADH, nicotinamide adenine dinucleotide (reduced); Ki, potassium ion; Nai, Sodium ion; Pi, inorganic phosphate; L_GC, L-glutamyl cysteine; GSH, glutathione (reduced); GSSG, glutathione (oxidized).
The metabolic network developed in this mathematical model covers a majority of the metabolic pathways: glycolysis, the pentose monophosphate shunt, the purine salvage pathway, glutathione metabolism, and Na+-K+-ATPase activity coupled to the leak of Na+ and K+. Membrane transporters for pyruvate, lactate, adenine, adenosine, hypoxanthine, and inosine are modeled, and the concentrations of all the extracellular metabolites, as well as intracellular glucose, are fixed at physiological concentrations. The rate equations for metabolic reactions are derived from previously published experimental data, and are largely based on the Mulquiney model [23, 33] and, in part, on other previous mathematical models [27, 36]. Most enzyme rate equations were derived using the King-Altman method for each of the enzyme-catalyzed reactions, with the exception of some reactions in purine metabolism and purine transport processes, which were described using Michaelis-Menten or first-order kinetics. Following the Mulquiney model, the following reactions are modeled as pH-dependent kinetics where intracellular pH is treated as an independent parameter: key enzymatic reactions in glycolysis, such as hexokinase (HK), phosphofructokinase (PFK), glyceraldehyde phosphate dehydrogenase (GAPDH), pyruvate kinase (PK), lactate dehydrogenase (LDH), the 2,3-BPG shunt reactions (2,3-BPG shunt), and binding of metabolites to Mg2+ and hemoglobin. The overall oxidization of glutathione in the cell and dephosphorylation of ATP except for Na+/K+-ATPase and kinases in above-mentioned major pathways are simplified into first-order reactions.
4.1.2. Hemoglobin Transition
The capacity of RBCs to deliver oxygen to tissues is fundamentally linked to the equilibrium between the two main states of hemoglobin structure, a tense state, “T” (or “deoxy”), and a relaxed state, “R” (or “oxy”), although the transition between the structures is not a simple two-state mechanism. The ability of oxygen to bind to hemoglobin is allosterically regulated by intracellular components such as 2,3-BPG and ATP. An increase in 2,3-BPG stabilizes the T-state of Hb, thereby facilitating O2 dissociation from the Hb. Since T-state hemoglobin has a higher affinity for 2,3-BPG and ATP than the R-state, stabilization of hemoglobin in the T-state by increasing the amount of 2,3-BPG binding reduces the amount of free 2,3-BPG and ATP, leading to a further acceleration of metabolism. Under these circumstances, consideration of the hemoglobin transition may exert considerable effects on RBC metabolism, especially on glycolysis, and vice versa.
The equation for the kinetics of oxygen saturation of hemoglobin, derived by Dash and Bassingthwaighte [39], was used to calculate the ratio of oxy-(R-) hemoglobin to total hemoglobin. The saturation kinetics is described by the Hill equation, with dependencies on the levels of pO2, pCO2, intracellular pH, concentration of 2,3-BPG, and temperature. In our model, pO2, pCO2, pH, and temperature are independent variables. Once the ratio between oxy-(R-) and deoxy-(T-) hemoglobin (oxygen saturation of hemoglobin, SHbO2) is determined under given conditions at a certain step in the simulation, the velocity of a reversible conversion of free T-state hemoglobin to free R-state hemoglobin in the step is derived as
| (1) |
where SHbO2 ranges from 0 to 1, Ttotal and Rtotal are the sum of all complexed forms and the free form of each state of hemoglobin, and k denotes the scaling constant, which is set to 1200 to be equal to those in other binding reactions. In our model, it is assumed that this transition occurs only between free form R/T-hemoglobins, even though Ttotal and Rtotal in the equation above are the “total hemoglobin” of each state. In our model, the P50 value, which is the partial pressure of oxygen at half-saturation of hemoglobin, is calculated to be 25-26 mmHg under virtual cell-free conditions with 5 mM 2,3-BPG. However, if the concentrations of the free forms of T-state and R-state hemoglobin are substituted for Ttotal and Rtotal, the P50 is calculated to be approximately 50 mmHg, which is significantly different from the physiological value.
4.1.3. Binding between Band 3 and Glycolytic Enzymes or Hemoglobin
Band 3 is the major anion transporter in RBCs and plays a role in chloride/bicarbonate exchange, as well as an important structural role as a plasma membrane protein that contributes to stabilization of the membrane surface by forming multiprotein complexes [40]. Band 3 has multiple cytoplasmic domains, including an N-terminal cytoplasmic domain, which binds to the glycolytic enzymes PFK, ALD and GAPDH. PK and LDH are also localized to the plasma membrane when intact RBCs were fixed in their oxygenated states, but there has been no evidence of direct association with band 3 [41]. The N-terminal cytoplasmic domain of band 3 also binds to Hb, with a greater affinity for T-state hemoglobin than R-state [42]. Recent observations have shown the enzymatic activities of PFK, ALD, and GAPDH are completely blocked by binding to band 3, whereas their activities are recovered upon dissociation from band 3. No changes in PK and LDH activities have been detected [43] and the membrane docking sites of PK and LDH have yet to be identified [44]. We were the first to incorporate the binding of band 3 to glycolytic enzymes/hemoglobins into the mathematical model of major RBC metabolism [11]. The binding affinity of the metabolite-hemoglobin complex to band 3 is assumed to be the same as that of the free form of hemoglobin (e.g., deoxyHb and deoxyHb-2,3BPG). We simply used the published association constants evaluated in vitro.
4.2. Application of the Model I: Analysis of the RBC Response to Hypoxia
ATP is required in the initial steps of glycolysis by HK and PFK to trigger glycolytic ATP synthesis, and a fraction of the intracellular ATP is released to the extracellular space under hypoxia to elicit hypoxia-induced vasodilation, although the amount of ATP released is small, compared to intracellular levels [45]. At the same time, RBCs are known to accelerate glucose consumption in response to exposure to hypoxia, which results from acceleration of glycolysis, as judged by the increase in 2,3-BPG [46]. This change leads to further T-state Hb stabilization and increases the supply of oxygen to hypoxic tissues. Taking into account these features, researchers have hypothesized that RBCs have appropriate mechanisms for responding quickly to hypoxia to upregulate de novo ATP synthesis and glycolytic flux, leading to the increase in 2,3-BPG. Historically, the hypoxic acceleration of glycolysis in RBCs was thought to be induced by alterations in pH or the metabolic compensation of ATP [47, 48]. However, recent evidence from studies with intact RBCs has shown that T-state hemoglobin triggers an increase in the activities of glycolytic enzymes that interact with band 3 and plays a central role in acceleration of glucose consumption to increase the synthesis of ATP and 2,3-BPG [49]. Our model was used to elucidate the mechanistic features of the coordination and dynamics of sequential glycolytic reactions and the outcomes, in terms of alterations in levels of intracellular metabolites upon hypoxia, in particular as a result of band 3 interactions. To mimic hypoxic conditions, the pO2 of the model, which was initially set to 100 mmHg, was reduced to 30 mmHg, a value consistent with capillary microvessels in vivo [50].
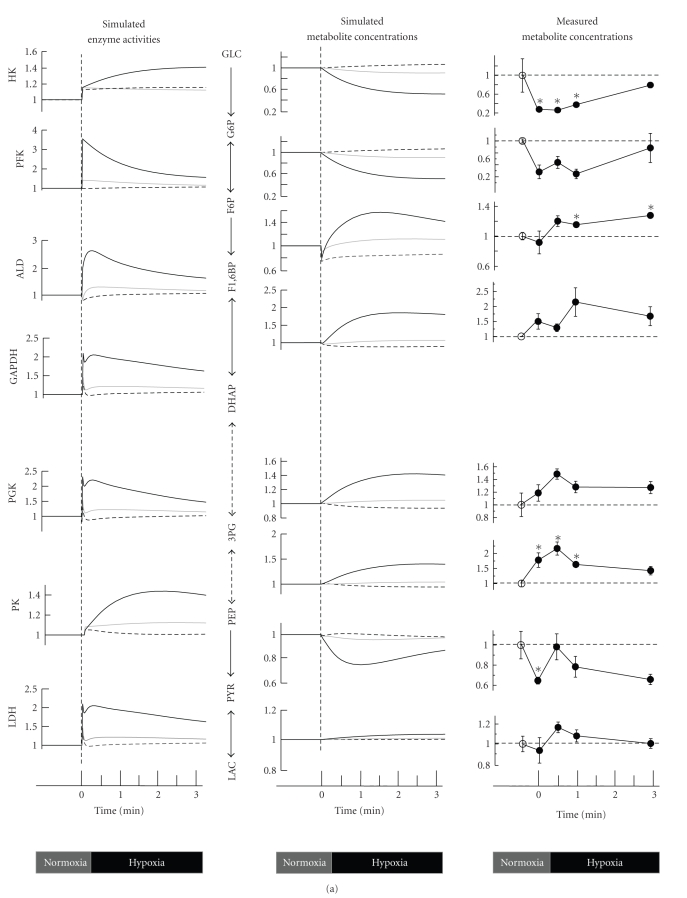
A comparison between three models in predicting temporal alterations of glycolysis during a 3-min virtual hypoxia is illustrated in Figure 3(a). Model A includes Band 3 interactions with hemoglobin and glycolytic enzymes (corresponding to the BIII(+) model described in [11]). Model B uses the same initial and normoxic steady-state conditions as model A, but omits interactions between Band 3 and hemoglobin/enzymes (corresponding to the BIII(−) model in the reference [11]). By comparing model A with model B, we can estimate the pure effect of hypoxia-induced glycolytic activation exerted by the accumulation of T-state hemoglobin. In Model C, all of the glycolytic enzymes exist in dissociated forms, even in the initial (PO2 = 100) state. Thus, the initial and steady-state conditions of model C are different from those of model A and model B. As shown in Figure 3(a), the overall activities of the glycolytic enzymes in model A are significantly accelerated relative to the other models. As expected, the activities of PFK, ALD, and GAPDH spiked immediately as a result of their release from band 3 upon hemoglobin binding, while these enzyme activities did not change significantly in model B. These differences in enzyme activities between the two models created a distinct metabolome profile: the glycolytic intermediates in model A displayed a pattern opposite to those in model B. In model A, G6P and F6P decreased by 50% and F1,6BP, DHAP, 3PG, and PEP increased by 40% versus the corresponding baseline levels (Figure 3(a)). The time-dependent alteration in levels of glycolytic intermediates predicted by model A is entirely consistent with results obtained from metabolome analyses using capillary electrophoresis mass spectrometry (CE-MS). These trends in time-variation in glycolytic fluxes/metabolite concentrations have also been reproduced in model C, where no band 3 interactions are considered. However, the alterations seen in model C exhibit much less variation than the experimental results. In both models (model A and model C), the activation of HK is caused by a decrease in the free form of 2,3-BPG, which is a strong inhibitor of HK; however, HK activity in model A exhibited greater activation. This difference appeared to result from a decrease in G6P and an increase in ATP, leading to a reduction in HK product inhibition. The distinct hypoxia-induced increase in glycolytic flux arising from hemoglobin transition and the consequent changes in band 3-interactions have been verified by several separate experiments. Messana et al. showed that the hypoxia-induced increase in glucose consumption disappeared in RBCs treated with 4,4′-diiso-thiocyanostilbene-2,2′-disulfonate (DIDS), which acts as an anion exchange inhibitor targeting band 3 [51]. We demonstrated that CO pretreatment of RBCs to stabilize hemoglobin in the R-state attenuated the hypoxia-induced acceleration in the conversion of 13C-glucose into 13C-lactate [11]. Furthermore, a recent study by Lewis et al. provided direct evidence for the role of band 3 in mediating metabolic shifts under more physiological conditions in intact RBCs, in which the metabolic fluxes were measured using (1)H-(13)C NMR and RBCs were treated with pervanadate, a reagent that blocks the interaction between band 3 and glycolytic enzymes [49].
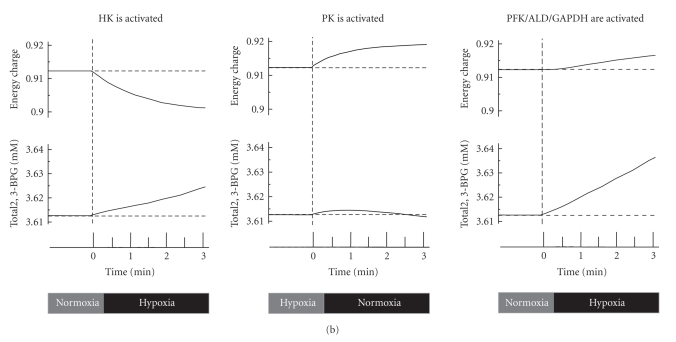
Figure 3.
Temporal alterations in glycolysis in response to hypoxia. (a) Predicted alterations in glycolytic enzyme activities under hypoxia and comparison of time-courses of glycolytic intermediates between the simulations and metabolome measurements using CE-MS. The simulation results of model A (including Band 3 interactions, corresponding to the solid red line in Figure 2 of [11]), model B (using the same initial steady-state conditions as A but neglecting further Band 3 interactions, corresponding to the dotted blue line in Figure 2 of [11]), and model C (with all enzymes dissociated from Band 3) are represented by the solid black (model A), dotted black (model B), and solid gray (model C) lines, respectively. See the main text for discussion of the models. In the metabolome data collected by CE-MS, closed circles indicate ratios of hypoxic metabolite concentrations to normoxic control concentrations, which are represented with open circles. Values are the means ± SE of four separate experiments. Asterisks: P < .05 versus the steady-state baseline values. (b) Results of model analysis to determine whether or not increasing the activity of a particular enzyme (HK, PK, or PFK+ALD+GAPDH) by 2-fold can promote a simultaneous increase in the energy charge and in 2,3-BPG under conditions of hypoxia.
Through the activation of glycolysis, the energy charge is greater in the band 3-implemented model than the other models. The basal energy charge under normoxic steady-state conditions was predicted to be 0.912, which is comparable to that reported in previous studies (ranging from 0.86 [52] to 0.935 [53]), and the value rose to 0.917 after three minutes in hypoxia. At the same time, the total amount of 2,3-BPG in model A was also predicted to be greater than the other two models, which is likely to contribute to a rapid increase in the formation of hemoglobin-2,3-BPG complexes that consequently lead to the release of residual hemoglobin-bound oxygen from the RBC. The calculated amount of oxygen released by increasing 2,3-BPG in three minutes of hypoxia was 6 μmol per L of RBC volume.
Another key finding of this simulation study is that PFK activation is a crucial step for the upregulation of both energy charge and 2,3-BPG generation, while activation of the other so-called rate-limiting enzymes, at the initial (e.g., HK) or final (e.g., PK) steps of the glycolytic pathway, fails to satisfy these requirements. HK activation resulted in a decrease in energy charge and an increase in 2,3-BPG, while PK activation increased energy charge without stimulating 2,3-BPG generation (Figure 3(b)).
Through these analyses, our model has demonstrated a pivotal role for band 3-intracellular protein interactions in enhancing activation of glycolysis in RBCs as part of a metabolic response to hypoxia, and in the consequent increase in both cell energetics and oxygen-carrying capacity. Taken together with the recent findings that the oxygenation-dependent assembly of glycolytic enzymes on the membrane is conserved in mammalian erythrocytes even in the absence of the intracellular band 3 binding site [44], the hypoxia induced glycolytic activation may be necessary for RBC viability over the long term and/or for temporally appropriate oxygen supply to the tissues.
4.3. Application of the Model II: Prediction of Metabolism of RBCs in Long-Term Storage
Whereas the kinetic metabolic models of human RBCs have led the simulation studies in the systems biology era, there has been no practical application of the RBC metabolic models for biotechnological use, at least to our knowledge. We intend to use our RBC model as a virtual experiment for optimization of RBC storage conditions. In the field of emergency medicine, it is critical to store RBCs in such a manner that their viability and capacity for oxygen delivery are retained after transfusion. These characteristics are strongly correlated with the intracellular metabolic status. Long-term cold storage reduced intracellular ATP and 2,3-BPG, which causes a reduction in deformability, oxygen-carrying capacity, and reduces energy sources for intracellular processes. In Japan, a mannitol-adenine-phosphate (MAP) solution is commonly used for storing RBCs (RC-MAP). In this solution, approximately 50% of ATP is retained after 42 days in storage, but 2,3-BPG is almost completely depleted after 2 weeks [54]. However, the large metabolic network of the RBC that underlies the depletion of these two metabolic indicators has not been considered.
To develop an “RC-MAP model” that can represent the long-term dynamics of cold MAP-stored RBC metabolism, concentrations of glucose and adenine of the basal model were set to the values in MAP-solution, and the extracellular sodium ion and inorganic phosphate concentrations were changed by calculating the content of sodium phosphate, sodium chloride and sodium citrate in the solution. The intracellular pH of RBCs in RC-MAP is lowered due to the large addition of citrate and gradually decreases further over time in storage due to the accumulation of lactate [55]. Thus, the initial pH and pH variation during cold storage were set according to the reported time-course pH data in the literature [54], where the decrease in intracellular pH is fitted by first-order kinetics. As the cold temperature stabilizes hemoglobin in the R-state, all hemoglobin was designated R-state in the RC-MAP model.
In the process of parameter estimation, we focused on those enzymatic activities that may be altered by physical changes in the storage conditions relative to the circulating conditions, the cold temperature and the low pH. All chemical reactions, including enzymatic reactions, chemical-binding reactions, and active transport processes, should be significantly reduced but not completely stopped at 4°C. In particular, the Na+/K+-ATPase pump is very sensitive to lowered temperature [56]. Moreover, there are many reports that enzymes involved in purine metabolism, including adenosine deaminase, adenosine monophosphate phosphohydrolase (AMPase), inosine monophosphatase, and adenosine monophosphate deaminase are optimized or activated at relatively low pH, such as in the RC-MAP storage conditions, but adenosine kinase and hypoxanthine-guanine phosphoryl-transferase are not. From these knowlodge, we grouped all reaction activities into three groups: Na+/K+ pump activity, purine metabolism enzyme activities, and all remaining enzymatic or binding activities. The three groups of reaction activities are then treated as “adjustable parameters” for the parameter estimation.
As a result of the GA (Genetic Algorithm) analysis, the three adjustable parameters were determined and their estimated values were supported by previous knowledge, as follows. (1) Na+/K+ pump activity was 0.6% of the basal values, since it decreases to 0.2–0.8% of the level at body temperature when the temperature is decreased to 5°C in most mammalian erythrocytes [56]. (2) The activity of purine metabolism enzymes was 24% of the basal model, which is considerably higher than that of other enzymatic processes. Furthermore, the loss of intracellular adenine within three weeks was reproduced only by the model when this parameter was 20–30% [12]. (3) The activity level of the other reactions was estimated to be 3.5%, since, in general, enzyme activities at 4°C are reduced to 1–5% of those at the normal body temperature [57]. The dynamics of the estimated RC-MAP model during cold storage was then compared with the experimentally measured glycolytic intermediates in MAP-stored RBCs using capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS), as previously described [12, 58] (Figure 4(b)).
Figure 4.
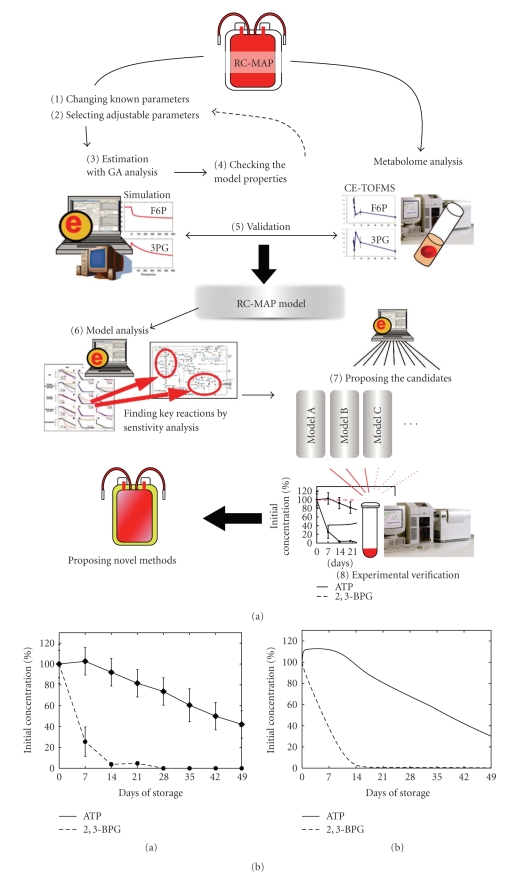
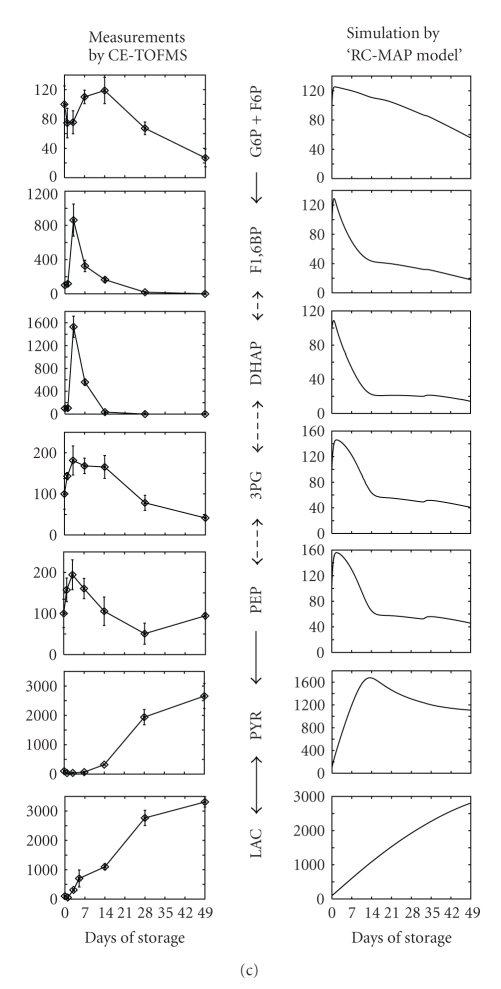
Modeling of metabolism in RBCs during long-term cold storage and model verification. (a) The work flow of developing and analyzing the “RC-MAP model”. (1) The values of external parameters in our E-Cell RBC model (basal model), which assumes normal circulating conditions, are modified to meet the conditions of the cold-MAP solution. (2) Adjustable parameters, which are thought to be changed by storage conditions, are determined. (3) Adjustable parameters are estimated using experimental data on metabolic alterations in RC-MAP. (4) The relevance and robustness of the parameter choices are checked. (5) The dynamics of metabolism are compared between the RC-MAP simulation and experimental treatment. (6) A sensitivity analysis of the model is conducted to find key reactions, metabolites, and parameters to maintain the energetics and oxygen-carrying capacity of stored RBCs. (7) Candidate components for optimized storage conditions are determined by computationally testing various combinations of factors that can be modified experimentally. (8) Finally, the candidate models are validated by a metabolome analysis. In (3), we employed the classical read number genetic algorithm within the E-Cell Simulation Environment to fit the model to the reported time-course data of ATP and 2,3-BPG concentration changes in RBCs held in cold RC-MAP for 49 days. (b) A comparison of time-related changes in ATP and 2,3-BPG levels between reported experiments (Shiba et al., 1991) and the “RC-MAP model” is presented. The time-course of ATP (solid black) and 2,3-BPG (broken black) in RC-MAP at 4°C for 49 days in (a) previously reported data and (b) the prediction of the estimated model derived by the Genetic Algorithm. Experimental values are shown as the mean ± S.D. of 19 separate experiments. Values are percentages of the initial concentrations. (c) Measured (left) and simulated (right) time-dependent alterations in glycolytic intermediates. In CE-TOFMS measurements, the RBC samples were suspended in cold-MAP for 49 days under laboratory conditions. Data represent the means ± S.D. of five separate experiments. “G6P + F6P” indicates the sum of the concentrations of G6P and F6P. Values are represented as percentages of the initial concentration of each metabolite.
In CE-TOFMS measurements, PYR, LAC, HX, and S7P were significantly increased, and all glycolytic intermediates, with the exception of PYR and LAC, were markedly decreased after 49 days. These measured alterations of intermediates were qualitatively reproduced in an estimated model that was fitted to the reported RC-MAP data. The models using random sets of adjustable parameters failed to predict these final increases and/or decreases in the concentrations of the glycolytic intermediates. However, the dynamics of the intermediates, such as the extraordinarily large increases in F1,6-BP and DHAP in the first week or the initial stagnation of PYR, could not be predicted by the estimated model. These gaps may be the result of both the level of simplicity of setting the adjustable parameters and the difference in experimental conditions in our lab relative to commercial RC-MAP usage. The early large peaks of F1,6-BP and DHAP did appear when the initial pH was set to lower values, but some mechanism of reduction in PK activity was needed to reproduce the initial stagnation of PYR (see the supporting material in [12]). More detailed refinement of the settings of the RC-MAP model using comprehensive metabolome data will be necessary to increase the predictive power of the model.
Based on the sensitivity analysis of the RC-MAP model, 2,3-BPG levels may be maintained when hemoglobin is shifted to the T-state. Reducing enzyme activities in purine metabolism would be very effective for maintaining ATP, whereas the ATP concentration control of enzymes in purine metabolism is small. Another interesting prediction of the model is that a slight activation of HK and PFK during storage can maintain both ATP and 2,3-BPG, despite the fact that all prior studies of blood storage have aimed to reduce enzymatic activities. These factors can serve as a possible target of the next round of experiments for improving RBC storage conditions.
However, many physicochemical aspects have yet to be incorporated in the model, such as the maintenance of cellular homeostasis through regulating intracellular pH and cell volume in connection with ion balance. In this study, the effects of additives in the MAP solution, such as mannitol and sodium chloride, as osmoregulatory substrates that prevent erythrocyte hemolysis, as well as the acidotic shift due to lactate accumulation, were ignored (or considered to be unchanged from normal circulating RBCs).
5. Discussion: Future Perspectives of RBC Metabolic Model
A mathematical model of the metabolic networks in human RBCs with precise enzyme kinetics and linkages between metabolism and oxygen pressure can provide us with a better understanding of the response to environmental stimuli that may occur in vitro or in storage conditions. Our model is the first to include the effects of intracellular protein-protein interactions (competitive binding to band 3 between hemoglobin and glycolytic enzymes) on RBC metabolism and to show the relevance of these interactions to the supply of ATP and 2,3-BPG over time.
Several factors must be considered in order for the model to provide an insight into the possible physiological importance of these intracellular events. First, a precise estimation of the energy demand of the cell, such as the energy required to maintain its biconcave shape, should be made [59]. Among all previously published mathematical models of RBC metabolism, the processes utilizing ATP are oversimplified into first-order kinetics with respect to the ATP concentration, with the exception of Na+-K+ pump ATPase activity in Jasimidi and Palsson's model [27, 28], and in our models [11, 36], despite the fact that both the glycolytic flux and the concentration of ATP are largely controlled by the ATP demand [19].
Secondly, it is important to examine the external effects of RBC metabolism, that is, the modulation of oxygen carrying capacity by altering the hemoglobin allostery by intracellular ATP and 2,3-BPG. Including these features requires knowing “when” and “to what extent” the environmental oxygen demand is raised, in order to obtain information at a systems level of behavior. In other words, the model would need to make a connection between the efficiency of carrying gas molecules in the RBC and the underlying metabolism, as well as the behavior of the RBC population in response to capillary geometry and blood flow. Bassingthwaights' group has begun to construct a multiscale model in their physiome project: A model of blood-tissue/tissue-capillary exchange of oxygen, carbon dioxide, which includes exchange of bicarbonate and hydrogen ion for considering Bohr and Haldane effects in RBC, intraerythrocytic adsorption of CO2 and O2 on hemoglobin, and extraerythrocytic tissue-dependent gas consumptions [58, 60]. Furthermore, a novel aspect of RBC metabolism is the recently observed release of intracellular ATP into the extracellular space in response to hypoxia, although the actual amount of ATP released is very small compared to levels within the cell [45]. The released ATP regulates blood flow by binding to P2Y purinergic receptors on the luminal surface of the endothelium, initiating the signaling events that result in vasodilation. A theoretical test of the contribution of the hypoxia-dependent ATP release by RBCs to an increase in vessel diameter in upstream arterioles has been carried out using a simplified theoretical blood-flow model [61]. An understanding of the physiological importance of temporal alterations in RBC metabolism could be accomplished by using these theoretical frameworks to develop models that connect the detailed metabolism in the RBC to higher level processes outside the cell.
Another intriguing addition to the model would be the incorporation of spatial or intracellular locus information into the metabolic model. Because RBCs do not contain any bound organelles in the cytoplasm, the intracellular system has been modeled as homogeneous in space. However, recent observations that glycolytic enzymes form a macromolecular complex [43] suggest the importance of considering spatial effects, even in models that focus on metabolic reactions. Additionally, another cytoplasmic domain of band 3 (C-terminal) binds carbonic anhydrase, which plays an important role in RBCs by catalyzing the hydration of carbon dioxide [62]. This interaction may improve the efficiency of Cl−/HCO3− exchange and, at the same time, may change the local pH significantly. Since the interactions between intracellular proteins and band 3 are strongly dependent on pH and ionic conditions [63], the local pH may control glycolytic flux both directly and indirectly. The intracellular compartmentalization and the diffusion of the glycolytic enzyme complexes and hemoglobin need to be taken into consideration for precise estimation of the functional requirements of the gas (oxygen or carbon dioxide) dependent assembly of these macromolecules, especially for the juxtamembrane localization of PK and LDH (whose activities are not masked by binding to the plasma membrane).
Acknowledgments
The authors thank Kazunari Kaizu and Takeshi Sakurada for helpful comments. This paper was supported by a grant from CREST, the Japan Science and Technology Agency (JST); a grant from Yamagata prefecture, Japan; and a Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Research and Development of the Next-Generation Integrated Simulation of Living Matter, which is part of the Development and Use of the Next-Generation Supercomputer Project of MEXT. A. Y-Kinoshita and T. Nishino are supported by the “Global-COE Program for Human Metabolomic Systems Biology”.
References
- 1.Kitano H. Computational systems biology. Nature. 2002;420(6912):206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 2.Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443(7111):527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- 3.Steuer R, Gross T, Selbig J, Blasius B. Structural kinetic modeling of metabolic networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):11868–11873. doi: 10.1073/pnas.0600013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomita M, Hashimoto K, Takahashi K, et al. E-CELL: software environment for whole-cell simulation. Bioinformatics. 1999;15(1):72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- 5.Tomita M. Whole-cell simulation: a grand challenge of the 21st century. Trends in Biotechnology. 2001;19(6):205–210. doi: 10.1016/s0167-7799(01)01636-5. [DOI] [PubMed] [Google Scholar]
- 6.Alves R, Antunes F, Salvador A. Tools for kinetic modeling of biochemical networks. Nature Biotechnology. 2006;24(6):667–672. doi: 10.1038/nbt0606-667. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Vel Arjunan SN, Tomita M. Space in systems biology of signaling pathways—towards intracellular molecular crowding in silico. FEBS Letters. 2005;579(8):1783–1788. doi: 10.1016/j.febslet.2005.01.072. [DOI] [PubMed] [Google Scholar]
- 8. E-Cell Project. http://www.e-cell.org/.
- 9.Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. Robustness of cellular functions. Cell. 2004;118(6):675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Kilpp E, Herwig R, Kowald A, Wierling C, Lehrach H. Systems Biology in Practice: Concepts, Implementation and Application. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. [Google Scholar]
- 11.Kinoshita A, Tsukada K, Soga T, et al. Roles of hemoglobin allostery in hypoxia-induced metabolic alterations in erythrocytes: simulation and its verification by metabolome analysis. Journal of Biological Chemistry. 2007;282(14):10731–10741. doi: 10.1074/jbc.M610717200. [DOI] [PubMed] [Google Scholar]
- 12.Nishino T, Yachie-Kinoshita A, Hirayama A, Soga T, Suematsu M, Tomita M. In silico modeling and metabolome analysis of long-stored erythrocytes to improve blood storage methods. Journal of Biotechnology. 2009;144(3):212–223. doi: 10.1016/j.jbiotec.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Hucka M, Finney A, Sauro HM, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 14.Hoops S, Gauges R, Lee C, et al. COPASI: a complex pathway simulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 15.Goryanin OD, Tobin F. Metabolic Engineering in the Post Genomic Era. Oxford, UK: Horizon Bioscience; 2004. Applications of whole cell and large pathway mathematical models in the pharmaceutical industry; pp. 321–356. [Google Scholar]
- 16.Moraru II, Schaff JC, Slepchenko BM, et al. Virtual cell modelling and simulation software environment. IET Systems Biology. 2008;2(5):352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauro HM, Hucka M, Finney A, et al. Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS. 2003;7(4):355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh HS, Jaffé ER. Electrophoretic and functional variants of NADH-methemoglobin reductase in hereditary methemoglobinemia. Journal of Clinical Investigation. 1971;50(1):196–202. doi: 10.1172/JCI106473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.du Preez FB, Conradie R, Penkler GP, Holm K, van Dooren FLJ, Snoep JL. A comparative analysis of kinetic models of erythrocyte glycolysis. Journal of Theoretical Biology. 2008;252(3):488–496. doi: 10.1016/j.jtbi.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Rapoport TA, Heinrich R, Jacobasch G, Rapoport S. A linear steady state treatment of enzymatic chains: a mathematical model of glycolysis of human erythrocytes. European Journal of Biochemistry. 1974;42(1):107–120. doi: 10.1111/j.1432-1033.1974.tb03320.x. [DOI] [PubMed] [Google Scholar]
- 21.Heinrich R, Rapoport TA. Mathematical analysis of multienzyme systems. II. Steady state and transient control. BioSystems. 1975;7(1):130–136. doi: 10.1016/0303-2647(75)90050-7. [DOI] [PubMed] [Google Scholar]
- 22.Ataullakhanov FI, Vitvitsky VM, Zhabotinsky AM, et al. The regulation of glycolysis in human erythrocytes. The dependence of the glycolytic flux on the ATP concentration. European Journal of Biochemistry. 1981;115(2):359–365. doi: 10.1111/j.1432-1033.1981.tb05246.x. [DOI] [PubMed] [Google Scholar]
- 23.Schauer M, Heinrich R, Rapoport SM. Mathematical modelling of glycolysis and of the adenine nucleotide metabolism of human erythrocytes. II. Simulation of the adenine nucleotide breakdown after glucose depletion. Acta Biologica et Medica Germanica. 1981;40(12):1683–1697. [PubMed] [Google Scholar]
- 24.Tosteson DC, Hoffman JF. Regulation of cell volume by active cation transport in high and low potassium sheep red cells. The Journal of General Physiology. 1960;44:169–194. doi: 10.1085/jgp.44.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brumen M, Heinrich R. A metabolic osmotic model of human erythrocytes. BioSystems. 1984;17(2):155–169. doi: 10.1016/0303-2647(84)90006-6. [DOI] [PubMed] [Google Scholar]
- 26.Holzhutter H-G, Jacobasch G, Bisdorff A. Mathematical modelling of metabolic pathways affected by an enzyme deficiency. A mathematical model of glycolysis in normal and pyruvate-kinase-deficient red blood cells. European Journal of Biochemistry. 1985;149(1):101–111. doi: 10.1111/j.1432-1033.1985.tb08899.x. [DOI] [PubMed] [Google Scholar]
- 27.Joshi A, Palsson BO. Metabolic dynamics in the human red cell. Part I. A comprehensive kinetic model. Journal of Theoretical Biology. 1989;141(4):515–528. doi: 10.1016/s0022-5193(89)80233-4. [DOI] [PubMed] [Google Scholar]
- 28.Joshi A, Palsson BO. Metabolic dynamics in the human red cell. Part II. Interactions with the environment. Journal of Theoretical Biology. 1989;141(4):529–545. doi: 10.1016/s0022-5193(89)80234-6. [DOI] [PubMed] [Google Scholar]
- 29.Jamshidi N, Edwards JS, Fahland T, Church GM, Palsson BO. Dynamic simulation of the human red blood cell metabolic network. Bioinformatics. 2001;17(3):286–287. doi: 10.1093/bioinformatics/17.3.286. [DOI] [PubMed] [Google Scholar]
- 30.Price ND, Reed JL, Papin JA, Wiback SJ, Palsson BO. Network-based analysis of metabolic regulation in the human red blood cell. Journal of Theoretical Biology. 2003;225(2):185–194. doi: 10.1016/s0022-5193(03)00237-6. [DOI] [PubMed] [Google Scholar]
- 31.Jamshidi N, Palsson BO. Top-down analysis of temporal hierarchy in biochemical reaction networks. PLoS Computational Biology. 2008;4(9) doi: 10.1371/journal.pcbi.1000177. Article ID e1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jamshidi N, Palsson BO. Using in silico models to simulate dual perturbation experiments: procedure development and interpretation of outcomes. BMC Systems Biology. 2009;3, article 44 doi: 10.1186/1752-0509-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulquiney PJ, Bubb WA, Kuchel PW. Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: in vivo kinetic characterization of 2,3-bisphosphoglycerate synthase/phosphatase using 13C and 31P NMR. Biochemical Journal. 1999;342(3):567–580. [PMC free article] [PubMed] [Google Scholar]
- 34.Mulquiney PJ, Kuchel PW. Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: equations and parameter refinement. Biochemical Journal. 1999;342(3):581–596. [PMC free article] [PubMed] [Google Scholar]
- 35.Mulquiney PJ, Kuchel PW. Model of the pH-dependence of the concentrations of complexes involving metabolites, haemoglobin and magnesium ions in the human erythrocyte. European Journal of Biochemistry. 1997;245(1):71–83. doi: 10.1111/j.1432-1033.1997.00071.x. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama Y, Kinoshita A, Tomita M. Dynamic simulation of red blood cell metabolism and its application to the analysis of a pathological condition. Theoretical Biology and Medical Modelling. 2005;2, article 18 doi: 10.1186/1742-4682-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita A, Nakayama Y, Kitayama T, Tomita M. Simulation study of methemoglobin reduction in erythrocytes: differential contributions of two pathways to tolerance to oxidative stress. FEBS Journal. 2007;274(6):1449–1458. doi: 10.1111/j.1742-4658.2007.05685.x. [DOI] [PubMed] [Google Scholar]
- 38.Hald B, Madsen MF, Danø S, Quistorff B, Sørensen PG. Quantitative evaluation of respiration induced metabolic oscillations in erythrocytes. Biophysical Chemistry. 2009;141(1):41–48. doi: 10.1016/j.bpc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Dash RK, Bassingthwaighte JB. Blood HbO2 and HbCO2 dissociation curves at varied O2, CO2, pH, 2,3-DPG and temperature levels. Annals of Biomedical Engineering. 2004;32(12):1676–1693. doi: 10.1007/s10439-004-7821-6. [DOI] [PubMed] [Google Scholar]
- 40.Peters LL, Shivdasani RA, Liu S-C, et al. Anion exchanger 1 (band 3) is required to prevent erythrocyte membrane surface loss but not to form the membrane skeleton. Cell. 1996;86(6):917–927. doi: 10.1016/s0092-8674(00)80167-1. [DOI] [PubMed] [Google Scholar]
- 41.Chu H, Low PS. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochemical Journal. 2006;400(1):143–151. doi: 10.1042/BJ20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuneshige A, Imai K, Tyuma I. The binding of hemoglobin to red cell membrane lowers its oxygen affinity. Journal of Biochemistry. 1987;101(3):695–704. doi: 10.1093/jb/101.3.695. [DOI] [PubMed] [Google Scholar]
- 43.Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(7):2402–2407. doi: 10.1073/pnas.0409741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campanella ME, Chu H, Wandersee NJ, et al. Characterization of glycolytic enzyme interactions with murine erythrocyte membranes in wild-type and membrane protein knockout mice. Blood. 2008;112(9):3900–3906. doi: 10.1182/blood-2008-03-146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellsworth ML, Forrester T, Ellis CG, Dietrich HH. The erythrocyte as a regulator of vascular tone. American Journal of Physiology. 1995;269(6):H2155–H2161. doi: 10.1152/ajpheart.1995.269.6.H2155. [DOI] [PubMed] [Google Scholar]
- 46.Lenfant C, Torrance J, English E, et al. Effect of altitude on oxygen binding by hemoglobin and on organic phosphate levels. Journal of Clinical Investigation. 1968;47(12):2652–2656. doi: 10.1172/JCI105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamasaki N, Asakura T, Minakami S. Effect of oxygen tension on glycolysis in human erythrocytes. Journal of Biochemistry. 1970;68(2):157–161. doi: 10.1093/oxfordjournals.jbchem.a129341. [DOI] [PubMed] [Google Scholar]
- 48.Rapoport I, Berger H, Rapoport SM, Elsner R, Gerber G. Response of the glycolysis of human erythrocytes to the transition from the oxygenated to the deoxygenated state at constant intracellular pH. Biochimica et Biophysica Acta. 1976;428(1):193–204. doi: 10.1016/0304-4165(76)90120-3. [DOI] [PubMed] [Google Scholar]
- 49.Lewis IA, Campanella ME, Markley JL, Low PS. Role of band 3 in regulating metabolic flux of red blood cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(44):18515–18520. doi: 10.1073/pnas.0905999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suganuma K, Tsukada K, Kashiba M, et al. Erythrocytes with T-state-stabilized hemoglobin as a therapeutic tool for postischemic liver dysfunction. Antioxidants and Redox Signaling. 2006;8(9-10):1847–1855. doi: 10.1089/ars.2006.8.1847. [DOI] [PubMed] [Google Scholar]
- 51.Messana I, Orlando M, Cassiano L, et al. Human erythrocyte metabolism is modulated by the O2 -linked transition of hemoglobin. FEBS Letters. 1996;390(1):25–28. doi: 10.1016/0014-5793(96)00624-2. [DOI] [PubMed] [Google Scholar]
- 52.Komarova SV, Mosharov EV, Vitvitsky VM, Ataullakhanov FI. Adenine nucleotide synthesis in human erythrocytes depends on the mode of supplementation of cell suspension with adenosine. Blood Cells, Molecules, and Diseases. 1999;25(3-4):170–179. doi: 10.1006/bcmd.1999.0243. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu T, Kono N, Kiyokawa H, et al. Erythrocyte glycolysis and its marked alteration by muscular exercise in type VII glycogenosis. Blood. 1988;71(4):1130–1134. [PubMed] [Google Scholar]
- 54.Shiba M, Mura T, Masuyama T, et al. Preparation and preservation of red blood cell concentrates in MAP solution by quadruple bag system. Japanese Journal of Transfusion Medicine. 1991;37:404–410. [Google Scholar]
- 55.Högman CF, Knutson F, Lööf H. Storage of whole blood before separation: the effect of temperature on red cell 2,3 DPG and the accumulation of lactate. Transfusion. 1999;39(5):492–497. doi: 10.1046/j.1537-2995.1999.39050492.x. [DOI] [PubMed] [Google Scholar]
- 56.Ellory JC, Willis JS. Kinetics of the sodium pump in red cells of different temperature sensitivity. Journal of General Physiology. 1982;79(6):1115–1130. doi: 10.1085/jgp.79.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacDonald JA, Storey KB. Temperature and phosphate effects on allosteric phenomena of phosphofructokinase from a hibernating ground squirrel (Spermophilus lateralis) FEBS Journal. 2005;272(1):120–128. doi: 10.1111/j.1742-4658.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 58.Dash RK, Bassingthwaighte JB. Simultaneous blood-tissue exchange of oxygen, carbon dioxide, bicarbonate, and hydrogen ion. Annals of Biomedical Engineering. 2006;34(7):1129–1148. doi: 10.1007/s10439-005-9066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Betz T, Lenz M, Joanny J-F, Sykes C. ATP-dependent mechanics of red blood cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(36):15320–15325. doi: 10.1073/pnas.0904614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bassingthwaighte JB, Raymond GM, Ploger JD, Schwartz LM, Bukowski TR. GENTEX, a general multiscale model for in vivo tissue exchanges and intraorgan metabolism. Philosophical Transactions of the Royal Society A. 2006;364(1843):1423–1442. doi: 10.1098/rsta.2006.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arciero JC, Carlson BE, Secomb TW. Theoretical model of metabolic blood flow regulation: roles of ATP release by red blood cells and conducted responses. American Journal of Physiology. 2008;295(4):H1562–H1571. doi: 10.1152/ajpheart.00261.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vince JW, Reithmeier RAF. Carbonic anhydrase II binds to the carboxyl terminus of human band 3, the erythrocyte Cl−/HCO3− exchanger. Journal of Biological Chemistry. 1998;273(43):28430–28437. doi: 10.1074/jbc.273.43.28430. [DOI] [PubMed] [Google Scholar]
- 63.Weber RE, Voelter W, Fago A, Echner H, Campanella E, Low PS. Modulation of red cell glycolysis: interactions between vertebrate hemoglobins and cytoplasmic domains of band 3 red cell membrane proteins. American Journal of Physiology. 2004;287(2):R454–R464. doi: 10.1152/ajpregu.00060.2004. [DOI] [PubMed] [Google Scholar]