Abstract
Coupling genetically encoded target sequences with specific and selective labeling strategies has made it possible to utilize fluorescence spectroscopy in complex mixtures to investigate the structure, function, and dynamics of proteins. Thus, there is a growing need for a repertoire of such labeling approaches to deploy based on a given application and to utilize in combination with one another by orthogonal reactivity. We have developed a simple approach to synthesize a fluorescent probe that binds to a poly-histidine sequence. The amino group of cysteine was converted into nitrilotriacetate to create a metal-chelating cysteine molecule, Cys-nitrilotriacetate. Two Cys-nitrilotriacetate molecules were then cross-linked using dibromobimane to generate a fluorophore capable of binding a His-tag on a protein, NTA2-BM. NTA2-BM is a potential fluorophore for selective tagging of proteins in vivo.
Keywords: cysteine, dibromobimane, fluorescence, His-tag, metal chelating, protein tagging
Selective labeling of proteins at defined positions provides a powerful tool to study their structure–function behavior. Protein labeling approaches exploit the chemical reactivity of the side chains of the amino acids and most often use amino-based or thiol-based chemistry. Reactions with labeled proteins are typically monitored using fluorescence spectroscopy, immunodetection, or mass spectrometry. One of the shortcomings of these methods is that labeling is restricted to purified proteins and not appropriate in a mixture of other proteins or in a cellular environment. Recently, there has been intensive effort to develop new probes to site-specifically tag a protein of interest present in a complex biochemical environment. These include the use of fusion constructs with fluorescent proteins (1,2), co-translational introduction of unnatural amino acids or modified amino acids (3–5) or targeting small molecule labels to specific sequences (5–11). The large size of fusion proteins may introduce complexities and limit their use. Moreover, their complex spectral properties can complicate data analysis, for example, distance measurements in fluorescence resonance energy transfer (FRET)-based experiments (12). Incorporation of unnatural amino acids into a growing polypeptide chain using suppressor tRNA technology or 4-base codon methods is one of the most efficient methods for selective labeling (13–15); however, the approach still remains challenging in practice. Small molecule labeling to a target sequence present in the protein of interest offers a clean and potentially straightforward method that can exploit the high affinity and specificity of well-designed labels. The target sequence is generally incorporated into the gene for the protein of interest. Metal affinity (9,10,16), intein-based semi-synthesis (11), enzyme-mediated labeling such as using biotin ligase (17), and arsenical reagent-based labeling of tetra-cysteine motifs (8,18) are a few of the methods introduced for specific small molecule labeling of proteins.
Multivalent transition metal ions bind relatively tightly and specifically to a cluster of cysteine and/or histidine residues in a protein through co-ordinate bonds (19,20). Such clusters of amino acids occur rather rarely in proteins, thus providing high selectivity for the metal ion. Such interactions have the advantage that co-ordinate bonds are strong, and in most cases these interactions are completely reversible upon addition of a competing ligand. The simple chemistry of many multivalent chelators enables synthesis of chelator molecules with different functionalities. A few of the applications based on metal chelation include the use of ligands like nitrilotriacetic acid (NTA) or imidodiacetic acid (IDA) together with metal ions-like nickel and cobalt for protein purification (21, 22), surface immobilization (16,23,24), and in vitro and in vivo protein labeling and detection (9,25). NTA creates stable (Ka > 1011), 1:1, octahedral complexes with nickel, in which four of the co-ordination sites are saturated by oxygen and nitrogen atoms from the chelator molecule, and the remaining two by water, which can be easily substituted by the imidazole nitrogens from histidine side chains.
Previous reports using metal chelation-mediated protein labeling indicate that the affinity of the poly-histidine tag (His-tag) for the Ni2+ metal ion is dependent not only on the number of histidine residues in the tag, but also on the number of NTA moieties in the probe (9,16). NTA (or IDA) chelator 'heads' have usually been created using an amino group of glutamic acid (26) or one of the free amino groups of lysine (21,27,28), leaving the remaining amino acid functional groups for further modification.
A clever strategy for specific labeling of proteins carrying a widely used and versatile genetically encoded tag is to link fluorescent dyes to nickel chelators that bind poly-histidine tags. Several groups have recently described synthetic approaches to realize this strategy. Linking a single fluorophore to multiple NTA groups has been achieved by using fluorophores carrying multiple functionalities as in the case of (NTA)2-Cy3/(NTA)2-Cy5 (9), by coupling two or four 'NTA synthons' by a dendrimer approach (16), or by adding three NTA groups to the imino groups of a cyclic scaffold followed by addition of the fluorophore to a free amino group on these multiple NTA head groups (16,29). These reports have provided a number of routes to linking fluorophores to chelators for specific binding to His-tags. In all cases, the NTA groups were coupled via a flexible linker, and significant synthetic effort was required in most of these approaches.
We sought to design a simpler synthesis strategy to conjugate two NTA head groups to a sensitive fluorogenic reporter group without the need for a flexible linker. Here, we report a novel approach to synthesize a fluorophore for labeling proteins carrying poly-histidine tags guided by metal affinity coupled with thiol chemistry. The primary amino group of cysteine was used to generate an NTA, chelator head group, and two such modified cysteines were linked to the reporter molecule, dibromobimane, using thiol-based chemistry. Details of the straightforward chemistry used to generate the probe, along with the potential applications of this probe for intramolecular FRET studies are discussed.
Materials and Methods
Reagents and analysis
All solvents were of ACS or high-performance liquid chromatography (HPLC) grade and were used directly without any further purification. H-Cys-(Trt)-OH and dibromobimane were purchased from EMD Biosciences (La Jolla, CA, USA), and t-butyl bromoacetate and diisopropylethamine (DIPEA) from Sigma (St. Louis, MO, USA). Lysozyme was obtained from ICN Biomedicals (Aurora, OH, USA).
Purification of the products was carried out using column chromatography on silica gel (grade 60, 230–400 mesh; EMD Chemicals Inc., Gibbstown, NJ, USA) or by reverse phase (RP)-HPLC on a preparative C18 column (22 × 250 mm, 10 µm, 300 Å; Vydac, South-borough, MA, USA). The purity of the products was assessed by thin layer chromatography (TLC) on aluminum sheets precoated with silica gel 60 F-254 (EMD Chemicals Inc.) using ethyl acetate (AcOEt):hexane (1:4, v/v) as the mobile phase (visualized using either a UV-lamp (λobs = 254 nm) or ninhydrin exposure) and by RP-HPLC analysis on an analytical C18 column (4.6 × 150 mm, 5 µm, 300 Å; Vydac) using appropriate acetonitrile elution gradient. Mass spectra were obtained using Esquire-LC Ion trap mass spectrometer (Bruker Daltonics, Billerica, MA, USA). NMR spectra were recorded using AVANCE 600 MHz instrument (Bruker Biospin, Billerica, MA, USA).
Synthesis
Nα,Nα-bis(t-butoxycarbonylmethyl)-S-trityl-l-cysteine (1)
About 3.64 g (10 mmol) of H-Cys(Trt)-OH and 10.45 mL (60 mmol) of DIPEA were added to 50 mL N,N′-dimethylformamide (DMF) and stirred at room temperature for 5 min. t-Butyl bromoacetate (5.4 mL, 40 mmol) was next added in one portion, and stirring was continued for 30 min at room temperature. The resulting yellowish solution was heated up to 40 °C overnight with continuous stirring. One molar equivalent of t-butyl bromoacetate was then added, the temperature was increased to 52 °C, and the reaction was continued for 6 h. Volatile products were removed under reduced pressure, and the crude product was purified using column chromatography on silica gel (200 g). Pure monoalkylated and dialkylated products were eluted with a hexane:AcOEt (5:1, v/v) solvent mixture at a flow rate of 1.5 mL/min. The appropriate fractions were collected and evaporated yielding 2.34 g of dialkylated and 3.77 g of monoalkylated products as thick oils. TLC: monoalkylated: Rf = 0.32, dialkylated: Rf = 0.43. 1H-NMR (p.p.m.; CDCl3) for dialkylated product (1): 1.38 (s, 18H, t-BuO-), 2.47–2.50 (m, 1H, Cβ-H, Cys), 2.66–2.69 (m, 1H, Cβ-H, Cys), 3.38 (s, 4H, N-CH2-CO), 4.10–4.14 (m, 1H, Cα-H, Cys), 7.17–7.20 (m, 3H, Trt), 7.25–7.32 (m, 6H, Trt), 7.41–7.42 (m, 6H, Trt).
Nα,Nα-bis(carboxymethyl)-l-cysteine [Cys(NTA)2] (2)
The dialkylated product was dissolved in 10 mL CH2Cl2, 4.85 mL (32.7 mmol) of triisopropylsilane, 0.662 mL (7.9 mmol) of ethane dithiol (EDT) and 25 mL trifluoroacetic acid (TFA) were added and the reaction mixture was stirred at room temperature for 8 h. About 5 mL of TFA was added after 6 and 7 h of reaction progress. Solvents were next evaporated to dryness, and the residue was dissolved in 100 mL 5% acetic acid and extracted 3 × 50 mL CHCl3. The aqueous phase was evaporated to dryness, and the residue was redissolved in 25 mL water and lyophilized. After lyophilization, 750 mg of a yellowish, sticky solid was obtained. It was dissolved in 40 mL water and additionally washed with diethyl ether (3 × 20 mL). Aqueous phase was again evaporated to dryness, redissolved in 5 mL water and lyophilized. RP-HPLC analysis revealed presence of two main products. About 100 mg of crude mixture was next purified on preparative RP-HPLC chromatography on C18 column. The second peak was found to correspond to the desired product. Yield: 42%. 1H-NMR (p.p.m.; D2O): 2.90–2.94 (m, 1H, Cβ-H, Cys), 3.00–3.04 (m, 1H, Cβ-H, Cys), 3.79 (s, 4H, N-CH2-CO), 3.90 (t, J = 7.2 Hz, 1H, Cα-H, Cys).
9,10-Dioxa-syn-{bis-[Nα,Nα-bis(carboxymethyl)-l-cysteinyl]}-bimane BM-[Cys (NTA)]2 (3)
About 0.1 mmol (23.8 mg) of Cys(NTA)2 was dissolved in 5 mL of acetonitrile in the presence of 87.1 µL (0.5 mmol) DIPEA, and 3.5 mL of 10 mm dibromobimane solution in acetonitrile was added dropwise (over 20 min) to the stirred reaction mixture protected against light. The reaction was allowed to proceed at room temperature in the dark for 3 h. Reaction progress was monitored by analytical RP-HPLC. After completion of the reaction (as indicated by consumption of both substrates), the reaction mixture was evaporated to dryness, redissolved in 5 mL 10% CH3CN/H2O mixture and lyophilized. The crude reaction mixture (yellow lyophilizate) was next purified by means of RP-HPLC on a preparative C18 column, using the following gradient: 10–20% B/10 min, 20–35% B/60 min, 35–95% B/20 min; flow 8 mL/min. The appropriate fractions were collected, concentrated in vacuo and lyophilized, yielding 7 mg of bimane derivative (BM)-[Cys(NTA)]2. The purity of final product, as assessed by RP-HPLC, was above 94%. 1H-NMR (D2O): 1.89 (s, 6H, CH3, BM), 3.02–3.05 (m, 2H, Cβ-H, Cys), 3.11–3.14 (m, 2H, Cβ-H, Cys), 3.72 (s, 8H, N-CH2-CO), 3.82 (t, J = 7.2 Hz, 2H, Cα-H, Cys), 4.07 (s, 4H, S-CH2-BM).
To further purify the product, gel filtration chromatography on a Sephadex G-25 column (2.54 × 10 cm) was performed. About 5.36 mg of product was loaded on the column. Fractions were eluted using water at flow rate of 0.5 mL/min. Fractions containing the desired products were identified using UV–VIS and fluorescence measurements, combined and lyophilized yielding 2.44 mg of pure BM-Cys(NTA)2.
NTA2-BM
Nickel complexes of BM-[Cys(NTA)]2, NTA2-BM, were prepared directly before labeling reactions by addition of a 10-fold molar excess of NiSO4 to the aqueous solution of dye and incubating the reaction at room temperature for 30 min.
Protein expression and purification
Cellular retinoic acid-binding protein I with a stabilizing mutation R131Q (CRABP I wt*) (30), cloned in pET16b for the N-terminal (His)10 extension, His10-CRABP I wt*, and in pET29b(+) for the C-terminal (His)6 extension, CRABP I wt*-His6, were expressed in BL21(DE3) cells. The split tetra-Cys CRABP I mutant, His10-CRABP I St1-2*, was generated by introducing the mutations T6C, K8C, H40C, and E42C using the QuikChange protocol (Stratagene, La Jolla, CA, USA) using His10-CRABP I wt* as a template plasmid. All the CRABP I proteins were purified from the soluble fraction of the cell extract on a Ni-NTA column (Qiagen, Valencia, CA, USA) as described (31).
In vitro labeling of His-tagged proteins
Purified proteins carrying varying lengths of His-tag, cellular retinoic acid-binding protein I with a N-terminal (His)10 extension (His10-CRABP 1 wt*) and with a C-terminal (His)6 extension (CRABP I wt*-His6), T7 RNA polymerase with a N-terminal (His)6 tag ((His)6-T7 RNA pol) (a gift from E. Esposito and C. Martin) and a protein component from prokaryotic signal recognition particle with a C-terminal (His)6 tag (Ffh-(His)6) (32). Lysozyme was used as a control protein to check specificity of the probe for His-tag. The labeling reaction was carried out with a 10-fold molar excess of NTA2-BM in 20 mm ammonium bicarbonate buffer, pH 7.4, for 30 min at room temperature. In the case of (His)6-T7 RNA Pol and Ffh-(His)6, 10% glycerol and 300 mm sodium chloride were required in the buffer for the stability of the proteins. Excess of the unreacted probe was removed by desalting the reaction mixture on a PD10 desalting column (Amersham Biosciences, Piscataway, NJ, USA). Fractions obtained from the desalting step (referred to as fractions henceforth) were analyzed by measuring absorbance at 280 nm for the protein content and at 385 nm for the presence of the dye. Fluorescence emission spectra were measured on a PTI QM1 spectrofluorometer (Photon Technologies International, Lawrenceville, NJ, USA) with the excitation set at 280 or 385 nm. Steady-state fluorescence anisotropy measurements were also made on the labeled protein fractions as described below.
Fluorescence anisotropy experiments
The affinity of the NTA2-BM probe for the His-tag (His10-CRABP I wt* and CRABP I wt*-His6) was determined using steady-state fluorescence anisotropy measurements. The reaction mixture, 150 µL containing 50 nm of NTA2-BM in 20 mm ammonium bicarbonate buffer, was titrated with varying concentration (0–12 µm)of His10-CRABP I and CRABP I-His6. Higher protein concentration (>15 µm) could not be used due to precipitation. Fluorescence anisotropy was measured after 30 min of incubation on a PTI QM1 spectrofluorometer. Excitation and emission wavelengths were set at 385 nm and 475 nm, respectively. Band widths used were 10 nm each. Anisotropy (R) was calculated using:
where Ivv and IVH are the fluorescence intensities with the excitation polarizer at a vertical position and the emission polarizer at vertical and horizontal positions, respectively, and G was calculated as the ratio of IHV to IHH (33).
The anisotropy of the ligand in bound and free forms was used to calculate per cent of ligand bound (% bound) using:
where R is the observed anisotropy, and Rb and Rf are the anisotropy of the ligand in bound and free form, respectively.
Data were then fit to obtain the dissociation constant using:
where Pt and Lt are the starting total concentration of the protein and the ligand (NTA2-BM), respectively, and Kd is the dissociation constant.
Labeling of bacterially expressed His-tagged protein in cell lysate
Escherichia coli BL21(DE3) cells expressing His-tagged proteins, His10-CRABP I wt* and CRABP I wt*-His6, were lysed using freeze-thaw and lysozyme followed by sonication (31). The lysed cells were pelleted to remove the insoluble fraction. Protein present in the soluble fraction (estimated to be 30–50 µm) was labeled with 100 µm NTA2-BM for 30 min at room temperature. The labeled protein mixture was desalted on PD10 column to remove the unreacted fluorophore, which showed fluorescence properties the same as the initial freshly prepared reagent. Fluorescence emission spectra of the various fractions were measured for the presence of BM using an excitation wavelength of 385 nm. BL21(DE3) cells were used as a negative control.
Double labeling of CRABP I St1-2*
His10-CRABP I St1-2* (23 µm), in 50 mm ammonium bicarbonate, pH 7.4, was labeled with FlAsH-EDT2 (25 µm) at room temperature for about 4 h. The reaction mixture was divided into two, and one of the aliquots was set up for labeling with NTA2-BM (100 µm). The NTA2-BM labeling reaction was also carried out at room temperature for 45 min. The single- and double-labeled proteins were desalted on a PD10 column to remove the excess of the fluorophores. The fractions were monitored for the presence of the fluorophores by measuring fluorescence emission spectra (λex 385 nm for BM, and 508 nm for FlAsH). The doubly-labeled protein fraction was further allowed to bind the 50 mm ammonium bicarbonate, pH 7.4, equilibrated Ni-NTA resin for 10 min to remove the singly FlAsH-labeled protein. The unbound fraction thus obtained was enriched in doubly-labeled protein. This protein was used further used for limited trypsinolysis experiments. The His-tag present in the protein contains a trypsin cleavage site and in a partial trypsinolysis, this results in the removal of the tag. Removal of the His-tag was also verified by analyzing the undigested and the digested proteins on an SDS-PAGE.
Monitoring internalization of NTA2-BM in mammalian cells
HeLa cells cultured in minimum essential medium (MEM) containing 10% fetal calf serum, 100 U/mL of penicillin and 100 µg/mL of streptomyocin were used to test the cell permeability of NTA2-BM. Cells were washed twice with 50 mm ammonium bicarbonate buffer supplemented with 100 mm sodium chloride. Cells were treated with about 100 µm of NTA2-BM at room temperature for 20 min. The cells were spun at 3000 rpm, further washed with buffer, and used in fluorescence measurements. Untreated cells were prepared similarly and used as a control. Fluorescence emission spectra probing for the presence of BM were carried out as described above.
Results and Discussion
Oligohistidine sequences, typically of six or 10 residues, are one of the most commonly used affinity tags to aid protein purification. A His-tag is typically placed at the N- or C-termini of the target protein and is non-interfering in most cases. Hence, these tags are attractive target sequences for selective labeling of proteins, including potential applications in vivo. By analogy to the use of chelated metal affinity chromatography for the purification of His-tagged proteins, we have chosen to use metal chelation to incorporate a fluorophore to a protein of interest.
Previous efforts to use metal chelation to generate fluorophores that bind His-tags (9,10), had the reporter group attached to the ε-amino group of lysine, which creates a longer spacer arm between the fluorophore and the attachment site. Using cysteine instead of lysine places the fluorophore closer to the NTA head. In order to increase affinity we attached two NTA head groups to a single fluorophore, thus narrowing our choice to thiol-based bifunctional cross-linking fluorophores.
Kosower et al. introduced 9,10-dioxobimane in 1979 (34) as a highly fluorescent, low molecular weight, neutral reagent. Its mono- and syn-dibromo derivatives show high reactivity toward thiol groups (35) and are less reactive to other nucleophilic reagents (36). These molecules are not fluorescent by themselves, but upon substitution form highly fluorescent derivatives, making them very attractive reagents for both labeling (37,38) and cross-linking (39–41). In addition, spectral properties of BM make it ideally suited for intramolecular FRET with fluorescein and its close analogues. As BM derivatives are also membrane permeable (34), they can be used in double fluorescent labeling of proteins in vivo, for example, as potential donors or acceptors for FRET studies. Thus, dibromobimane was used to cross-link two Cys-NTA groups to yield the fluorescent His-tag labeling fluorophore, NTA2-BM.
Synthesis and stability of NTA2-BM
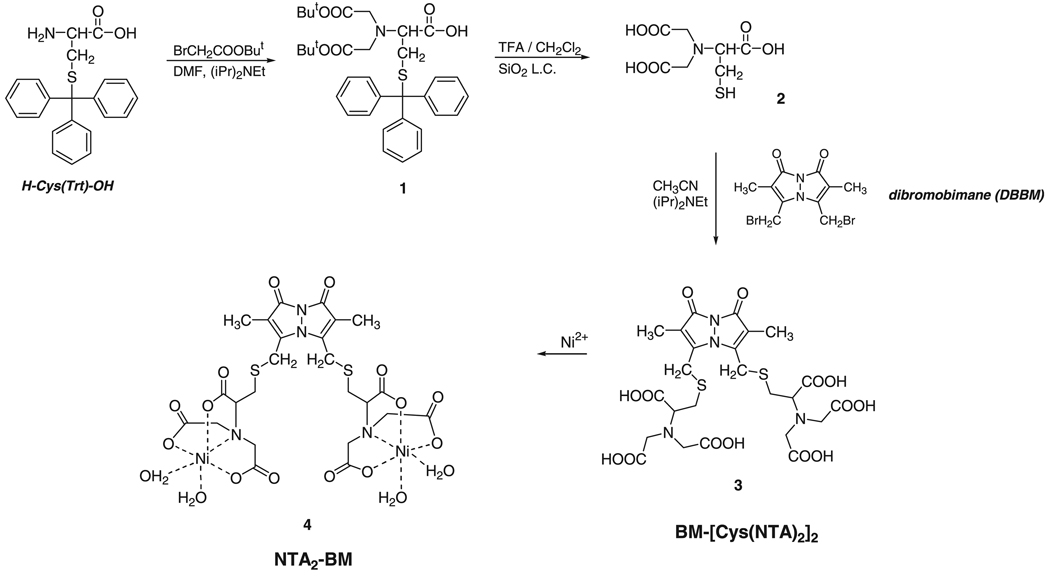
We originally intended to generate the NTA group by exhaustive N-alkylation of the amino groups of cystine followed by the disulphide bond reduction coupled with the fluorophore addition. However, this design was unsuccessful due to the difficulty of disulfide reduction even with the use of a combination of reductants. Additionally, we observed incomplete alkylation of both amino groups of cystine, presumably because of steric hindrance and/or electrostatic repulsion between the carboxylate groups. We therefore decided to use a protected derivative of cysteine and bromoacetic acid. Straightforward bis-alkylation of the α-amino group of S-trityl-protected cysteine generated the NTA head group of the probe. The use of t-butyl bromoacetate for the alkylation reaction allowed removal of the protecting groups in a single step (Scheme 1). Deprotected Cys-NTA showed some tendency to decompose with time, even when stored at low temperature as an oil obtained after lyophilization, and hence, was used within a few days of synthesis to generate the final fluorophore. Application of anhydrous conditions limited the scope of side reactions (e.g. hydrolysis of dibromobimane), resulting in a reasonable yield and purity of the final product. RP-HPLC analysis of the purified compound along with 1H-NMR analysis, as described in Materials and Methods, confirmed the synthesis of the desired product at each stage. The analysis of the final product indicated the presence of some contaminants that could not be easily removed by RP-HPLC. In our initial protein labeling experiments with this product, separation of the excess fluorophore from the labeled protein using a Sephadex G25 matrix indicated separation of two different types of small molecules with one of the species eluting only after a column volume. Further application of the free fluorophore on the Sephadex G25 column resulted in a clear separation of the contaminant molecule from the desired fluorophore (Figure 1A). We assume that non-specific interaction between the matrix and the impurity is the basis of the separation process. Therefore, the final purification was carried out on a Sephadex G25 column. The purity of the final product was >95% and was also confirmed based on the differences in spectroscopic properties of the compounds, higher absorbance at 385 nm and lower fluorescence of the desired product relative to the impurity, and the ability of only the desired product to bind nickel (Figure 1). Formation of the active fluorophore by Ni2+ co-ordination was monitored by quenching of BM fluorescence. Addition of Ni2+ ions causes the BM fluorescence to decrease by nearly 60% [cf. a reported 75% decrease in the fluorescence intensity of Cy5-NTA conjugates (9)]. Steady-state anisotropy measurements of the probe in the absence and presence of Ni2+ yielded anisotropy values of 0.006 and 0.065, respectively. An increase in the anisotropy of the probe upon Ni2+ co-ordination indicates formation of a more rigid complex due to cross-linking of two NTA groups. Mass spectrometry of the final product also confirmed its purity and the presence of a single metal ion per NTA group.
Scheme 1.
Synthesis of NTA2-BM. Trityl-protected cysteine was alkylated to generate Cys-NTA (1) and two of the deprotected molecules (2) were cross-linked by reaction of dibromobimane with free sulfhydryl groups to yield the final fluorophore, NTA2-BM (3).
Figure 1.
Purification of NTA2-BM on a PD10 column. (A) Absorbance profile at 385 nm for the various fractions collected from the PD10 column during final step of purification of NTA2-BM. The peaks corresponding to (I) and (II) represent the desired product and the contaminant, respectively. (B) Fluorescence of the corresponding fractions, as shown in (A), on a UV-transilluminator.
NTA2-BM is stable when stored at low temperature (4 °C) in the solid state for at least a few months. In contrast, HPLC analysis of the compound that was stored in aqueous solution at room temperature for 2 weeks revealed significant (>50%) decomposition, indicating that the half-life is <2 weeks under these conditions. Decomposition of the dye was further stimulated in the presence of nickel ions. Nonetheless, HPLC analysis of the Ni2+–NTA2-BM complex indicated that it was stable for a couple of hours, and our typical incubation time required for labeling was less than an hour. We suggest that the active fluorophore should be prepared fresh by the addition of NiSO4 to an aqueous solution of NTA2-BM. In contrast to many highly fluorescent probes, NTA2-BM is water-soluble, a desirable feature for labeling biomolecules. A less bulky ester such as the methyl group in comparison with the t-butyl ester used in our approach might improve the yield of dialkylated product; however, the additional deprotection step would result in similar final yields.
NTA2-BM binds site-specifically to His-tagged proteins
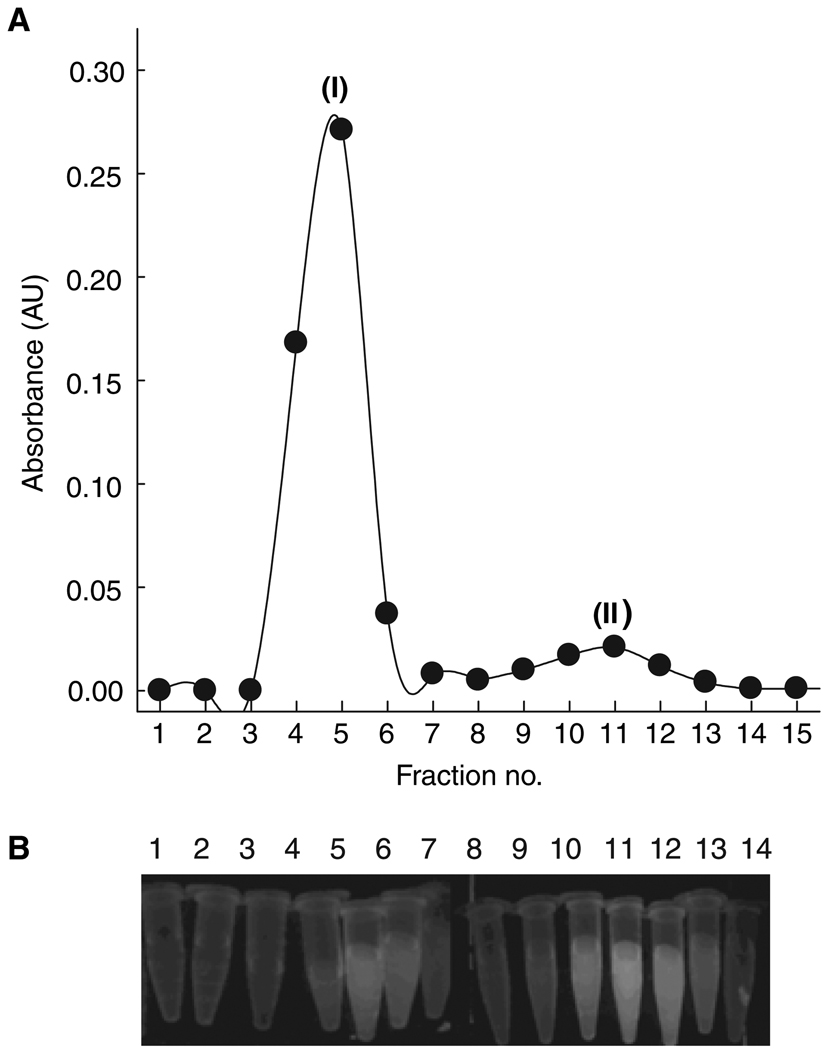
Five different proteins carrying varying lengths of His-tag either at their N- or C-termini (listed in Materials and Methods), along with non-His-tagged lysozyme as a control, were used to establish specific labeling of the protein at oligohistidine sequences. Nickel ion concentration was restricted to a fivefold molar excess over the NTA concentration, as excess metal ion resulted in precipitation of some of the test proteins. Moreover, in order to avoid precipitation of the Ni2+ ion itself, we used a carbonate-containing buffer, as NiCO3 is the most soluble of the Ni2+ salts. Hence, all of our labeling experiments were carried out in ammonium bicarbonate buffer including salt and glycerol if required. The first level of labeling specificity was evident upon inspection of fluorescence of fractions obtained from desalting the labeling reaction mixture under a UV-transilluminator and more quantitatively from the absorbance profile (Figure 2). Bimane absorbance (at 385 nm) was observed along with protein absorbance (at 280 nm) in all the early fractions eluted in the void volume of the desalting column, indicating presence of BM on the protein. In the case of lysozyme, fractions containing protein did not have BM absorbance, indicative of no labeling of the non-His-tagged protein.
Figure 2.
Specificity of NTA2-BM labeling of a His-tag. Absorbance profile of desalted fractions of NTA2-BM-labeled lysozyme (circle), NTA2-BM-labeled His10-CRABP I wt* (triangle), and free NTA2-BM (square). Filled and open circles represent the protein absorbance at 280 nm and bimane absorbance at 385 nm, respectively.
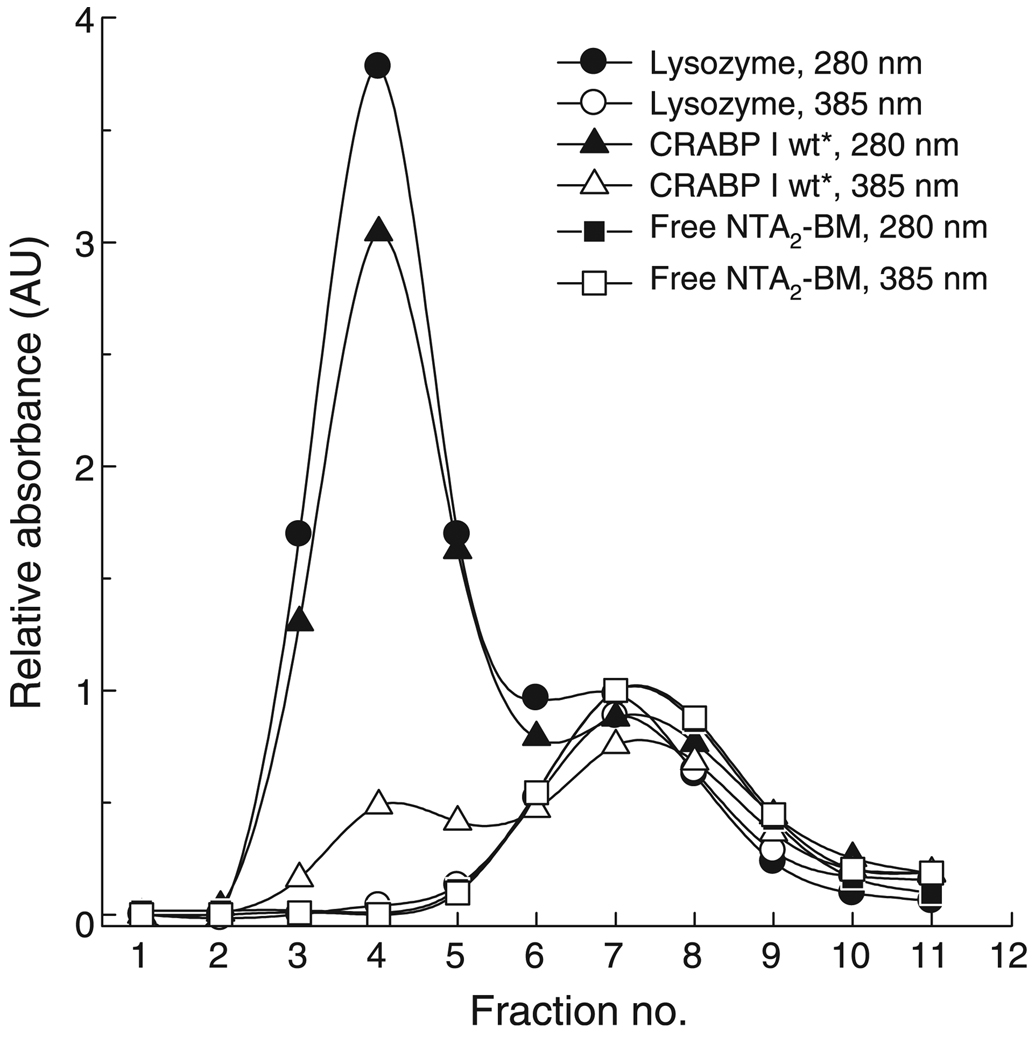
Labeled protein fractions were further subjected to fluorescence measurements to assess NTA2-BM binding. For each of our test proteins, binding of the probe to the His-tag resulted in a 3–5 nm blue shift in the BM fluorescence (to a maximum emission wavelength of 470 nm) compared with the free probe, and this shift could be used as an indication of binding (Figure 3). An increase in the fluorescence anisotropy (R) of BM in the fractions containing proteins from R = 0.065 for the free probe to R = 0.34 for the bound probe accompanied the attachment of the fluorophore to the protein. In order to confirm that the probe attachment on the protein was metal ion-mediated, the labeling reaction was carried out in the absence of the Ni2+ ion, i.e. with the metal ion-free probe. Absorbance and fluorescence analysis showed no labeling, indicating the absence of any non-specific interaction of the probe with the protein (data not shown). NTA2-BM also formed a similar complex with Co2+ ion and bound to the His-tagged protein as a Co2+ complex. Addition of ethylenediaminetetraacetic acid easily reversed the binding of NTA2-BM to the protein as reported by the decrease in the anisotropy of the probe (data not shown). Mass spectral analysis of the NTA2-BM-labeled protein using either ESI-MS or MALDI was unsuccessful due to dissociation of the dye from the protein.
Figure 3.
Fluorescence spectra of NTA2-BM-labeled His-tagged proteins in vitro (A) and in cell lysate (B). (A) Labeling proteins with varying lengths of His-tag: (His)10-CRABP I wt* (dashed-dotted line), (His)6-T7 RNA pol (dashed line), and (His)9-CRABP I mutant (dotted line), with NTA2-BM resulted in a blue shift of nearly 5 nm in the bimane fluorescence compared with that of free NTA2-BM (solid line). (B) The soluble fraction of cells expressing His10-CRABP I wt* and CRABP I wt*-His6 was used for labeling with NTA2-BM as described in Materials and Methods section. BL21(DE3) cells were treated in a similar fashion as a control to assess background labeling. Labeling of the protein was also accompanied by a blue shift of nearly 5 nm compared with fluorescence of free fluorophore in cell lysate.
Specificity of NTA2-BM binding to a His-tag was also demonstrated by labeling His-tagged proteins expressed in the soluble fraction of cell lysate. The blue shift observed upon labeling protein in cell lysate was comparable with that seen in the in vitro labeling reactions (Figure 3B).
The affinity of NTA2-BM for a His-tag
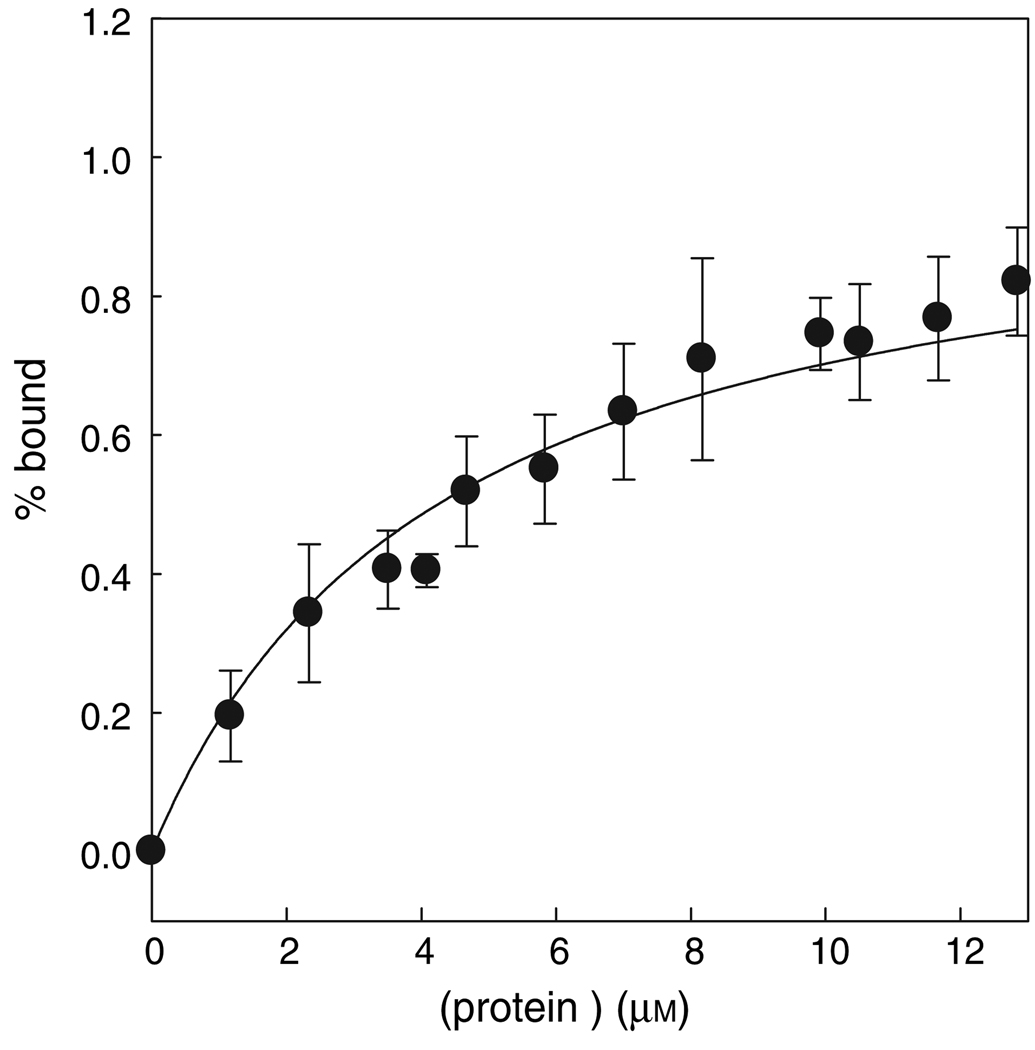
The significant difference in the anisotropy of the free versus the bound probe was used to determine the affinity of NTA2-BM for (His)6 and (His)10 tags using His10-CRABP I wt* and CRABP I wt*-His6 as test proteins. The affinity of the probe was observed to be in the micromolar range with a Kd of 5 µm for the deca-histidine tag (Figure 4) and a Kd of nearly 40 µm for the hexa-histidine tag. Good agreement between the data and the fit also supports the assumption of 1:1 stoichiometry of the probe to the protein used in the data analysis. In the case of hexa-histidine-tagged protein, the measurements could not be made with higher protein concentration because of precipitation, and hence the observed Kd value represents an approximate affinity.
Figure 4.
Determination of the dissociation constant (Kd) using fluorescence anisotropy measurements. Affinity of the probe for a His10-tag was estimated using CRABP I wt* protein. The error bar represents the standard error between two independent measurements at each concentration of the protein. Data were fit as described in Materials and Methods section, yielding a Kd of 5.0 µm.
Although the apparent affinity of NTA2-BM is in the micromolar range, probe binding to the His-tag is quite kinetically stable. The protein labeled with NTA2-BM can be stored at 4 °C for at least a week as indicated by the lack of change in its fluorescence spectrum. We also assessed the extent of dissociation of the fluorophore over time using a Ni-NTA agarose matrix in a competition-based assay. NTA2-BM-labeled His10-CRABP I cannot bind the Ni-NTA group on the agarose affinity matrix and elutes in the unbound fraction. We checked the BM fluorescence of the eluate after three rounds of binding the same labeled protein to the Ni-NTA resin and found little change, indicating minimal dye dissociation. In a control experiment, a similar concentration of unlabeled protein applied to the Ni-NTA resin was >90% retained on the resin. In the case of the unlabeled protein, tryptophan fluorescence was used as a measure to calculate the binding efficiency. Thus, fluorophore binding cannot be competed by the Ni-NTA matrix, arguing for high kinetic stability of the fluorophore attachment to the protein (data not shown).
The Ni2+–NTA co-ordination complex has four of the six ligand-binding sites of the Ni2+ ion occupied by the NTA molecule with the remaining two sites filled by two water molecules. The affinity of Ni2+ for the NTA molecule is lowered by a few orders of magnitude when the water molecules are replaced with the nitrogens of the histidine side chains of the His-tag. The affinity of a (His)6-tagged protein for Ni-NTA matrix is reported to be 10−13 m (22). However, all reports, including our study, indicate that the metal chelation-based fluorophores have micromolar affinity for a His-tag. This difference in the affinity is likely due to the 'Velchro effect', i.e. a high local effective concentration of the Ni-NTA head groups on the NTA-agarose beads. Additionally, the orientation of the histidine side chains on a His-tag on a protein may not be optimal. As the His-tag is a part of a large and charged macromolecule, steric effects could also play a role in reducing the ability of the probe to approach the histidine residues optimally. Therefore, the affinity of the NTA-based fluorophore might be improved by optimizing the geometry of the histidine residues, for instance by using a His-tag with alternating histidine residues (42) or increasing the length of the His-tag. Nevertheless, the selectivity in the labeling, even in a complex mixture such as cell lysate, along with the reversibility of binding, make it a promising candidate for both in vitro and in vivo studies.
NTA2-BM as a FRET donor to fluorescein-based acceptors
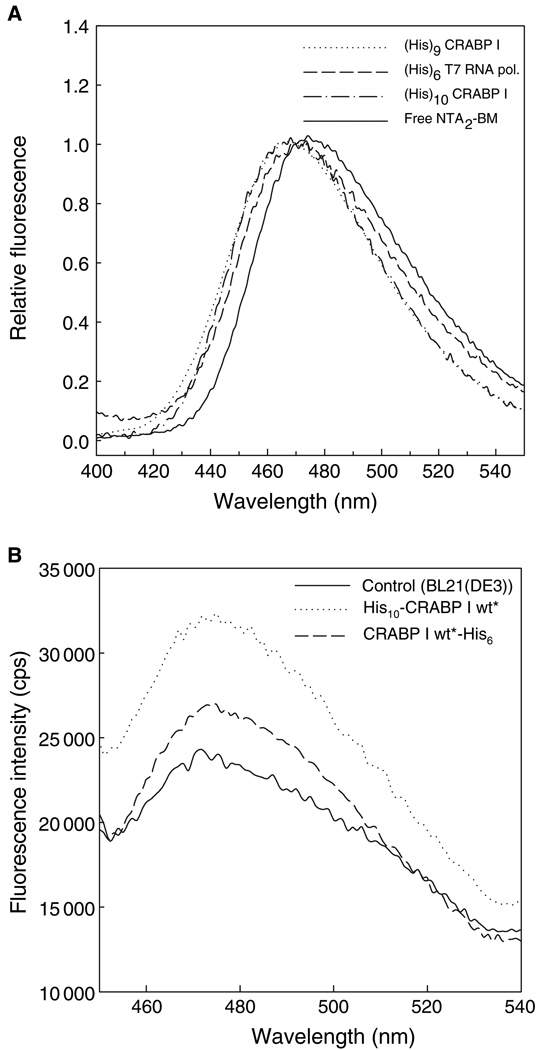
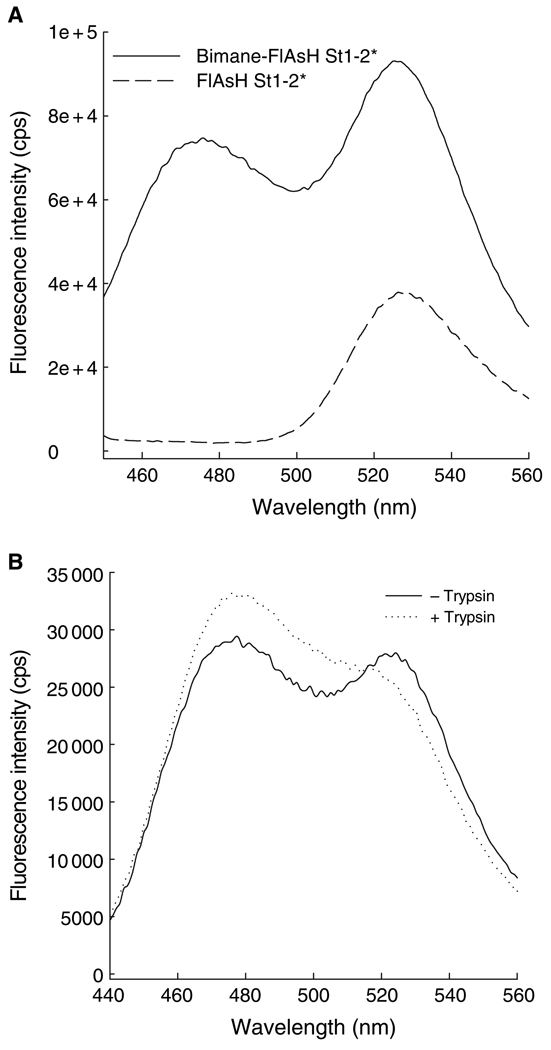
In vitro double-labeling experiments were carried out using NTA2-BM as a donor and fluorescein or FlAsH-EDT2 as an acceptor fluorophore in order to test the usefulness of NTA2-BM in FRET measurements. His-tagged CRABP I mutants carrying a tetra-Cys motif (Cys-Cys-X-X-Cys-Cys) either at the C-terminus or internally in one of the loops (omega-loop) could not be efficiently labeled with FlAsH-EDT2 if they were first labeled with NTA2-BM. Interchanging the order of modification and carrying out labeling of the protein first with FlAsH-EDT2 and later with NTA2-BM resulted in a decrease in the FlAsH fluorescence, indicating that the NTA2-BM partially displaced the bound FlAsH as a result of its higher affinity for the tetra-Cys motif. However, we have designed a CRABP I mutant carrying two cysteines on strand 1 (T6C and K8C) and two cysteines on strand 2 (H40C and E42C) such that in the native protein the four cysteine residues are in close proximity to form a 'split' FlAsH-binding tetra-Cys motif (unpublished data), referred to as His10-CRABP I St1-2*. Presumably because of the lower affinity of the split tetra-Cys motif for the NTA2-BM label, this protein could be doubly labeled with both fluorophores. In this case, energy transfer from BM to FlAsH was indicated by comparing the FlAsH fluorescence of the protein with only FlAsH labeling with that of the doubly-labeled protein upon excitation at 385 nm, the λmax for BM excitation (Figure 5). Both proteins exhibit a similar FlAsH fluorescence when FlAsH is directly excited at 508 nm. Energy transfer was further confirmed by a restoration of the BM fluorescence upon cleavage of the protein by partial trypsinolysis (Figure 5). Compatibility in double-labeling protein with NTA2-BM and fluorescein was also verified by labeling a single cysteine mutant of another test protein (the E. coli protein Ffh, harboring a His-tag and a Ser366 to Cys mutation), with NTA2-BM followed by labeling with fluoroscein iodoacetamide (data not shown). It should also be noted that the use of any metal chelation-based fluorophores is restricted to proteins that are not affected by the presence of metal ions as, for example, proteins rich in cysteines tend to precipitate in the presence of metal ion. Presence of excess of thiol-based reducing agents can also lead to precipitation and hence should be avoided.
Figure 5.
FRET between NTA2-BM and FlAsH. (A) Fluorescence emission spectra of His10-CRABP I St1-2* either FlAsH-labeled (dashed line) or doubly labeled (with both FlAsH-EDT2 and NTA2-BM; solid line). An increase in the FlAsH fluorescence in the doubly-labeled protein arises from energy transfer from donor, bimane, to acceptor, FlAsH. (B) Subjecting the doubly-labeled protein to trypsinolysis resulted in an increase in the bimane (donor) fluorescence of the trypsin-treated protein compared with that of the untreated protein due to the loss of the acceptor.
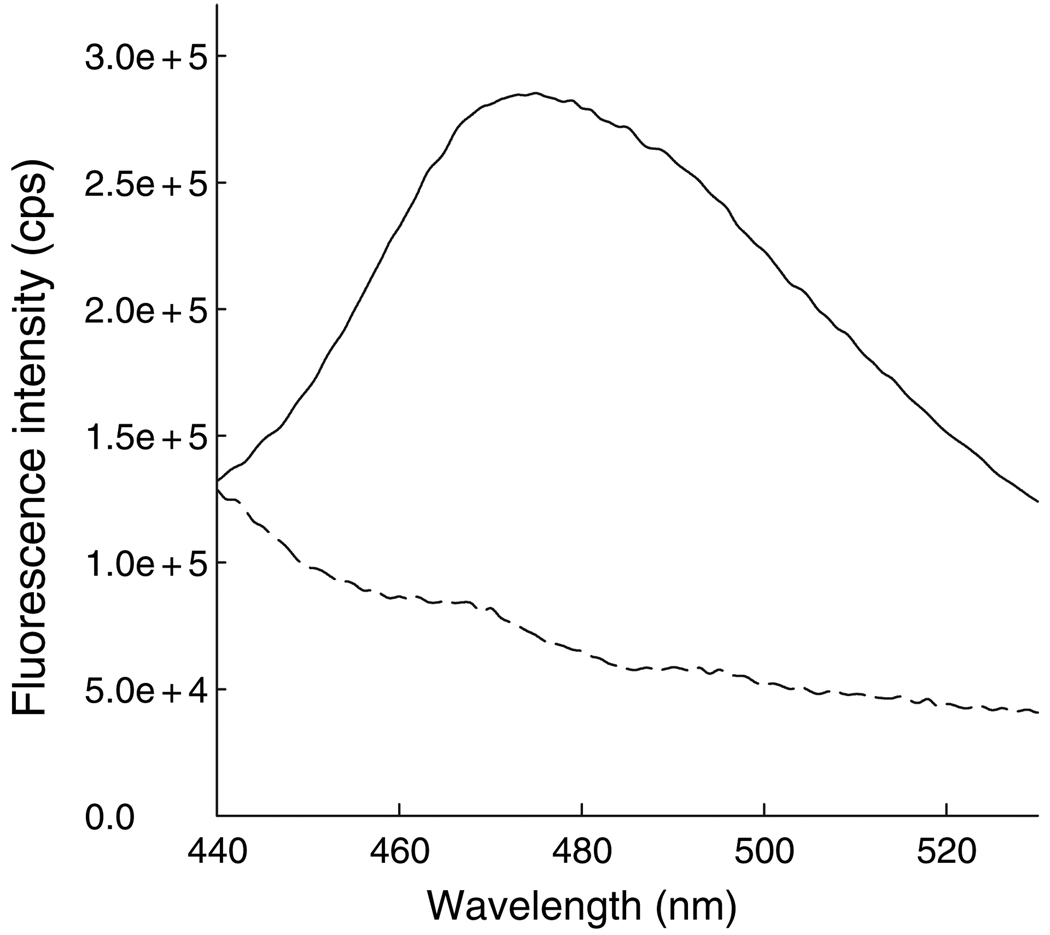
The NTA head (10) and BM are both membrane permeable, and therefore it can be expected that NTA2-BM will most likely be internalized into mammalian cells. Indeed, fluorescence measurements of HeLa cells treated with NTA2-BM clearly indicated presence of the fluorophore inside the cells (Figure 6).
Figure 6.
Fluorescence emission spectra of untreated (dashed line) and NTA2-BM-treated (solid line) HeLa cells. Bimane fluorescence indicates that NTA2-BM is permeable to the mammalian cell membrane.
Conclusions and Future Directions
Using the α-amino group of cysteine as a template for synthesis of an NTA 'head' and side chain thiol groups for reaction with dibromobimane yielded a new, smaller, neutral, and economical fluorescent reagent, NTA2-BM, with affinities toward His-tags comparable with similar fluorophores built from lysine residues previously reported in the literature (9,16,29). The dye enters rapidly (<20 min) into mammalian cells. Thus, NTA2-BM can be used for in vivo labeling of proteins expressed with a His-tag. We expect that extracellularly disposed His-tags on surface proteins would be labeled very rapidly upon treatment of cells with dye, and that this dye, like others reported previously (10,29), could be used for labeling of surface receptors, although rapid dye removal would be necessary to minimize background fluorescence from intracellular dye. Our in vitro double-labeling experiments with FlAsH-EDT2 suggest that NTA2-BM can complement FlAsH labeling in vivo as well as be used for intramolecular FRET experiments. However, it should be noted that use of EDT in the in vivo FlAsH-labeling experiments to reduce background fluorescence is not desirable in combination with any Ni2+ chelation-based labeling as EDT can form a co-ordination complex with Ni2+ ions. This limitation can most likely be overcome by using excess nickel ions. Furthermore, one could extrapolate the strategy described here to incorporate various other fluorophores with higher quantum yields such as Cy5-bis-C5-maleimide (43), bis-[(N-iodoacetyl) piperazinyl]sulfonerhodamine, or BODIPY® (Invitrogen Corp., Carlsbad, CA, USA) FL bis-(methyleneiodoacetamide). Another application of the new reagent could be to develop more economical His-tag staining reagents for gel chromatography. The NTA2 dye offers higher affinity than comparable fluorophores linked via a single NTA head (16,27).
Acknowledgments
We thank Joanna Swain and Ken Rotondi for help with the NMR measurements, Eddie Esposito and Craig Martin for providing His-tagged T7 RNA polymerase, Jim Cormier and Dan Hebert for providing HeLa cells, and Steve Eyles for help with mass spectrometry. We appreciate critical reading of the manuscript by Joanna Swain, Zoya Ignatova, and Eugenia Clerico. This work was supported by the National Institute of Health grants GM027616 and GM034962.
References
- 1.March JC, Rao G, Bentley WE. Biotechnological applications of green fluorescent protein. Appl Microbiol Biotechnol. 2003;62:303–315. doi: 10.1007/s00253-003-1339-y. [DOI] [PubMed] [Google Scholar]
- 2.Davis TN. Protein localization in proteomics. Curr Opin Chem Biol. 2004;8:49–53. doi: 10.1016/j.cbpa.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Xie J, Schultz PG. Adding amino acids to the genetic repertoire. Curr Opin Chem Biol. 2005;9:548–554. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Stromgaard A, Jensen AA, Stromgaard K. Site-specific incorporation of unnatural amino acids into proteins. Chembiochem. 2004;5:909–916. doi: 10.1002/cbic.200400060. [DOI] [PubMed] [Google Scholar]
- 5.Yeo DS, Srinivasan R, Uttamchandani M, Chen GY, Zhu Q, Yao SQ. Cell-permeable small molecule probes for site-specific labeling of proteins. Chem Commun (Camb) 2003;23:2870–2871. doi: 10.1039/b309196a. [DOI] [PubMed] [Google Scholar]
- 6.Miller LW, Sable J, Goelet P, Sheetz MP, Cornish VW. Methotrexate conjugates: a molecular in vivo protein tag. Angew Chem Int Ed Engl. 2004;43:1672–1675. doi: 10.1002/anie.200352852. [DOI] [PubMed] [Google Scholar]
- 7.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 8.Griffin BA, Adams SR, Jones J, Tsien RY. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- 9.Kapanidis AN, Ebright YW, Ebright RH. Site-specific incorporation of fluorescent probes into protein: hexahistidinetag-mediated fluorescent labeling with (Ni(2+):nitrilotriacetic acid (n)-fluorochrome conjugates. J Am Chem Soc. 2001;123:12123–12125. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- 10.Guignet EG, Hovius R, Vogel H. Reversible site-selective labeling of membrane proteins in live cells. Nat Biotechnol. 2004;22:440–444. doi: 10.1038/nbt954. [DOI] [PubMed] [Google Scholar]
- 11.Tan LP, Yao SQ. Intein-mediated, in vitro and in vivo protein modifications with small molecules. Protein Pept Lett. 2005;12:769–775. doi: 10.2174/0929866054864193. [DOI] [PubMed] [Google Scholar]
- 12.Zeng W, Seward HE, Malnasi-Csizmadia A, Wakelin S, Woolley RJ, Cheema GS, Basran J, Patel TR, Rowe AJ, Bagshaw CR. Resonance energy transfer between green fluorescent protein variants: complexities revealed with myosin fusion proteins. Biochemistry. 2006;45:10482–10491. doi: 10.1021/bi060943u. [DOI] [PubMed] [Google Scholar]
- 13.Taki M, Matsushita J, Sisido M. Expanding the genetic code in a mammalian cell line by the introduction of four-base codon/anticodon pairs. Chembiochem. 2006;7:425–428. doi: 10.1002/cbic.200500360. [DOI] [PubMed] [Google Scholar]
- 14.Olejnik J, Gite S, Mamaev S, Rothschild KJ. N-terminal labeling of proteins using initiator tRNA. Methods. 2005;36:252–260. doi: 10.1016/j.ymeth.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 16.Lata S, Reichel A, Brock R, Tampe R, Piehler J. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J Am Chem Soc. 2005;127:10205–10215. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 17.Tirat A, Freuler F, Stettler T, Mayr LM, Leder L. Evaluation of two novel tag-based labelling technologies for site-specific modification of proteins. Int J Biol Macromol. 2006;39:66–76. doi: 10.1016/j.ijbiomac.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 19.Regan L. The design of metal-binding sites in proteins. Annu Rev Biophys Biomol Struct. 1993;22:257–287. doi: 10.1146/annurev.bb.22.060193.001353. [DOI] [PubMed] [Google Scholar]
- 20.Barondeau DP, Getzoff ED. Structural insights into protein-metal ion partnerships. Curr Opin Struct Biol. 2004;14:765–774. doi: 10.1016/j.sbi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Hochuli E, Dobeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt J, Hess H, Stunnenberg HG. Affinity purification of histidine-tagged proteins. Mol Biol Rep. 1993;18:223–230. doi: 10.1007/BF01674434. [DOI] [PubMed] [Google Scholar]
- 23.Ataka K, Giess F, Knoll W, Naumann R, Haber-Pohlmeier S, Richter B, Heberle J. Oriented attachment and membrane reconstitution of His-tagged cytochrome c oxidase to a gold electrode: in situ monitoring by surface-enhanced infrared absorption spectroscopy. J Am Chem Soc. 2004;126:16199–16206. doi: 10.1021/ja045951h. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson AL, Hughes AD, Tufail U, Baumann CG, Scott DJ, Hoggett JG. Interaction of σ70 with Escherichia coli RNA polymerase core enzyme studied by surface plasmon resonance. FEBS Lett. 2000;481:281–284. doi: 10.1016/s0014-5793(00)02028-7. [DOI] [PubMed] [Google Scholar]
- 25.Ueda EK, Gout PW, Morganti L. Current and prospective applications of metal ion-protein binding. J Chromatogr A. 2003;988:1–23. doi: 10.1016/s0021-9673(02)02057-5. [DOI] [PubMed] [Google Scholar]
- 26.Saito M, Yamamoto T, Ikuya S. Preparation of Aminodicarboxylic acid N,N-diacetic acids as Biodegradable Chelating Agents to be used as Substitutes for EDTA. No. JKXXAF JP 10077253 A2 19980324. Tokyo, Japan: Japan Patent Office; Japan Patent. 1998
- 27.McMahan SA, Burgess RR. Single-step synthesis and characterization of biotinylated nitrilotriacetic acid, a unique reagent for the detection of histidine-tagged proteins immobilized on nitrocellulose. Anal Biochem. 1996;236:101–106. doi: 10.1006/abio.1996.0137. [DOI] [PubMed] [Google Scholar]
- 28.Hutschenreiter S, Neumann L, Radler U, Schmitt L, Tampe R. Metal-chelating amino acids as building blocks for synthetic receptors sensing metal ions and histidine-tagged proteins. Chembiochem. 2003;4:1340–1344. doi: 10.1002/cbic.200200455. [DOI] [PubMed] [Google Scholar]
- 29.Lata S, Gavutis M, Tampe R, Piehler J. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J Am Chem Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Liu ZP, Jones TA, Gierasch LM, Sambrook JF. Mutating the charged residues in the binding pocket of cellular retinoic acid-binding protein simultaneously reduces its binding affinity to retinoic acid and increases its thermostability. Proteins. 1992;13:87–99. doi: 10.1002/prot.340130202. [DOI] [PubMed] [Google Scholar]
- 31.Clark PL, Weston BF, Gierasch LM. Probing the folding pathway of a β-clam protein with single-tryptophan constructs. Fold Des. 1998;3:401–412. doi: 10.1016/s1359-0278(98)00053-4. [DOI] [PubMed] [Google Scholar]
- 32.Cleverley RM, Zheng N, Gierasch LM. The cost of exposing a hydrophobic loop and implications for the functional role of 4.5 S RNA in the Escherichia coli signal recognition particle. J Biol Chem. 2001;276:19327–19331. doi: 10.1074/jbc.M011130200. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz JR. 2nd edn. New York: Kluwer Academic/Plenum Publishers; 1999. Principles of Fluorescence Spectroscopy (Chapter 10) [Google Scholar]
- 34.Kosower NS, Kosower EM, Newton GL, Ranney HM. Bimane fluorescent labels: labeling of normal human red cells under physiological conditions. Proc Natl Acad Sci U S A. 1979;76:3382–3386. doi: 10.1073/pnas.76.7.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosower NS, Kosower EM. Thiol labeling with bromobimanes. Methods Enzymol. 1987;143:76–84. doi: 10.1016/0076-6879(87)43015-2. [DOI] [PubMed] [Google Scholar]
- 36.Kosower NS, Kosower EM, Zipser Y, Faltin Z, Shomrat R. Dynamic changes of red cell membrane thiol groups followed by bimane fluorescent labeling. Biochim Biophys Acta. 1981;640:748–759. doi: 10.1016/0005-2736(81)90105-x. [DOI] [PubMed] [Google Scholar]
- 37.Danielsohn P, Nolte A. Bromobimanes–fluorescent labeling agents for histochemical detection of sulfur containing neuropeptides in semithin sections. Histochemistry. 1987;86:281–285. doi: 10.1007/BF00490259. [DOI] [PubMed] [Google Scholar]
- 38.Newton GL, Fahey RC. Determination of biothiols by bromobimane labeling and high-performance liquid chromatography. Methods Enzymol. 1995;251:148–166. doi: 10.1016/0076-6879(95)51118-0. [DOI] [PubMed] [Google Scholar]
- 39.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J Mol Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Sinz A, Wang K. Mapping spatial proximities of sulfhydryl groups in proteins using a fluorogenic cross-linker and mass spectrometry. Anal Biochem. 2004;331:27–32. doi: 10.1016/j.ab.2004.03.075. [DOI] [PubMed] [Google Scholar]
- 41.Giron-Monzon L, Manelyte L, Ahrends R, Kirsch D, Spengler B, Friedhoff P. Mapping protein-protein interactions between MutL and MutH by cross-linking. J Biol Chem. 2004;279:49338–49345. doi: 10.1074/jbc.M409307200. [DOI] [PubMed] [Google Scholar]
- 42.Arnau J, Lauritzen C, Petersen GE, Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif. 2006;48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, Graber P. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]