Abstract
Inducible gene expression appears to be an essential event that couples light to entrainment of the master mammalian circadian clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus. Recently, we reported that light triggers phase-dependent activation of the mammalian target of rapamycin (mTOR) signaling pathway, a major regulator of protein synthesis, in the SCN, thus raising the possibility that mTOR-evoked mRNA translation contributes to clock entrainment. Here, we used a combination of cellular, molecular, and behavioral assays to address this question. To this end, we show that the in vivo infusion of the mTOR inhibitor rapamycin led to a significant attenuation of the phase-delaying effect of early-night light. Conversely, disruption of mTOR during the late night augmented the phase-advancing effect of light. To assess the role of mTOR signaling within the context of molecular entrainment, the effects of rapamycin on light-induced expression of PERIOD1 and PERIOD2 were examined. At both the early- and late-night time points, abrogation of mTOR signaling led to a significant attenuation of light-evoked PERIOD protein expression. Our results also reveal that light-induced mTOR activation leads to the translation of mRNAs with a 5′-terminal oligopyrimidine tract such as eukaryotic elongation factor 1A and the immediate early gene JunB. Together, these data indicate that the mTOR pathway functions as potent and selective regulator of light-evoked protein translation and SCN clock entrainment.
Introduction
In mammals, the master circadian clock is localized in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus. Through both synaptic and paracrine mechanisms, the SCN pacemaker imparts rhythms to an array of biochemical, physiological, and behavioral processes. At a molecular level, the SCN clock timing mechanism is derived from the workings of several interlocking transcription/translation feedback loops (for review, see Reppert and Weaver, 2002; Ko and Takahashi, 2006). A key feature of the SCN clock is that its phasing is tightly regulated by the 12 h light/dark (LD) cycle. Light entrainment of the clock is mediated via a photic input pathway from the retina. Thus, in response to light, melanopsin-expressing retinal ganglion cells release glutamate and pituitary adenylate cyclase-activating polypeptide (PACAP) (for review, see Hannibal, 2002). During the night time domain, these two transmitters work in combination to elicit a rapid and irreversible resetting of the clock. Although the precise molecular mechanism by which photic input drives clock entrainment have not been resolved, a number of studies support a model in which the rapid induction of gene expression, and in particular, the expression of the circadian clock genes Period1 (Per1) and Period2 (Per2), drive the resetting process (Akiyama et al., 1999; Albrecht et al., 2001; Wakamatsu et al., 2001).
With respect to light-evoked gene expression, studies examining the transregulatory events have revealed a number of light-activated kinase pathways and transcription factors (Golombek et al., 2003). Along these lines, the 42/44 mitogen-activated protein kinase (MAPK) pathway is activated by light in a phase-restricted manner, and couples photic information to activation of cAMP response element-binding protein (CREB), a key effector of light-actuated clock gene transcription (Obrietan et al., 1998, 1999; Coogan and Piggins, 2003; Tischkau et al., 2003; Butcher et al., 2005). Given that gene expression is also regulated at a translational level, there has been significant interest in examining the role inducible translation plays in clock entrainment. Initial studies using a variety of model systems have shown that clock entrainment requires protein synthesis (Johnson and Nakashima, 1990; Raju et al., 1990; Murakami et al., 1995; Zhang et al., 1996). More recently, specific translation regulatory mechanisms have been identified. For example, we reported that light-evoked microRNA expression functions as a feedback regulator of clock entrainment (Cheng et al., 2007). In addition, the clock-regulated immediate early gene nocturnin, a deadenylase, which regulates mRNA stability, has been shown to regulate rhythmic gene expression (Baggs and Green, 2003; Garbarino-Pico et al., 2007). Furthermore, we recently reported on another potential mechanism of inducible gene expression in the SCN: the mammalian target of rapamycin (mTOR) signaling pathway (Cao et al., 2008).
mTOR is a Ser/Thr kinase that serves as the central component of the multiprotein mTOR complex 1 (mTORC1). A large number of studies have revealed a key role for mTORC1 as an integration center, regulating inducible mRNA translation to match changes in the metabolic/physiologic state of the cell (for review, see Sarbassov et al., 2005; Wullschleger et al., 2006; Proud, 2007, 2009). Actuation of mTORC1 triggers bifurcating signal transduction pathways. One arm, which is activated by p70 S6 kinase (p70 S6K) has been implicated in the translation of mRNAs with a 5′-terminal oligopyrimidine tract (5′-TOP mRNAs), a subset of which encode for translation factors that increase ribosomal processivity (Jastrzebski et al., 2007). The second arm is activated by eukaryotic initiation factor 4E-binding protein 1 (4E-BP1), which stimulates cap-dependent translation initiation (Hay and Sonenberg, 2004; Tee and Blenis, 2005). Our previous work revealed that photic stimulation triggers robust activation of both arms of the mTOR-dependent signaling pathway in the SCN (Cao et al., 2008). Here we further these studies by examining the potential contribution of the mTOR-dependent signaling pathway to SCN clock entrainment. The data presented here reveal that the disruption of mTOR signaling leads to an attenuation of the early-night light-evoked phase delay, whereas late night mTOR abrogation enhances the phase-advancing effect of light. The modulatory effects of mTOR were paralleled by data showing that mTOR alters the capacity of light to couple to the core clock timing mechanism. Together, these data reveal a key role for the mTOR-signaling pathway in sculpting the phase-specific entraining effects of lights.
Materials and Methods
Light-treatment paradigm and tissue processing.
Adult (8- to 10-week-old) C57BL/6 mice were entrained to a 12 h light/dark (LD) cycle (100 lux) for at least 2 weeks and then transferred to total darkness for two consecutive 24 h cycles. After dark adaptation, animals received a single light exposure (400 lux, 15 min) either during the early subjective night, circadian time (CT) 15, or the late subjective night (CT20). CT was calculated based on Zeitgeber time (ZT) and the tau for C57BL/6 mice under free-running conditions (23.77 h) (Schwartz and Zimmerman, 1990), with ZT 0 denoting lights on and ZT 12 denoting lights off. At the indicated time points after light exposure, animals were anesthetized with an intraperitoneal injection of ketamine hydrochloride (140 mg/ml) and xylazine (13 mg/ml) under dim red light (<10 lux). Opaque black tape was placed over the eyes, and mice were transcardially perfused with cold PBS, pH 7.4, followed by 4% paraformaldehyde in PBS. Next, brains were postfixed in 4% paraformaldehyde for 4 h at 4°C and cryoprotected with 30% sucrose in PBS. The “no light” control animal groups underwent the same handling procedures (except for light exposure) and were killed at the same time points. All procedures were in accordance with Ohio State University animal welfare guidelines and approved by the Institutional Animal Care and Use Committee.
Cannulation and infusion.
Mice were cannulated in the lateral ventricles using the techniques described by Cao et al. (2008). Briefly, animals were anesthetized with ketamine hydrochloride (91 mg/ml) and xylazine (9 mg/ml) and placed in a stereotaxic apparatus (Cartesian Research). The coordinates (posterior, 0.34 mm from bregma; lateral, 0.90 mm from the midline; dorsoventral, −2.15 mm from bregma) were used to place the tip of a 24 gauge guide cannula into the lateral ventricle. Cannulae were held in place with dental cement and a 30 gauge stylus was secured in the cannula to ensure patency. After surgery, animals were housed individually and allowed to recover for at least 2 weeks under a standard 12 h LD cycle. For the infusion, animals were restrained by hand under dim red light, and the infusate was delivered at a rate of 1 μl/min. To disrupt light-evoked PACAPergic and ionotropic glutamatergic neurotransmission, mice were infused with the PAC1 receptor antagonist, PACAP 6-38 (Bachem) and the ionotropic glutamate antagonists CNQX (Sigma) and APV (Sigma) in a total volume of 4 μl (diluted in physiological saline) 15 min before photic stimulation (400 lux, 15 min) at CT15. Inhibitor concentrations are noted in Figure 2. To activate mTOR signaling, PACAP (200 μm; Bachem) and glutamate (2.5 mm) were diluted in physiological saline (4 μl) and infused at CT14.5. Control animals were infused with the same volume of physiological saline at CT14.5, and mice were killed 30 min later. To inhibit mTOR signaling, rapamycin (2 μl, 100 μm; Cell Signaling Technology) was infused 30 min before light treatment. Control animals were infused with an equivalent volume of vehicle (DMSO). To inhibit gene transcription, 1 μl of actinomycin D (2 μg/μl; Sigma) was infused 30 min before light treatment.
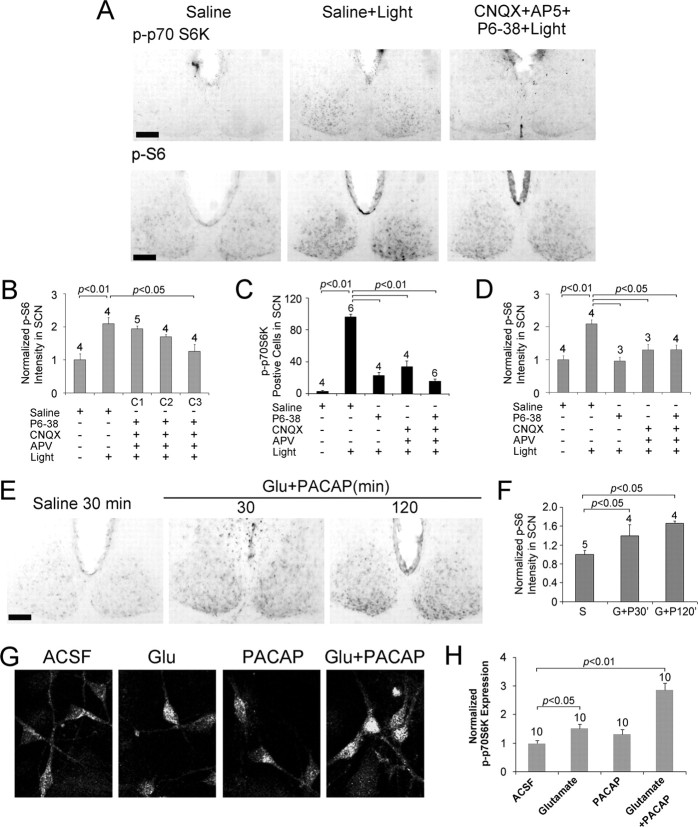
Figure 2.
Glutamatergic and PACAPergic neurotransmission couples light to mTOR activation in the SCN. A, Representative immunohistochemical labeling for control (Saline) and light-evoked (400 lux, 15 min, CT15) p-p70S6K and p-S6 after infusion of physiological saline (Saline+Light) or a mixture of CNQX (2.5 mm), AP5 (25 mm), and PACAP 6-38 (P6-38, 1 mm; CNQX+AP5+P6–38+Light). Compared to the Saline+Light condition (middle), the infusion of PACAP and glutamate receptor antagonists led to an attenuation of p70S6K and S6 phosphorylation (right). Scale bars, 100 μm. B, Quantitative analysis of p-S6 expression under three dosages of the inhibitor cocktail. C1, CNQX (150 μm), AP5 (1.6 mm) and PACAP 6-38 (60 μm) plus light; C2, CNQX (600 μm), AP5 (6 mm), and PACAP 6-38 (240 μm) plus light; C3, CNQX (2.5 mm), AP5 (25 mm), and PACAP 6-38 (1 mm) plus light. The light-stimulus paradigm is described in A. C, D, Quantitative analysis of p-p70S6K (C) and p-S6 (D) expression under the five treatment conditions indicated below each panel. Mice were infused with PACAP 6-38 (P6-38, 1 mm) and/or CNQX (2.5 mm) and AP5 (25 mm) at CT14.5. The light-stimulus paradigm is described in A. E, Representative immunohistochemical labeling for p-S6 after the ventricular infusion of saline or PACAP (200 μm) and glutamate (2.5 mm) at CT14.5. Animals were killed 30 or 120 min after infusion. Under the control condition, low p-S6 levels were detected in the SCN. PACAP and glutamate infusion increased S6 phosphorylation. Scale bar, 100 μm. F, Histogram quantifying p-S6 under the three conditions outlined in D. S, Saline; G+P30′, killed 30 min after glutamate and PACAP infusion; G+P120′, killed 120 min after glutamate and PACAP infusion. G, Representative confocal images of p70 S6K phosphorylation in cultured SCN neurons. Cultures were stimulated (30 min) with glutamate (10 μm), PACAP (200 nm), or glutamate and PACAP (10 μm and 200 nm, respectively) and then fixed and immunolabled. Relative to mock stimulation (ACSF), glutamate or PACAP alone induced a moderate increase in p-p70 S6K expression. However, the coadministration of glutamate and PACAP induced robust p70 S6K activation. H, Quantitative analysis of p-70S6K expression in SCN neurons under the conditions described in G. For all histograms, data were normalized to the saline (or ACSF) condition, which was set to a value of 1. Please see Materials and Methods for a detailed description of the quantitation methods.
SCN neuron culture and stimulation.
For neuronal culture, SCN-enriched tissue was collected from embryonic day 19–21 Sprague Dawley rat pups. Brains were placed in a sterile Petri dish containing cold oxygenated dissociation media (DM) [containing the following (in mm): 90 Na2SO4, 30 K2SO4, 16 MgCl2, 0.25 CaCl2, and 32 HEPES, and 0.01% phenol red (Sigma), pH 7.7], and using the optic chiasm and optic nerves as landmarks, the region containing the SCN was microdissected from the ventral surface of the brain. SCN tissue was the washed in DM and incubated in digestion solution [100 U/ml papain latex (Worthington) and 4.5 mg of cysteine (Sigma) in DM] for 30 min at 37°C. Tissue was then transferred to standard culture medium (Minimal Essential Medium; Invitrogen) containing 1% fetal bovine serum (Invitrogen), 2% B27 (Invitrogen), and 100 U/ml penicillin/streptomycin (Invitrogen), and triturated into a single-cell suspension. Cells were plated onto poly-d-lysine-coated 9 mm2 glass coverslips at a density of 50,000 cells/cm2. Media was changed 1 h after plating, and cells were maintained in a Napco 6100 incubator (37°C, 5.5% CO2) for 10 d. Media was replenished every third day. Four hours before experimental stimulation, tissue culture media was replaced with artificial CSF (ACSF) buffer [containing the following (in mm): 137 NaCl, 25 glucose, 10 HEPES, 5 KCl, 1 MgCl2, and 3 CaCl2, pH 7.4]. Cells were treated for 30 min with glutamate (10 μm) and/or PACAP (200 nm). Stimulated cells were washed with ACSF and fixed (15 min) with paraformaldehyde (4% w/v, 25°C) followed by cold methanol (−20°C). Phospho-p70 (p-p70) S6K immunofluorescent labeling was performed as described below.
Behavioral rhythm recording.
Cannulated mice were individually housed in polycarbonate cages equipped with running wheels. Wheel rotation was detected via the closure of a magnetic switch and recorded using ActiView software (MiniMitter). Mice were entrained to a 12 h LD cycle (100 lux) for at least 10 d, and then transferred to continuous darkness (DD). Rapamycin (100 μm, 2 μl) or vehicle (DMSO, 2 μl) was infused 30 min before light treatment (100 lux, 15 min) at CT15 or CT22 using the technique described above. Mice were then returned to their home cages for at least 14 d. After this period, mice received a second infusion and light pulse: half of the mice were infused with rapamycin first; the other half received DMSO first. Control “no light” animals received the same two-infusion paradigm (rapamycin and DMSO at CT14.5 or CT21.5) but no light treatment.
For core body temperature recoding, animals were anesthetized with ketamine hydrochloride (91 mg/ml) and xylazine (9 mg/ml) and implanted with transmitters (E-Mitters; MiniMitter) in the intraperitoneal cavity. After surgery, animals were housed singly in cages placed on receiving platforms (ER-4000 receiver) that communicated with the E-Mitter: data were collected every 60 s. The signal was acquired via the VitalView software program (MiniMitter) and analyzed via the ActiView software program. To clearly visualize the circadian rhythm of body temperature in an actograph format, a threshold was set to 2∼2.5°C below the maximum recorded temperature during the entire experimental period, and only signals above this threshold temperature were displayed.
Assessment of light-induced phase shifts.
The linear regression method described by Daan and Pittendrigh (1976) was used to assess light-induced phase shifts. Specifically, the difference in activity onset before and after the day of light exposure was determined by a least-squares method. Thus, a line calculating the activity onset for a period of at least 6 d preceding light treatment was calculated. This line was extended to project to when activity onset should occur during the period after light exposure. A second regression line was generated to determine activity onset after light administration. Wheel running activity 3–10 d after light treatment was used to generate this line. The difference in the projected versus the actual activity onset after light treatment was the phase shift. Group data (DMSO, DMSO plus light, rapamycin, and rapamycin plus light) are expressed as mean phase shift ± SEM. Significance was assessed using one-way ANOVA analysis followed by the Student–Newman–Keuls (SNK) test. A value of p < 0.05 was accepted as statistically significant.
Immunohistochemistry.
For immunohistochemical staining, brains sections (500 μm thick) were prepared using a vibratome, and SCN-containing sections were then thin-cut (40 μm) on a sliding microtome. Sections containing the central SCN were washed in PBS containing 2 mm sodium azide and 3 mm NaF, pH 7.4, and then treated with 0.3% H2O2 and 20% methanol in PBS for 10 min to deactivate endogenous peroxidases and permeabilize the tissue. Sections were then blocked for 1 h in 10% goat serum/PBS and incubated (overnight, 4°C) in one of the following antibodies: rabbit monoclonal anti-phospho-mTOR (p-mTOR, Thr-2448, 1:500; Cell Signaling Technology); mouse monoclonal anti-p-p70 S6 kinase (Thr-389, 1:1000; Cell Signaling Technology); rabbit polyclonal anti-phospho-S6 ribosomal protein (p-S6; Ser-240/244, 1:1000; Cell Signaling Technology); mouse monoclonal anti-S6 ribosomal protein (S6; 1:100; Cell Signaling Technology); rabbit anti-phospho-MSK1 (Ser-360, 1:500; Cell Signaling Technology); rabbit anti-mPER1 (1:3000; a generous gift from Dr. Steven Reppert, University of Massachusetts, Worcester, MA); mouse anti-mPER2 (1:500; Alpha Diagnostic); mouse eukaryotic elongation factor 1A (eEF1A) monoclonal antibody (1:1000; Millipore); mouse monoclonal anti-JunB (1:2000; Santa Cruz Biotechnology); or rabbit polyclonal anti-c-Fos (1:3000; Calbiochem). Next, tissue was incubated for 1.5 h at room temperature in biotinylated anti-mouse or rabbit IgG (1:200; Vector Laboratories) and then placed in an avidin/biotin HRP complex (Vector Laboratories) for 1 h, following the manufacturer's instructions. The signal was visualized using nickel-intensified DAB substrate (Vector Laboratories), and sections were mounted on gelatin-coated slides with Permount media (Fisher Scientific). Sections were washed in PBS (three times, 10 min per wash) between each labeling step.
For immunofluorescent labeling, both tissue sections and cultured neurons were permeabilized with PBS with 1% Triton X-100 (PBST) for 30 min, blocked as described above and then incubated (overnight, 4°C) in 5% goat serum/PBS with the following antibodies: mouse monoclonal anti-p-p70 S6 kinase (Thr-389, 1:300; Cell Signaling Technology) and/or rabbit polyclonal anti-p-S6 (Ser-240/244, 1:300; Cell Signaling Technology). The following day, sections were incubated (3 h, room temperature) in Alexa Fluor-594-conjugated goat anti-rabbit IgG antibody (1:500; Invitrogen) and Alexa Fluor-488-conjugated goat anti-mouse IgG antibody (1:500; Invitrogen). Sections were mounted on slides with Cytoseal 60 (Richard-Allan Scientific). Tissues were washed in PBS (three times, 10 min per wash) between each labeling step.
Bright-field photomicrographs were captured using a 16 bit digital camera (Micromax YHS 1300; Princeton Instruments) mounted on an inverted Leica microscope (DM IRB). Images were acquired with MetaMorph software (Molecular Devices). Confocal fluorescence images were captured using a Zeiss 510 Meta confocal microscope. All confocal parameters (pinhole, contrast, brightness, etc.) were held constant for all data sets from the same experiment.
Western blot analysis.
Mice were dark adapted for 2 d and then exposed to light at CT15 (400 lux, 15 min). Brains were isolated, and 500 μm sections were cut with a vibratome. The SCN was excised using a 700 μm tissue punch and frozen on dry ice. Tissue was pooled from six animals per condition, and then lysed in 100 μl radioimmunoprecipitation assay buffer [50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate, 1 mm sodium vanadate, 1 mm NaF, and 1× protease inhibitor cocktail (Roche)]. Protein extracts (24 μl/lane) were electrophoresed into an 8% SDS-PAGE gel and then transblotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore). Membranes were blocked in 10% bovine serum albumin (BSA) (Fisher Scientific) and then incubated (overnight, 4°C) in PBST (with 5% BSA) with the p-p70 S6K (Thr-389, 1:1000 dilution), p-mTOR (Ser-2448, 1:500 dilution), or p-S6 (Ser-240/244, 1:1000 dilution) antibodies. These are the same antibodies used for our immunolabeling analysis. Next, membranes were incubated in PBST (with 5% milk) with a goat anti-mouse or anti-rabbit IgG horseradish peroxidase or alkaline phosphatase-conjugated antibodies (1:2000 dilution; PerkinElmer Life Sciences). As a protein loading control, membranes were probed for total ERK expression using a goat polyclonal anti-ERK antibody (1:1000 dilution; Santa Cruz Biotechnology) followed by a donkey anti-goat IgG antibody conjugated to horseradish peroxidase (1:2000 dilution; Rockland Immunochemicals). The signal was visualized using the Western Lighting Chemiluminescence light-emitting system (PerkinElmer Life Sciences) and a Blue Lite Autorad film (ISC BioExpress). Between each antibody treatment, membranes were washed a minimum of three times (10 min per wash) in PBST.
Data analysis.
All data were quantified using Adobe Photoshop software (Adobe Systems). For the p-S6 and PER2 intensity analysis, images of the SCN were acquired (10×) and digitally outlined, and the mean pixel values were determined. Next, a digital oval (150 × 200 pixels) was placed on the adjacent lateral hypothalamus, and this mean value was subtracted from the adjacent SCN signal to provide a normalized SCN intensity value. For PER1 and eEF1A immunolabeling, SCN images were acquired at 40×, a digital circle (ϕ1000 pixels) was placed in the ventral SCN, and the mean labeling intensity was determined. A digital oval (150 × 200 pixels) was then placed over the adjacent lateral hypothalamus, and the mean value was subtracted from the SCN value to generate a normalized SCN intensity value. Of note, the lateral hypothalamic immunolabeling values were not altered by light or rapamycin infusion. For p-p70 S6K-, p-MSK1-, c-Fos-, and JunB-positive cell counting, SCN images were acquired at 40× and a digital circle (ϕ1000 pixels) was placed in the ventral SCN. An intensity threshold filter was applied to eliminate nonspecific background labeling. Then, the number of detectable signals above threshold (now defined as positive cells) was tabulated. For all experiments, data were averaged from three central SCN sections per animal, and these values were pooled to generate mean values for treatment groups.
For p-p70 S6K expression analysis in SCN cell culture, neuronal images were acquired at 40×, the cell soma were digitally outlined, and the mean pixel values were determined. Next, a digital oval (150 × 200 pixels) was placed in an adjacent empty area, and the mean value within this region was subtracted from the adjacent cellular signal to provide a background-corrected p-p70 S6K intensity value. For statistical analysis, the p-p70 S6K expression level of all cells within a visual field (mean, six cells per field) was averaged. Data from 10 visual fields were statistically analyzed for each treatment group. Group statistical analysis was performed via one-way ANOVA followed by an SNK test. A value of p < 0.05 was accepted as statistically significant. The values are presented as mean ± SEM. All statistical analysis was performed using SPSS software.
Results
Light-evoked mTOR activation in the SCN
We begin our study with an examination of the light-evoked activation and temporal regulation of mTOR signaling. As an initial validation of our light-stimulus paradigm, we monitored mTOR phosphorylation (p-mTOR) at Ser-2448, a widely used marker of mTOR activity (Hou and Klann, 2004; Tsokas et al., 2005; Hoeffer et al., 2008). For these studies, mice were dark adapted for 2 d, exposed to light (400 lux, 15 min) at CT15, and then killed 5 min after light cessation. Immunohistochemical labeling and Western blot analysis of SCN extracts revealed that light triggered an increase in p-mTOR within the SCN (Fig. 1A,C,E). Using the same stimulus conditions, immunofluorescent labeling detected a marked increase in the Thr-389 phosphorylated form of p70 S6K (Fig. 1A). Phosphorylation of p70 S6K is mediated by mTOR, and thus can be used to validate mTOR activation (Jefferies et al., 1997; Burnett et al., 1998; Weng et al., 1998). Of note, this antibody also recognizes p85 S6K when phosphorylated at Thr-412, a site analogous to Thr-389 in p70 S6K. The specificity of the antibody was tested in our previous study (Cao et al., 2008) and in the published studies of several other groups (Tsokas et al., 2005; Cota et al., 2006; Hoeffer et al., 2008). Western blot analysis presented here detected a light-induced increase in an ∼70 kDa band, which is likely to be p70 S6K (Fig. 1D). To avoid confusion, we will specifically refer to p70 S6K when describing results generated using this antibody. Next, double labeling revealed a light-evoked increase in the active (i.e., Ser-240 and Ser-244 phosphorylated) form of S6 (p-S6) (Fig. 1A,C). The two phosphorylation sites examined here are specifically targeted by p70 S6K. Thus, given that S6 catalytic activity is regulated by p70 S6K, S6 phosphorylation can be used to monitor the relative activation of the p70 S6K/S6 arm of the mTOR pathway. Along these lines, merging the p-p70 S6K and p-S6 signals revealed colocalized expression of the antigens (Fig. 1A). A light-evoked increase in p-mTOR and p-S6 was also detected using Western blot analysis of SCN extracts (Fig. 1E). Together, these data support and extend our prior findings (Cao et al., 2008) that light triggers activation of the mTOR/p70 S6K/S6 signaling cassette in the SCN.
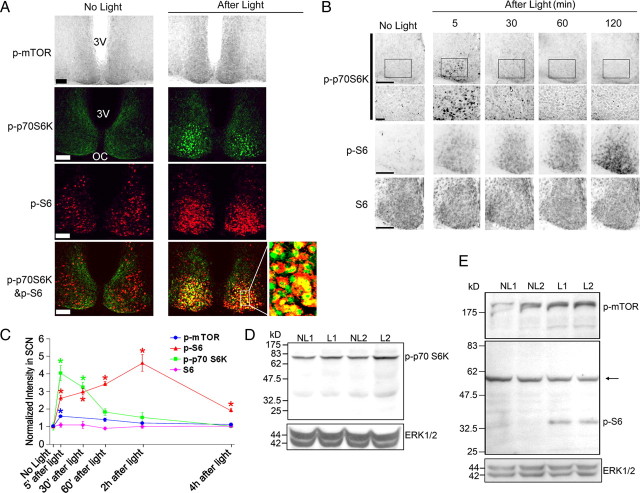
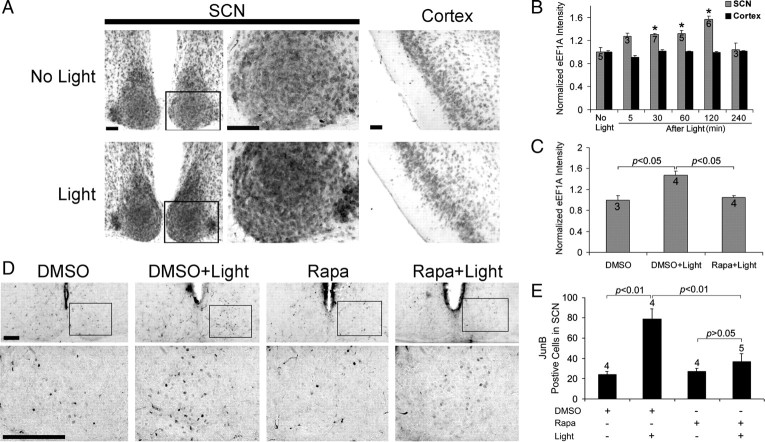
Figure 1.
Light-induced mTOR activation in the SCN. A, Top, Representative micrographs of tissue labeled for the Thr-2448-phosphorylated form of mTOR. Relative to the control condition (No Light), photic stimulation (400 lux, 15 min, CT15) triggered an increase in mTOR phosphorylation. Mice were killed 5 min after cessation of the light pulse. Middle, Representative confocal images of SCN tissue double labeled for the expression of Thr-389-phosphorylated p70 S6K (p-p70 S6K; green) and Ser-240/244 phosphorylated S6 (p-S6; red). Relative to the control condition (No Light), light triggered a marked increase in the phosphorylated forms of both proteins. Bottom, Merging of the two signals revealed cellular colocalization of the two antigens. The framed area is magnified to the right. 3V, Third ventricle; OC, optic chiasm. Scale bars, 100 μm. B, Top, Middle rows, Temporal profile of light-induced p70 S6K and S6 phosphorylation. Mice were killed at the noted times after light exposure (400 lux, 15 min) at CT15. Control mice (No Light) were killed at CT15. Bottom, Immunohistochemical profile of total S6 expression after light exposure. Scale bars, 100 μm. C, Quantification of light-induced p-mTOR, p-p70 S6K, p-S6, and S6 expression. *p < 0.05 compared to the “no light” control value, which was normalized to a value of 1. Data were collected from four or five animals for each group. D, E, Control and light-pulsed animals (CT15, 400 lux, 15 min) were killed at CT15.25, and SCN tissue was examined via Western blot analysis. Probing with a p-p70 S6K antibody (D) revealed that light treatment (L1 and L2; biological replicates) stimulated an increase in the phosphorylation of an ∼70 kDa band relative to the control “no light” condition (NL1, NL2; biological replicates). Relative to the “no light” condition, an increase in p-mTOR and pS6 expression (E) was also detected. The molecular weights of these bands are consistent with the sizes of mTOR (∼289 kDa) and S6 (∼32 kDa). The arrow indicates a nonspecific band. As loading controls, blots were probed for total ERK expression.
To gain insight into the regulation of mTOR signaling, we examined the temporal profile of light-evoked mTOR, p70 S6K, and S6 phosphorylation (Fig. 1B,C). For these experiments, mice were treated to the same light-stimulus paradigm described above, and then returned to darkness for 5 min to 4 h before being killed. Immunolabeling revealed that mTOR and p70 S6K phosphorylation was rapidly activated, reaching a peak phosphorylation level by 30 min after light exposure and declining quickly thereafter. However, light led to a robust and persistent increase in p-S6: maximal expression was detected at 120 min after light, and a significant increase in expression could still be detected at 4 h after light treatment. In contrast, the level of total S6 within the SCN was not affected by light (Fig. 1B,C).
Next, we examined the proximal transmitter systems that actuate light-induced mTOR activation in the SCN. Toward this end, we focused on the two principal neurotransmitters of the retinohypothalamic tract (RHT): PACAP and glutamate. PACAP has been shown to regulate the clock via the PACAP type 1 (PAC1) receptor subtype (Bergström et al., 2003), whereas glutamate functions at both NMDA and AMPA/kainate subclasses of ionotropic glutamate receptors to affect clock timing (Colwell and Menaker, 1992; Rea et al., 1993). Thus, to antagonize signaling through both pathways, we infused a mixture of the PAC1 receptor antagonist PACAP 6-38 (1 mm), the NMDA receptor antagonist APV (25 mm), and the AMPA/kainate receptor antagonist CNQX (2.5 mm) 15 min before photic stimulation (400 lux, 15 min) at CT15. Abrogation of PACAP and ionotropic glutamate receptor activation potently suppressed light-induced p70 S6K and S6 phosphorylation (Fig. 2A–D). These effects were dose dependent (Fig. 2B). Interestingly, infusion of either PACAP 6-38 or CNQX and APV also significantly decreased light-induced p70 S6K and S6 phosphorylation (Fig. 2C,D), indicating that coactivation of PACAPergic and glutamatergic signaling is required for robust mTOR activation. In the absence of light, the infusion of PACAP 6-38, CNQX, and APV did not markedly affect basal p70 S6K and S6 phosphorylation (data not shown). To confirm these results, we attempted to activate mTOR signaling in the SCN via the exogenous administration of glutamate (2.5 mm) and PACAP (200 μm). Animals were killed at 30 or 120 min after neurotransmitter microinjection at CT14.5. Relative to the injection of drug vehicle (saline), the infusion of glutamate and PACAP led to an increase in p-S6 expression at both time points (Fig. 2E,F). We also used cultured SCN neurons to test the capacity of glutamate (10 μm) and PACAP (200 nm) to evoke p70 S6K activation. Application of glutamate or PACAP induced relatively modest p70 S6K phosphorylation. However, a combined application of the two transmitters induced robust p70 S6K phosphorylation (Fig. 2G,H). In conclusion, these data indicate that coordinate PACAPergic and glutamatergic neurotransmission is required to drive marked mTOR activation in the SCN.
mTOR signaling couples light to SCN clock entrainment
Light exposure during the early subjective night induces a phase delay of the circadian clock, whereas late-night light induces a phase advance of the SCN clock timing process. Our prior work showing that mTOR signaling is activated by both early- and late-night light (Cao et al., 2008) led us to investigate the potential role of this pathway in clock entrainment.
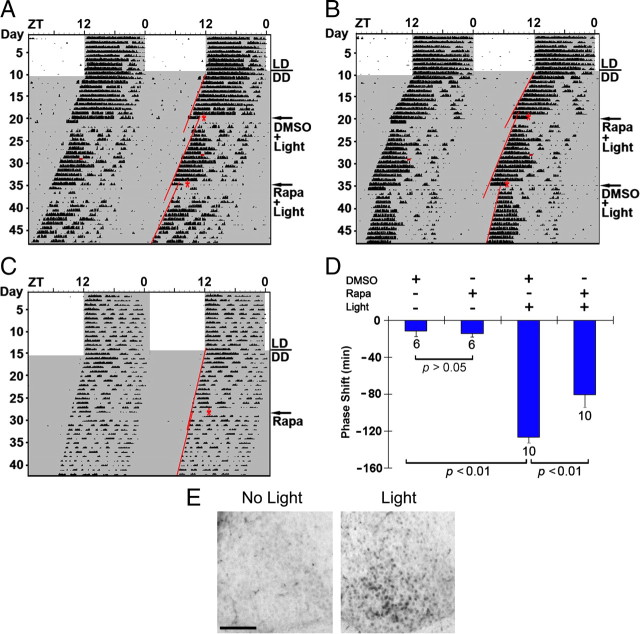
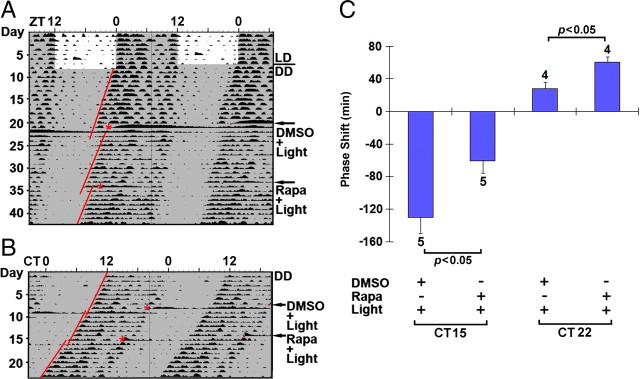
To test this idea, we used a ventricular infusion approach to transiently disrupt mTOR signaling and assess its effect on clock entrainment via an examination of overt wheel-running activity. For these experiments, cannulated mice were dark adapted for at least 10 d and then infused with the mTOR inhibitor rapamycin (100 μm) or vehicle (DMSO) 30 min before light (100 lux, 15 min) exposure at CT15 or CT22. We showed recently (Cao et al., 2008) that this rapamycin-infusion paradigm results in potent repression of light-evoked mTOR kinase activity in the SCN; furthermore, the 100 lux photic stimulation paradigm used here led to a statistically significant (p < 0.01, Student's t test) increase in p70 S6K activation (light, 89 ± 14 SEM immunopositive cells vs control, 7 ± 2 immunopositive cells; n = 4 animals for each condition) (Fig. 3E). After light treatment, animals were returned to darkness for ∼14 d and then received a counterbalanced infusion of rapamycin or vehicle: half of the animals received rapamycin first, and the other half received vehicle first. Additionally, to test for the effects of rapamycin and drug vehicle on clock phasing or period, control “no light” animals received counterbalanced infusions of rapamycin and DMSO vehicle. As expected, in vehicle-infused mice, light exposure at CT15 triggered a significant phase delay in activity onset. Interestingly, pretreatment with rapamycin significantly attenuated (∼50 min or 36%) this phase-delaying effect of light (Fig. 3A,B,D). Importantly, in the absence of light, the early-night infusion of rapamycin did not significantly affect clock phasing (Fig. 3C,D), indicating that the transient disruption of mTOR signaling specifically affected the capacity of light to entrain the clock.
Figure 3.
Disruption of mTOR signaling attenuates light-induced phase delaying of circadian locomotor activity. A, B, Representative double-plotted actographs of wheel-running activity. Initially, mice were entrained on a 12 h LD cycle and then transferred to total darkness. After ∼10 d under DD, mice were infused with DMSO vehicle (A) or the mTOR inhibitor rapamycin (100 μm; B) 30 min before light exposure (100 lux, 15 min) at CT15 (red asterisks). Animals free ran for 14 d and then received a second infusion of rapamycin (A) or DMSO (B) followed by light treatment. Regression lines approximate the phase-delaying effects of light. The small horizontal red bars in the activity records denote an “off-line” period when wheel-running activity was not recorded. C, Representative actograph shows that rapamycin (100 μm) infusion at CT15 did not markedly affect clock timing or phasing. D, Statistical representation of the early-night phase-shifting data. Of note, the light-evoked phase delay was significantly attenuated by rapamycin. Numbers above the bars denote the number of animals examined for each condition. E, Light-evoked p70 S6K phosphorylation. To test whether the light intensity (100 lux, 15 min) used in the behavioral experiments evokes mTOR activation, mice were exposed to light (100 lux, 15 min) at CT22 and killed immediately thereafter. Immunohistochemical labeling revealed a light-evoked increase in p-p70 S6K, relative to control mice (no light). Scale bar, 100 μm.
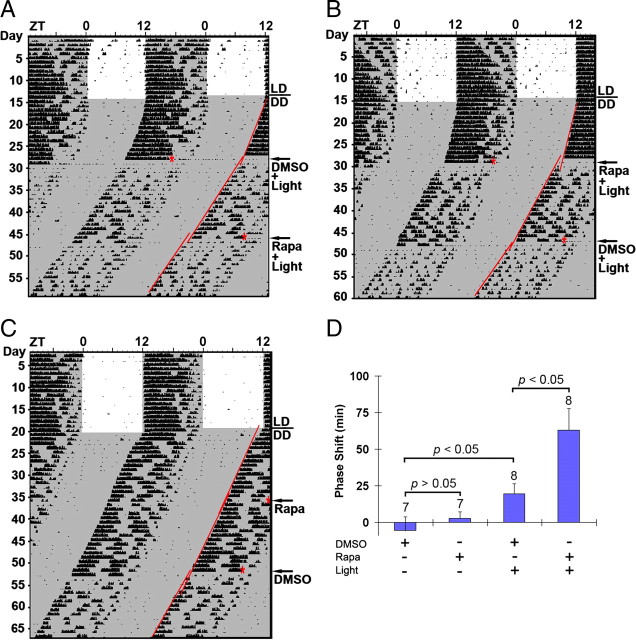
Next, we examined whether mTOR signaling affects the phase-advancing effects of late-night light. As expected (Schwartz and Zimmerman, 1990), in vehicle-treated mice, light exposure at CT22 triggered a relatively modest phase advance (Fig. 4A,B,D). Interestingly, disruption of mTOR signaling led to a significant lengthening of the light-induced phase advance. Thus, relative to vehicle infusion, the infusion of rapamycin triggered an ∼2.2-fold increase in the phase advance. Given that rapamycin infusion in the absence of light did not significantly alter clock phase (Fig. 4C,D), these data indicate that light-actuated mTOR signaling represses the phase-advancing effect of late-night light.
Figure 4.
mTOR functions as a negative regulator of late-night light-evoked phase advancing of the circadian clock. A, B, Representative double-plotted actographs of wheel-running activity. Mice were dark adapted and infused with DMSO vehicle (A) or rapamycin (100 μm; B) 30 min before light (100 lux, 15 min) exposure at CT22 (red asterisks). Mice were allowed to free run under DD for 19 d, and then they received a second infusion of rapamycin (A) or DMSO (B) followed by light exposure. Regression lines approximate the light-evoked phase shifts. C, A representative actograph shows that rapamycin infusion at CT22 did not markedly affect clock timing or phasing. D, Statistical analysis of late-night phase shifting. Of interest, the phase-advancing effect of light was significantly enhanced by rapamycin infusion. Numbers above the bars denote the number of animals analyzed for each condition.
To complement the wheel-running experiments, we analyzed the effects of rapamycin on light-evoked entrainment of core body temperature rhythms. To this end, mice were implanted with radio transmitters, entrained to a 12 h LD cycle, and then transferred to DD. After ∼10 d under DD, mice were infused with DMSO vehicle or the mTOR inhibitor rapamycin (100 μm) 30 min before light exposure (100 lux, 15 min) at CT15 or CT22. By applying a threshold, which was set to be 2–2.5°C below the maximum recorded temperature during the entire experiment, a circadian temperature oscillation could be clearly displayed in the actogram (Fig. 5A,B). Similar to the results of the wheel-running experiments, pretreatment with rapamycin significantly attenuated (∼69 min or 53%) the phase-delaying effect of light at CT15 (Fig. 5A,C) but led to a significant lengthening (∼32 min or 114%) of the light-induced phase advance at CT22 (Fig. 5B,C). Together, these behavioral rhythms studies suggest that mTOR functions as a time-of-day-specific regulator of the clock, facilitating early-night light-induced phase delays and repressing late-night phase advances.
Figure 5.
Disruption of mTOR signaling attenuates light-induced phase delaying but enhances light-induced phase advancing of the circadian core body temperature rhythm. A, B, Representative double-plotted actographs of core body temperature recordings. Initially, cannulated mice were entrained on a 12 h LD cycle and then transferred to total darkness. After ∼10 d under DD, mice were infused with DMSO vehicle or the mTOR inhibitor rapamycin (Rapa; 100 μm) 30 min before light exposure (100 lux, 15 min) at CT15 (A) or CT22 (red asterisks) (B). Animals free ran for 7–14 d and then received a second infusion of rapamycin or DMSO followed by light treatment. Regression lines approximate the phase-delaying (A) or phase-advancing (B) effect of light. C, Statistical representation of the early-night (CT15) and late-night (CT22) data sets. Of note, the light-evoked phase delay was significantly attenuated by rapamycin. However, the light-evoked phase advance was significantly enhanced by rapamycin. Numbers above the bars denote the number of animals examined for each condition.
mTOR modulates light-induced PERIOD protein expression
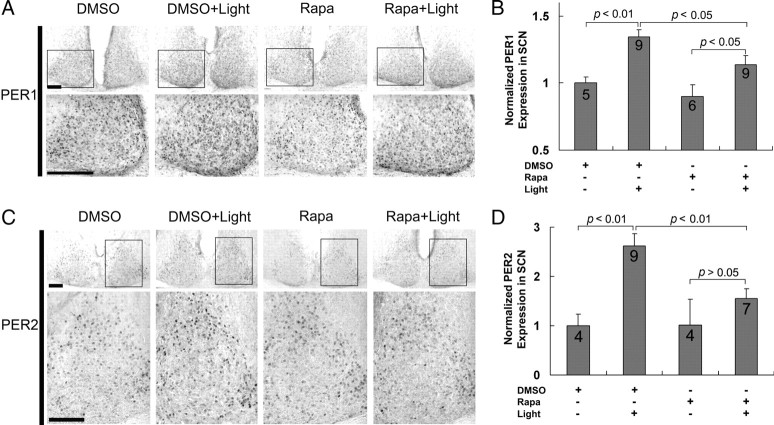
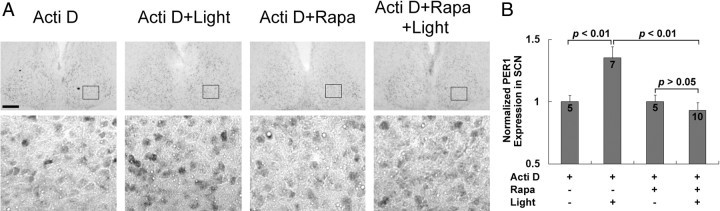
A good number of studies support the idea that light-induced Period1 and Period2 gene expression are key events in the clock entrainment process (Akiyama et al., 1999; Albrecht et al., 2001; Wakamatsu et al., 2001). These findings, coupled with our work showing that mTOR regulates clock entrainment, led us to examine whether mTOR influences light inducible PERIOD (PER) protein expression. To this end, we used the ventricular infusion technique described above to deliver rapamycin (100 μm) or vehicle (DMSO) 30 min before a light pulse (400 lux, 15 min) at CT15. Animals were then returned to darkness and killed 4 h later. Sampling schedules were based on a recent study (Yan and silver, 2004) showing that maximum PER protein induction within the core region of the SCN was observed 4–6 h after photic stimulation. Immunohistochemical labeling revealed that photic stimulation triggered an increase in PER1 and PER2 protein expression in the SCN, relative to control animals (no light) (Fig. 6). Similar to the expression pattern reported by Yan and Silver (2004), light-evoked PER1 was mainly detected within the central SCN (Fig. 6A), whereas the increase in PER2 expression was predominately located in the dorsal and lateral regions of the SCN (Fig. 6C). Interestingly, light induction of both PER1 (Fig. 6A,B) and PER2 (Fig. 6C,D) was significantly attenuated by rapamycin infusion. In the absence of light, rapamycin did not significantly alter basal PER1 and PER2 expression, thus indicating that mTOR facilitates light-evoked clock protein expression. Control experiments revealed that rapamycin infusion did not significantly alter the capacity of light to trigger MSK1 phosphorylation (data not shown), thus suggesting that rapamycin does not affect kinase signaling events that have been shown to be stimulate CREB-dependent Per1 transcription (Butcher et al., 2005). To test whether this repression of PER1 protein expression is specifically mediated at the level of mRNA translation, we performed a parallel set of experiments, where the transcription inhibitor actinomycin D (2 μg/μl, 1 μl) was infused before light treatment. Previous work has shown that the infusion of this concentration of actinomycin D effectively inhibits transcription (Daniels, 1971; Frey et al., 1996). Interestingly, even in the absence of transcriptional activation (i.e., actinomycin D treatment), light still induced moderate PER1 expression in a subset of SCN cells (Fig. 7A,B). Under this condition, the infusion of rapamycin significantly attenuated light-evoked PER1 expression (Fig. 7A,B). Together, these data indicate that photic input stimulates PER1 induction via mTOR-meditated mRNA translation.
Figure 6.
mTOR facilitates early-night light-induced PER1 and PER2 expression in the SCN. A, C, Representative immunohistochemical labeling for PER1 (A) and PER2 (C) protein expression in the SCN. Cannulated mice were dark adapted for 2 d and then infused (CT14.5) with rapamycin (100 μm) or DMSO vehicle and exposed to light (400 lux, 15 min) at CT15. After light exposure, animals were returned to darkness for 4 h and then killed at CT19. In addition, two “no light” control groups were infused with DMSO or rapamycin at CT14.5 and killed at CT19. Immunolabeling revealed that light (DMSO+Light) evoked an increase in PER1 and PER2 expression relative to the control condition (DMSO). PER1 induction was mainly located in the ventral SCN (A), whereas the increase in PER2 expression was predominantly located in the lateral and dorsal regions of the SCN (C). For each representative section, the boxed area is magnified and shown below. Scale bars, 100 μm. B, D, Quantification of light-induced PER1 (B) and PER2 (D) expression in the SCN. Of note, the light-induced increase in PER1 and PER2 expression was significantly attenuated by rapamycin. Under basal conditions, rapamycin led to a modest but statistically insignificant decrease in PER1 and PER2 protein expression. Numbers in the bars denote the number of animals analyzed for each condition. PER1 and PER2 expression data were normalized to the DMSO infusion condition, which was set to a value of 1.
Figure 7.
mTOR facilitates light-induced PER1 expression in the SCN in the presence of actinomycin D. A, Cannulated mice were dark adapted for 2 d and then infused (CT14.5) with actinomycin D (Acti D; 2 μg/μl, 1 μl) and rapamycin (Rapa; 100 μm, 2 μl) or DMSO vehicle and exposed to light (400 lux, 15 min) at CT15. After light exposure, animals were returned to darkness for 4 h and then killed (CT19). In addition, two “no light” control groups were infused with actinomycin D and DMSO or rapamycin at CT14.5 and killed at CT19. Immunolabeling revealed that in the presence of actinomycin D, light (Acti D+Light) evoked a moderate increase of PER1 expression relative to the control condition (Acti D). Infusion of rapamycin attenuated the light-evoked increase (Acti D+Rapa+Light) in PER1 expression. For each representative section, the boxed area is magnified and shown below. Scale bars, 100 μm. B, Quantification of the data set depicted in A. Of note, the light-evoked, actinomycin D-insensitive increase in PER1 expression was significantly attenuated by rapamycin. Numbers on the bars denote the number of animals analyzed for each condition. The PER1 expression data were normalized to the actinomycin/no light infusion condition, which was set to a value of 1. See Materials and Methods for a detailed description of the quantitation methods.
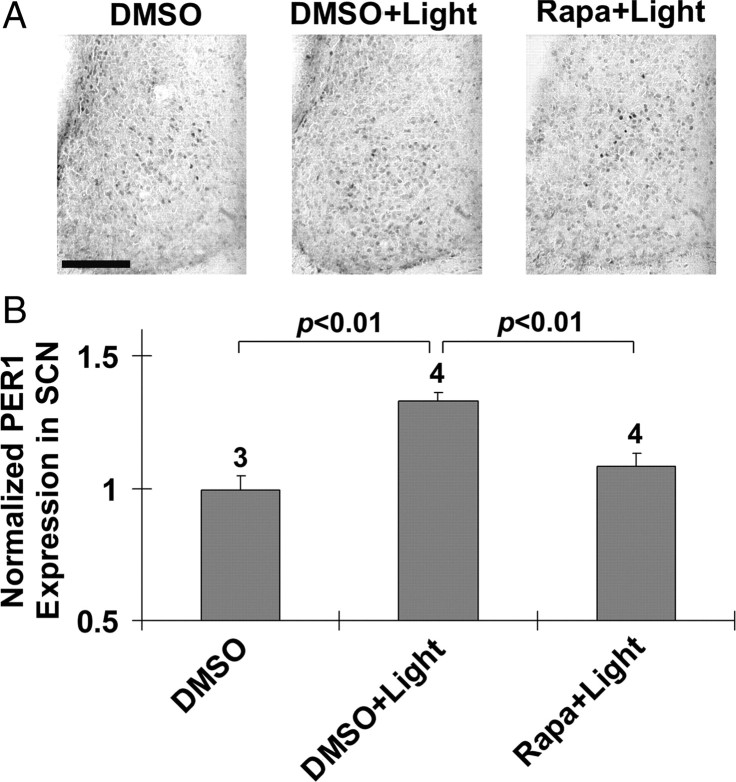
Finally, similar to the early-night data, infusion of rapamycin 30 min before a late night (CT20) light pulse (400 lux, 15 min) triggered a significant reduction in light-induced PER1 expression relative to vehicle-infused animals (Fig. 8A,B). Given its limited light inducibility during the late night (Albrecht et al. 1997; Zylka et al., 1998; Yan and Silver, 2002, 2004), PER2 expression was not examined.
Figure 8.
mTOR facilitates late-night light-induced PER1 expression in the SCN. A, Representative immunohistochemical labeling for PER1 protein expression. Cannulated animals were dark adapted for 2 d and then infused at CT19.5 with rapamycin (100 μm) or DMSO vehicle. Mice were exposed to light (400 lux, 15 min) at CT20, returned to darkness, and killed 6 h later (CT2). Control mice were infused with DMSO as described above, and killed at CT2. Immunolabeling revealed that light induced a moderate increase in PER1 expression in the SCN (DMSO+Light vs DMSO). The light-evoked increase in PER1 was attenuated by rapamycin infusion (Rapa+Light). The presentation of high-magnification images of a single SCN was necessitated by the relatively modest light-evoked PER1 induction pattern. Scale bar, 100 μm. B, Quantification of PER1 expression. Numbers above the bars denote the number of animals analyzed for each condition. PER1 expression data were normalized to the DMSO infusion condition, which was set to a value of 1. See Materials and Methods for a detailed description of the quantitation methods.
mTOR and 5′-TOP mRNA translation in the SCN
Next, we turned to possible mechanisms by which mTOR affects PERIOD protein expression. Along these lines, the p70 S6K arm of the mTOR pathways has been shown to stimulate the translation of 5′-TOP mRNA transcripts. A subset of these mRNAs encode for translational machinery, including ribosomal proteins and elongation factors (e.g., eEF1A and PABP1) (Jefferies et al., 1994, 1997; Peterson and Schreiber, 1998), and the increased expression of these proteins has been shown to correlate with enhanced mRNA translation (Giustetto et al., 2003; Huang et al., 2005; Tsokas et al., 2005; Antion et al., 2008). To examine whether light-activation of mTOR results in increased elongation factor expression, we profiled eEF1A protein levels in the SCN. Using an immunohistochemical labeling approach, we detected a marked increase in eEF1A expression after a brief light pulse (400 lux, 15 min) at CT15 (Fig. 9A,B). Induction was observed within 5 min of light treatment and peaked 2 h after light treatment (Fig. 9B). Induction appeared be specific to the SCN, since light did not significantly alter eEF1A expression within the piriform cortex (Fig. 9A,B). To test the idea that eEF1A expression in the SCN is dependent on mTOR, we used the ventricular infusion approach described above to transiently disrupt mTOR signaling. Analysis of mice killed 120 min after light exposure (CT15) revealed that the disruption of mTOR activity completely repressed EF1A expression (Fig. 9C).
Figure 9.
Light induces mTOR-dependent eEF1A and JunB expression in the SCN. A, Representative immunohistochemical images of eEF1A expression in the SCN. Mice were dark adapted for 2 d and then exposed to light (400 lux, 15 min) at CT15. Animals were killed at CT17. Control animals (No Light) were also killed at CT17. Relative to control animals, light exposure triggered a moderate increase in eEF1A expression in the SCN; the boxed regions of the SCN are magnified and shown to the right. In contrast to the SCN, light did not affect eEF1A expression in the piriform cortex (Cortex). Scale bars: Low magnification, 100 μm; high magnification, 50 μm. B, Quantification of light-induced eEF1A expression in the SCN and piriform cortex (Cortex). Animals were killed 5, 30, 60, 120, and 240 min after light exposure at CT15. Control animals (No Light) were killed at CT15. The eEF1A expression in the SCN increased as a function of time after light exposure (up to 2 h after light). Numbers on the bars denote the number of animals analyzed for each condition. *p < 0.05 versus the “no light” control. eEF1A expression data were normalized to the no light conditions, which were set to a value of 1. C, Rapamycin infusion abolished light-induced eEF1A expression in the SCN. Rapamycin (100 μm) was infused 30 min before light treatment at CT15. Animals were killed 2 h after light exposure. Data are presented as described in B. D, Representative immunohistochemical labeling for JunB protein expression in the SCN. Mice were dark adapted for 2 d and then infused with rapamycin (100 μm) or DMSO vehicle 30 min before light (400 lux, 15 min) exposure at CT15. Animals were killed 0–10 min after termination of the light stimulus. Immunolabeling revealed that the light-induced increase in JunB expression (DMSO+Light) was attenuated by rapamycin (Rapa+Light). Scale bars, 100 μm. E, Quantification of light-induced JunB expression. Numbers above the bars denote the number of animals analyzed for each condition. See Materials and Methods for a detailed presentation of the cell-counting method.
In addition to eEF1A, the mRNA of AP-1 binding protein JunB also belongs to the 5′-TOP mRNA family (Staber et al., 2007; Vesely et al., 2009). Given its marked light-inducibility, we examined whether JunB protein expression is modulated by mTOR. In these experiments, mice were killed 0–10 min after termination of a brief light pulse (400 lux, 15 min) at CT15. The goal of this short time course was to examine rapid mRNA translation (e.g., translation of the mRNA pool that was transcribed before the light pulse), and to minimize the contribution of light-evoked junB transcription to the evoked JunB signal. The data presented in Figure 9, D and E, reveal that light-evoked JunB protein expression was significantly repressed by rapamycin. Together, these data indicate that 5′-TOP mRNA expression within the SCN is regulated by light in an mTOR-dependent manner.
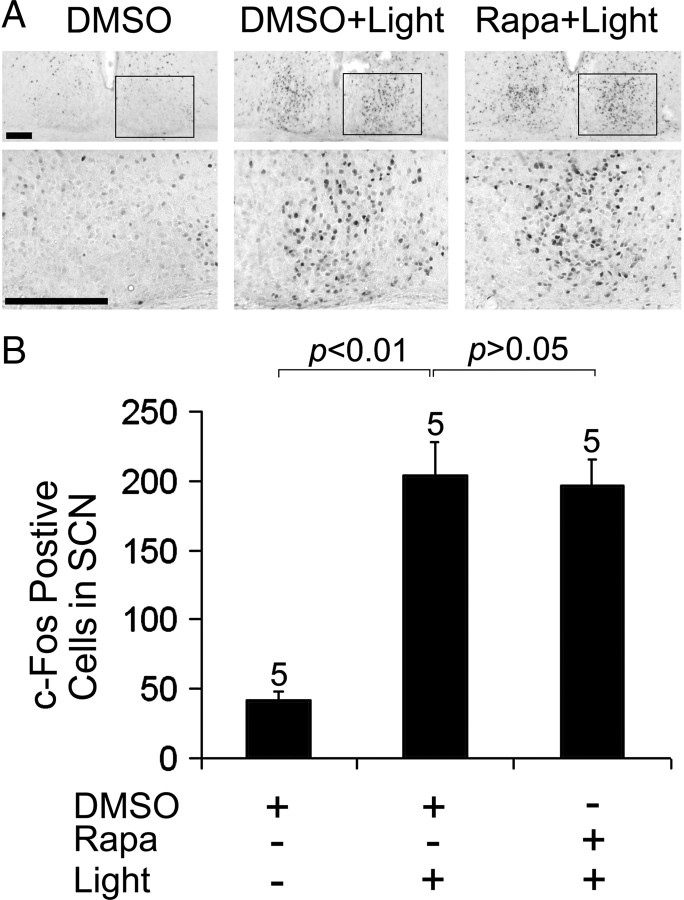
Finally, the expression of the immediate early gene c-fos has been shown to be translated via an mTOR-independent mechanism. Along these lines, c-fos mRNA does not belong to the 5′-TOP mRNA family, and it can be translated via a cap-independent mechanism (Vesely et al., 2009). Thus, to test the specificity of the protein expression findings described above, we examined whether rapamycin infusion affects light-evoked c-Fos protein expression. Using the same infusion and photic stimulation paradigm described above for JunB, we found that c-Fos expression was not significantly altered by disruption of mTOR (Fig. 10A,B). Together, these data indicate that the mTOR pathway functions as selective regulator of light-evoked protein translation and, in turn, SCN clock entrainment.
Figure 10.
mTOR does not regulate light-induced c-Fos expression in the SCN. A, Representative immunohistochemical labeling for c-Fos protein. Mice were dark-adapted for 2 d and then infused with rapamycin (100 μm) or DMSO vehicle 30 min before light (400 lux, 15 min) at CT15. Animals were killed 0–10 min after termination of the light stimulus. The robust light-evoked increase in c-Fos expression (DMSO+Light) was not affected by rapamycin treatment (Rapa+Light). Framed areas of the SCN are magnified and shown below. Scale bars, 100 μm. B, Quantification of c-Fos protein expression in the SCN. Numbers above the bars denote the number of animals analyzed for each condition. See Materials and Methods for a detailed presentation of the cell-counting method.
Discussion
Our previous work identified a light-responsive mTOR cascade in the SCN (Cao et al., 2008). In this study, we examined the role of mTOR as a potential regulator of clock entrainment. The results reported here reveal that the mTOR cascade modulates the responsiveness of the clock to light, facilitating early-night phase delays and attenuating late night phase advances. These effects were paralleled by data showing that mTOR alters the capacity of light to couple to PER1 and PER2 expression. The data also show that light-induced 5′-TOP mRNA translation is dependent on mTOR signaling. Together, these results indicate that mTOR-regulated mRNA translation plays a role shaping the phase-specific entraining effects of light.
The mTOR signal transduction pathway is a key effector of translational machinery (e.g., ribosomes and translation factors) biogenesis and mRNA translation (Sarbassov et al., 2005; Wullschleger et al., 2006; Proud, 2007, 2009). Within the CNS, a number of recent studies have revealed a key role for mTOR in neuronal physiology. Central to this are studies showing that mTOR plays an essential role in translational control of long-term synaptic plasticity and memory (Costa-Mattioli et al., 2009; Richter and Klann, 2009). Furthermore, hypothalamic mTOR functions as a cellular energy sensor, which plays a critical role in regulating food intake and energy balance (Cota et al., 2006; Mori et al., 2009). These findings, coupled with our prior work showing that photic stimulation leads to mTOR activation in the SCN, led us to test the contribution of mTOR to clock entrainment.
As a starting point for this examination, we monitored the time course of light-induced mTOR, p70 S6K, and S6 activation. These data indicate that a short light pulse (15 min) led to a rapid (within 5 min) and long-lasting (up to 4 h) activation in mTOR-dependent signaling events. As noted, p70 S6K and S6 have been implicated in regulating the translation of 5′-TOP mRNAs, a subclass of which code for translation factors and ribosomal proteins. The increased expression of translation machinery would, in turn, increase the translational capacity of the cell (Proud, 2007, 2009). To assess the ability of light to elicit the induction of 5′-TOP mRNAs, we monitored the expression of eEF1A, an elongation factor that drives the movement of charged aminoacyl-tRNA to the ribosome A site, a key event in mRNA translation/peptide elongation (Sonenberg, 1993; Proud, 1994). Our data revealed that light induced a rapid (within 5 min) and sustained (>2 h) upregulation of eEF1A in the SCN. This relatively long time course paralleled the profile of light-induced S6 phosphorylation, and thus raises the interesting prospect that light actuates a long-lasting period of enhanced mRNA translation. Rapid inducible expression (within minutes) of eEF1A has also been reported in other model systems (Tsokas et al., 2005). Interestingly, eEF1A mRNA has been shown to be docked to activated synapses, and primed for rapid translation in response to increased neuronal activity (Moon et al., 2008). In addition to eEF1A, junB also belongs to the 5′-TOP mRNA family (Staber et al., 2007; Vesely et al., 2009). Here, we also showed that the mTOR pathway contributes to the well-characterized light induction of JunB expression in the SCN (Kornhauser et al., 1992). The specificity of these findings is supported by work showing that c-Fos protein expression was not affected by rapamycin infusion. Importantly, previous studies have shown that rapamycin did not affect stimulus-induced junB (Staber et al., 2007) or eEF1A gene transcription (Huang et al. 2005; Tsokas et al., 2005). Together, these studies support the idea that rapamycin affects 5′-TOP mRNA expression via a translational-dependent mechanism within the SCN.
As part of this study, we also examined the transmitter events that couple light to mTOR activation. To this end, the infusion of a mixture of ionotropic glutamate receptor and PACAP receptor antagonists led to a marked suppression of light-evoked mTOR activation. This observation indicates that the canonical glutamatergic/PACAPergic photic input pathway drives mTOR activation. Simultaneous activation of glutamate and PACAP receptors appears to synergize at the level of mTOR activation. One possible explanation for this effect is that PACAP augments glutamate receptor-evoked neuronal excitation. Along these lines, recent work has shown that PACAP receptor activation increases the magnitude of both NMDA and AMPA-evoked currents, and enhances L-type Ca2+ calcium channel transients (Dziema and Obrietan, 2002; Michel et al., 2006). Downstream of glutamate and PACAP, the MAPK pathway appears to be a key intermediate that couples receptor activation to mTOR in the SCN. Along these lines, we recently reported that light triggers the colocalized cellular expression of activated ERK (a component of the MAPK signaling cassette) and p-p70 S6K, and that the in vivo disruption of MAPK signaling blocks light-induced mTOR activity in the SCN (Cao et al., 2008). Thus, these data, coupled with the work presented here, provide another route by which the MAPK pathway affects the clock timing process.
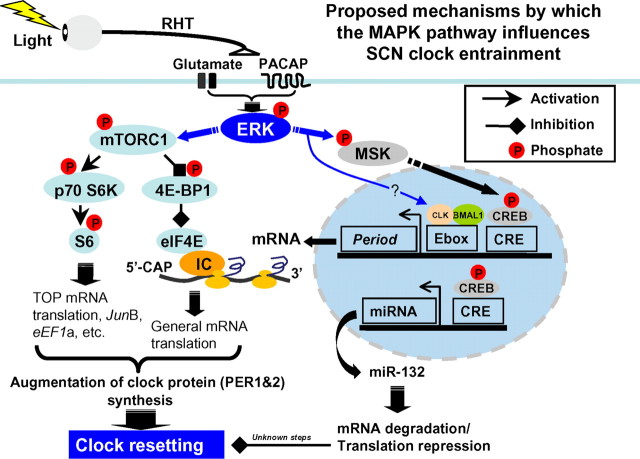
The MAPK cascade is a complex and context-specific regulator of a vast array of physiological processes. Thus, MAPK signaling has been shown to affect the cellular protein profile via the regulation of gene transcription, mRNA translation, and posttranslational protein stability (Adams and Sweatt, 2002; Carrière et al., 2008). In the earliest and most parsimonious examination of MAPK signaling and clock entrainment, Butcher et al. (2002) and Coogan and Piggins (2003) reported that the pharmacological inhibition of MAPK signaling led to a marked repression of light-induced clock entrainment. Given that MAPK signaling functions via multiple downstream effectors, a good amount of effort has been directed toward understanding specific routes by which the MAPK pathway regulates clock entrainment. For example, we have shown that activation of the MAPK cascade is required for light-evoked, MSK-dependent, CRE-mediated transcription in the SCN (Obrietan et al., 1999; Butcher et al., 2005). Interestingly, inducible PER1 transcription is mediated by MAPK signaling via the CREB/CRE pathway, thus providing one transcriptionally dependent mechanism by which the MAPK pathway regulates the clock (Travnickova-Bendova et al., 2002; Butcher et al., 2005). In addition, our recent work has revealed a role for MAPK signaling in mRNA translation regulation within the SCN. Along these lines, the MAPK pathway was shown to couple light to miR-132, thus leading to a modulation of clock entrainment (Cheng et al., 2007). Studies have also raised the possibility that MAPK signaling affects the clock through posttranslational (i.e., phosphorylation) processes. For example, MAPK signaling has been shown to evoke the phosphorylation of CLOCK (a component of the core clock timing mechanism) and affect the transactivation potential of the CLOCK/CYCLE transcriptional complex (Weber et al., 2006). Importantly, the current work reveals a new translation-dependent mechanism (i.e., the mTOR signaling pathway) by which the MAPK couples to the molecular clock. To place these data into a larger context, we have included a diagram outlining the potential signaling pathways by which MAPK signaling influences clock entrainment (Fig. 11).
Figure 11.
Schematic overview of MAPK-regulated processes that are thought to couple light to the SCN clock during the early night. Photic input from the RHT drives the release of the excitatory amino acid glutamate and the neuropeptide PACAP. Postsynaptic receptors trigger actuation of the MAPK signaling cassette, ultimately leading to the phosphoactivation of the effector kinase ERK. Two principal ERK-mediated signaling events are depicted: activation of MSK and the mTORC1 signaling complex. A potential signaling pathway from ERK to CLOCK (CLK) is also depicted (Weber et al., 2006). Signaling through MSK leads to activation of transcription factors such as CREB (Vermeulen et al., 2009), which in turn drives the induction of early response genes, including c-fos and the clock gene Period1, and potentially Period2 (Travnickova-Bendova et al., 2002). CREB also stimulates the expression of the microRNA miR-132, which works through an as yet unidentified mechanism to limit the resetting effects of light (Cheng et al., 2007). ERK-dependent activation of mTORC1 causes phosphorylation-dependent activation of a p70 S6K/S6 signaling cassette, which stimulates TOP mRNA translation, and a 4E-BP1 and eEF1A signaling cassette, which increases CAP-dependent translation. These two arms of the mTOR pathway work in conjunction to enhance the rate of mRNA processivity. With respect to clock entrainment, we posit that the ERK-dependent transcription (via MSK/CREB) and ERK-dependent translation facilitation (via the mTOR pathway) lead to a robust induction of the PERIOD protein expression. As a state variable of the clock (Reppert and Weaver, 2002), the induction of PERIOD leads to a rapid resetting of the molecular oscillator.
To gain insight into the potential role of mTOR in clock physiology, we used a pharmacological approach (i.e., rapamycin) to acutely disrupt mTOR signaling in the SCN. Rapamycin specifically inhibits mTOR as part of mTORC1, a multiprotein complex that drives mRNA translation via p70 S6K and 4E-BP1 (Sarbassov et al., 2005; Wullschleger et al., 2006; Proud, 2007, 2009). This drug delivery method proved to be very effective, completely blocking mTOR signaling in the periventricular region for at least 90 min (data not shown), which is a sufficient period of time to uncouple a 15 min light pulse from mTOR activation. Using a standard Ashoff type 1 light-stimulus paradigm, we found that the early-night phase-delaying effect of light was depressed by rapamycin. In contrast to the effects during the early night, late-night phase advances were significantly lengthened by disruption of mTOR. Of note, in the absence of light, infusion of rapamycin did not significantly affect the free-running rhythm, indicating that transient suppression of mTOR-dependent mRNA translation does not significantly alter the inherent pacemaker capacity of the SCN. Collectively, these data support the idea that protein expression mediated by both inducible transcription and mRNA translation are key components of clock entrainment. In some respects, these data are analogous to work showing that long-lasting forms of LTP are dependent on a bipartite gene transcription and mTOR-dependent mRNA translation process.
A good deal of work has implicated inducible gene expression as a key event in the actuation of clock entrainment. Here, we addressed whether the mTOR pathway may be contributing to this process. For these studies, we focused on Per gene expression, which is thought to be a key event in the light entrainment process (Shigeyoshi et al., 1997; Akiyama et al., 1999; Albrecht et al., 2001). Our data reveal that the infusion of rapamycin led to a significant decrease in early- and late-night light-evoked PER expression, indicating that PER protein synthesis is under the influence of the mTOR pathway. Of note, relative to control conditions, significant PER induction was still detected after rapamycin infusion, thus indicating that Per mRNA translation occurs in the absence of light-actuated mTOR. One possible reason for PER expression in the absence of functional mTOR (within the mTORC1 complex) is the presence of residual 4E-BP1 and p70 S6K activity, which was actuated before the pharmacological inhibition of mTOR. Additionally, it should be noted that mTOR functions as an auxiliary signaling pathway, augmenting constitutive mRNA translation, and thereby allowing the translation capacity of the cell to match increased physiological demand. Thus, in the absence of mTOR activity, the basal translational capacity of SCN neurons may be sufficient to drive moderate levels of light-evoked PER protein expression.
Several issues regarding mTOR regulation of light-evoked PER expression and clock entrainment should be noted. With respect to the early-night time point, the results showing a rapamycin-induced reduction in PERIOD protein expression are consistent with the data showing a reduction in the light-induced phase shift in behavioral rhythms. Consistency is based on work showing that PER proteins (i.e., PER1 and PER2) are state variables of the clock, and as such, attenuated expression would be expected to decrease the light-evoked phase shift (Albrecht et al., 1997; Akiyama et al., 1999). Conversely, at the late-night time point, the rapamycin-induced attenuation of PERIOD expression is not mechanistically consistent with the observed augmentation of the light-induced phase advance. There may be a number of reasons for this apparently discordant result. One potential explanation is that mTOR regulates the expression of proteins that enhance the phase-advancing effects of light. These effects would likely be independent of the effects on PERIOD proteins, and work in opposition to the decrease in PERIOD expression. Conceivably, late night disruption of mTOR translation could give rise to a transient light-evoked alteration in clock speed, which could result in an amplification of the phase shift. This explanation is based on work showing that clock speed contributes to the amplitude of the light-evoked phase shift (Daan and Pittendrigh, 1976). Clearly, additional work will be required to reveal how mTOR shapes the light-evoked proteomic profile and, in turn, clock entrainment.
In conclusion, these research findings reveal a key role for the mTOR-signaling pathway in regulating the entraining effects of light. These effects appear to be mediated by a rapid and sustained increase mTOR-dependent mRNA translation within the SCN. With these data, we can now add mTOR-dependent translation to the list of cellular signaling events that drive clock entrainment.
Footnotes
This work was supported by an National Science Foundation Grant IBN-0090974, National Institutes of Health Grants MH62335 and NS067409, and Ohio State Neuroscience Center Core Grant 5P30NS045758.
References
- Adams J, Sweatt JD. Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu Rev Pharmacol Toxicol. 2002;42:135–163. doi: 10.1146/annurev.pharmtox.42.082701.145401. [DOI] [PubMed] [Google Scholar]
- Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–1121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. mPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Antion M, Hou L, Wong H, Hoeffer CA, Klann E. mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice. Mol Cell Biol. 2008;28:2996–3007. doi: 10.1128/MCB.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs J, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Bergstrom AL, Hannibal J, Hindersson P, Fahrenkrug J. Light-induced phase shift in the Syrian hamster (Mesocricetus auratus) is attenuated by the PACAP receptor antagonist PACAP6-38 or PACAP immunoneutralization. Eur J Neurosci. 2003;18:2552–2562. doi: 10.1046/j.1460-9568.2003.03000.x. [DOI] [PubMed] [Google Scholar]
- Burnett P, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher G, Dziema H, Collamore M, Burgoon PW, Obrietan K. The p42/44 mitogen-activated protein kinase pathway couples photic input to circadian clock entrainment. J Biol Chem. 2002;277:29519–29525. doi: 10.1074/jbc.M203301200. [DOI] [PubMed] [Google Scholar]
- Butcher G, Lee B, Cheng HY, Obrietan K. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J Neurosci. 2005;25:5305–5313. doi: 10.1523/JNEUROSCI.4361-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Lee B, Cho HY, Saklayen S, Obrietan K. Photic regulation of the mTOR signaling pathway in the suprachiasmatic circadian clock. Mol Cell Neurosci. 2008;38:312–324. doi: 10.1016/j.mcn.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrière A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Cheng H, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Menaker M. NMDA as well as non-NMDA receptor antagonists can prevent the phase-shifting effects of light on the circadian system of the golden hamster. J Biol Rhythms. 1992;7:125–136. doi: 10.1177/074873049200700204. [DOI] [PubMed] [Google Scholar]
- Coogan A, Piggins HD. Circadian and photic regulation of phosphorylation of ERK1/2 and Elk-1 in the suprachiasmatic nuclei of the Syrian hamster. J Neurosci. 2003;23:3085–3093. doi: 10.1523/JNEUROSCI.23-07-03085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in noctural rodents, II The variability of phase response curves. J Comp Physiol. 1976;106:253–266. [Google Scholar]
- Daniels D. Effects of actinomycin D on memory and brain RNA synthesis in an appetitive learning task. Nature. 1971;231:395–397. doi: 10.1038/231395a0. [DOI] [PubMed] [Google Scholar]
- Dziema H, Obrietan K. PACAP potentiates L-type calcium channel conductance in suprachiasmatic nucleus neurons by activating the MAPK pathway. J Neurophysiol. 2002;88:1374–1386. doi: 10.1152/jn.2002.88.3.1374. [DOI] [PubMed] [Google Scholar]
- Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490:703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci U S A. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek DA, Ferreyra GA, Agostino PV, Murad AD, Rubio MF, Pizzio GA, Katz ME, Marpegan L, Bekinschtein TA. From light to genes: moving the hands of the circadian clock. Front Biosci. 2003;8:s285–93. doi: 10.2741/1038. [DOI] [PubMed] [Google Scholar]
- Hannibal J. Neurotransmitters of the retino-hypothalamic tract. Cell Tissue Res. 2002;309:73–88. doi: 10.1007/s00441-002-0574-3. [DOI] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hoeffer C, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, Klann E. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. The mRNA for elongation factor 1alpha is localized in dendrites and translated in response to treatments that induce long-term depression. J Neurosci. 2005;25:7199–7209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebski K, Hannan KM, Tchoubrieva EB, Hannan RD, Pearson RB. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors. 2007;25:209–226. doi: 10.1080/08977190701779101. [DOI] [PubMed] [Google Scholar]
- Jefferies H, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies H, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Nakashima H. Cycloheximide inhibits light-induced phase shifting of the circadian clock in Neurospora. J Biol Rhythms. 1990;5:159–167. doi: 10.1177/074873049000500207. [DOI] [PubMed] [Google Scholar]
- Ko C, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kornhauser J, Nelson DE, Mayo KE, Takahashi JS. Regulation of jun-B messenger RNA and AP-1 activity by light and a circadian clock. Science. 1992;255:1581–1584. doi: 10.1126/science.1549784. [DOI] [PubMed] [Google Scholar]
- Michel S, Itri J, Han JH, Gniotczynski K, Colwell CS. Regulation of glutamatergic signalling by PACAP in the mammalian suprachiasmatic nucleus. BMC Neurosci. 2006;7:15. doi: 10.1186/1471-2202-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon I, Cho SJ, Lee H, Seog DH, Jung YW, Jin I, Walikonis R. Upregulation by KCl treatment of eukaryotic translation elongation factor 1A (eEF1A) mRNA in the dendrites of cultured rat hippocampal neurons. Mol Cells. 2008;25:538–544. [PubMed] [Google Scholar]
- Mori H, Inoki K, Münzberg H, Opland D, Faouzi M, Villanueva EC, Ikenoue T, Kwiatkowski D, MacDougald OA, Myers MG, Jr, Guan KL. Critical role for hypothalamic mTOR activity in energy balance. Cell Metab. 2009;9:362–374. doi: 10.1016/j.cmet.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami N, Nishi R, Katayama T, Nasu T. Inhibitor of protein synthesis phase-shifts the circadian oscillator and inhibits the light induced-phase shift of the melatonin rhythm in pigeon pineal cells. Brain Res. 1995;693:1–7. doi: 10.1016/0006-8993(95)00633-2. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Peterson R, Schreiber SL. Translation control: connecting mitogens and the ribosome. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- Proud C. Peptide-chain elongation in eukaryotes. Mol Biol Rep. 1994;19:161–170. doi: 10.1007/BF00986958. [DOI] [PubMed] [Google Scholar]
- Proud C. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Proud C. mTORC1 signalling and mRNA translation. Biochem Soc Trans. 2009;37:227–231. doi: 10.1042/BST0370227. [DOI] [PubMed] [Google Scholar]
- Raju U, Yeung SJ, Eskin A. Involvement of proteins in light resetting ocular circadian oscillators of Aplysia. Am J Physiol. 1990;258:R256–R262. doi: 10.1152/ajpregu.1990.258.1.R256. [DOI] [PubMed] [Google Scholar]
- Rea MA, Buckley B, Lutton LM. Local administration of EAA antagonists blocks light-induced phase shifts and c-fos expression in hamster SCN. Am J Physiol. 1993;265:R1191–R1198. doi: 10.1152/ajpregu.1993.265.5.R1191. [DOI] [PubMed] [Google Scholar]
- Reppert S, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Richter J, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- Sarbassov D, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Schwartz W, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. Translation factors as effectors of cell growth and tumorigenesis. Curr Opin Cell Biol. 1993;5:955–960. doi: 10.1016/0955-0674(93)90076-3. [DOI] [PubMed] [Google Scholar]
- Staber P, Vesely P, Haq N, Ott RG, Funato K, Bambach I, Fuchs C, Schauer S, Linkesch W, Hrzenjak A, Dirks WG, Sexl V, Bergler H, Kadin ME, Sternberg DW, Kenner L, Hoefler G. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- Tee A, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem. 2003;278:718–723. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, Berghe WV, Beck IM, De Bosscher K, Haegeman G. The versatile role of MSKs in transcriptional regulation. Trends Biochem Sci. 2009;34:311–318. doi: 10.1016/j.tibs.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Vesely P, Staber PB, Hoefler G, Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat Res. 2009;682:7–12. doi: 10.1016/j.mrrev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Wakamatsu H, Takahashi S, Moriya T, Inouye ST, Okamura H, Akiyama M, Shibata S. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. NeuroReport. 2001;12:121–131. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- Weber FH, Maurer C, Kay SA. Second messenger and Ras/MAPK signalling pathways regulate CLOCK/CYCLE-dependent transcription. J Neurochem. 2006;98:248–257. doi: 10.1111/j.1471-4159.2006.03865.x. [DOI] [PubMed] [Google Scholar]
- Weng Q, Kozlowski M, Belham C, Zhang A, Comb MJ, Avruch J. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur J Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Silver R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur J Neurosci. 2004;19:1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Takahashi JS, Turek FW. Critical period for cycloheximide blockade of light-induced phase advances of the circadian locomotor activity rhythm in golden hamsters. Brain Res. 1996;740:285–290. doi: 10.1016/s0006-8993(96)00900-6. [DOI] [PubMed] [Google Scholar]
- Zylka M, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]