Abstract
Exposure to bisphenol-A (BPA) has been observed to alter developmental pathways and cell processes, at least in part, through epigenetic mechanisms. This study sought to investigate the effect of BPA on microRNAs (miRNAs) in human placental cells. miRNA microarray was performed following BPA treatment in three immortalized cytotrophoblast cell lines and the results validated using quantitative real-time PCR. For functional analysis, overexpression constructs were stably transfected into cells that were then assayed for changes in proliferation and response to toxicants. Microarray analysis revealed several miRNAs to be significantly altered in response to BPA treatment in two cell lines. Real-time PCR results confirmed that miR-146a was particularly strongly induced and its overexpression in cells led to slower proliferation as well as higher sensitivity to the DNA damaging agent, bleomycin. Overall, these results suggest that BPA can alter miRNA expression in placental cells, a potentially novel mode of BPA toxicity.
Keywords: placenta, microRNA, microarray, bisphenol A, epigenetics
1. Introduction
Bisphenol A (BPA), an industrial plasticizer, is a key monomer in the production of polycarbonate plastic, a fungicide, a flame retardant, and a component of epoxy resins used as food-contact surface coating for a variety of objects including cans and metal jar lids [1]. Because of the increased use of both polycarbonate plastics and epoxy resins in the coatings of food cans and plastic water bottles, the frequency of human exposure to BPA has steadily increased in recent years [2].
Exposure of the developing fetus to BPA is of particular concern as the compound readily crosses the placental barrier and accumulates both in the placenta and in the fetus [3, 4]. The placenta, comprised of both fetal tissue derived from the chorionic sac and maternal tissue from the endometrium, is the first complex mammalian organ to form in pregnancy and is required for the early development of all viviparous species [5]. It not only allows the transport of nutrients and waste products between the mother and the fetus, but also provides a barrier protecting the fetus against harmful substances in the mother’s circulation. Importantly, the placenta also serves as an endocrine organ, providing peptide and steroid hormones, which influence placental, fetal, and maternal metabolism and development.
BPA’s toxicity has been linked to interference with endogenous estrogens and thyroid hormone, and to the disruption of reproductive, immune, and central nervous systems [6–10]. These functions may be mediated through BPA’s binding to the estrogen receptor, though they may also be related to nuclear-receptor independent activation of cell signaling systems [11]. Evidence is weaker regarding BPA’s potential as a carcinogen, although literature on the association of BPA with increased cancer risk in adult life through fetal exposures or across generations suggests that BPA may elicit epigenetic effects [12, 13]. Further, experiments in mice have demonstrated that BPA exposure can cause altered DNA methylation patterns in cell signaling genes, suggesting that BPA can exert its effects through epigenetic mechanisms [13–15]
MicroRNAs (miRNAs) are approximately 22 nucleotide long non-coding RNAs capable of regulating gene expression post-transcriptionally through imperfect complementarity with a target mRNA, and are considered a mode of epigenetic regulation [16]. As they potentially target up to one third of human mRNAs, miRNAs regulate myriad cellular processes, playing pivotal roles in developmental timing and patterning [16, 17]. Our group and others have demonstrated that miRNAs are susceptible to cellular stresses and environmental exposures, such as folate deficiency, exposure to arsenic and nicotine, and ionizing radiation, and diesel exhaust particles [18–21]. Several studies have suggested that miRNA expression modulation may be a mechanism through which toxicants can exert both developmental and carcinogenic effects [22, 23].
The effect of BPA on miRNA expression has yet to be studied and the high prevalence of fetal exposure to BPA from maternal sources makes the placenta a particularly important model. The specific risks associated with fetal exposures and the key role of the placenta in mediating the potential toxic effects of BPA make the study of molecular alterations at different stages of fetal development particularly important. Here we have used an in vitro cell culture system to examine the effect of BPA exposure on alterations in miRNA expression, and our results suggest that changes in miRNA expression may be critical mediators of BPA toxicity.
2. Materials and Methods
2.1. Cell Lines and BPA exposure conditions
Human SV40 transformed placental cell lines 3A, TCL-1, and HTR-8 were cultured in phenol-free RPMI 1640 (Invitrogen, Inc., Gaithersburg, MD). Phenol-free growth medium was supplemented with 10% charcoal-stripped fetal bovine serum (FBS; Invitrogen) and 1% penicillin-streptomycin for both 3A and TCL-1 cell lines. HTR-8 cell line growth medium was supplemented with 5% charcoal stripped fetal bovine serum (FBS; Invitrogen) with no penicillin-streptomycin. For the microarray experiment, 3A, TCL-1, and HTR-8 cells were treated over a six-day period with 25 ng/µL of BPA, a dose which causes only minimal disruption of cell proliferation as determined by a dose-response experiment (data not shown) and which was within the range of doses previously used to study the effects of BPA on placental cell lines [24]. These three placental cell lines represent three different stages and aspects of placental development and were selected to examine the effects of BPA in models of different time-points during pregnancy. 3A cells are first-trimester villous cells, HTR-8s are first trimester extravillous cells and TCL-1s are third trimester extravillous cells. For BPA exposure experiments (microarray and quantitative real-time PCR validation), 105 cells were seeded in 10 cm plates in normal growth medium. After 24 hours, the medium was replaced with medium containing BPA prepared to final concentrations ranging from 0.25 to 25.0 ng/µL in DMSO (0.1% final concentration) or vehicle alone (0.1% DMSO). Cells were cultured for 6 days, with exposure medium refreshed on days 2 and 4. The 6-day period of exposure was chosen to mimic a long-term chronic exposure, but to avoid sub-culturing of the cells, and has been previously described [18]. All experimental and control conditions were performed in triplicate.
2.2. RNA isolation, microarray processing, and analysis
Following appropriate incubation times, the cells were harvested by trypsinization and centrifugation, the medium discarded, and the pelleted cells washed with cold PBS. Total RNA was isolated immediately from 106 cells with the mirVANA RNA Isolation Kit (Ambion, Inc., Austin, TX) according to manufacturer’s protocol. RNA was quantified using the ND-1000 spectrophotometer (Nanodrop, Wilmington, DE), aliquoted, and stored at −80°C until needed. An aliquot of each sample was examined for RNA integrity and quality using the Agilent Bioanalyzer, following manufacturer’s protocol. Samples for miRNA profiling studies were processed by NCode Rapid miRNA Expression Profiling Services (Invitrogen, San Diego, CA) according to the standard operating procedures of the company. Each RNA sample was spiked with 1µL of NCode Multi-Species miRNA MicroArray Controls, and Poly-A tailing reactions were set up following procedures recommended in the manual. Poly-A tailed reactions were ligated to labeled DNA polymers by using the 6× Alexa Fluor 3 (A3) or 5 (A5) Rapid Ligation Kit, following protocol. The two reactions were combined into one tube and the volume was reduced by half in a SpeedVac Concentrator. BSA (50mg/µL) was then added for a total volume of 28.5 µL. The sample was incubated with 28.5 µL of 2× enhanced hybridization buffer at 65°C for 10 minutes and then loaded onto NCode Human miRNA Microarrays V3. The arrays were mounted with the Maui Mixer SL and hybridized overnight at 52°C with constant mixing. The following day, arrays were washed following standard protocol and scanned using a GenePix 4000B microarray scanner. The arrayed images were annotated and analyzed using GenePix software and the .GAL files containing the array list for human samples. Results from the microarray analysis are being deposited in the GEO archive.
2.3. Statistical Analysis
A latin squares or loop design for pairing of samples on each array was utilized, to allow for appropriate controlling of dye labeling and fluorescence efficiency, and to allow for immediate comparison across treatments [25]. This model-based analysis allows for simultaneous normalization and differential marker detection. P-values describing the difference in miRNA expression between treated and control groups were calculated through bootstrapping of the residuals of the model. To control for the multiple comparisons performed in each analysis, false discovery rate (FDR) analysis was applied to these P-values, following the methods of Benjamini and Hochberg [26] and Storey and Tibshirani [27]. FDR was implemented using the QValue software, in the R statistical package, setting the lambda range from 0 to 0.95, and the smoother methods of [27] for πO determination. miRNAs with an FDR of less than 20% (q-value <0.2) were considered for follow-up of their differential expression. All hierarchical clustering analyses were carried out using Cluster 3.0 (Stanford University) with Euclidian distance as the distance metric and complete linkage between clusters.
2.4. Quantitative reverse-transcription real-time PCR
TaqMan miRNA Assays (Applied Biosystems) were used to quantify mature miRNA. cDNA was synthesized by priming with miRNA-specific looped primers for the miRNAs of interest and RNU48, as a universally-expressed endogenous control (Applied Biosystems). 1 ng/µl was used for each reverse transcription (RT) reaction along with other RT components, per manufacturer’s specifications. 15 µL reactions were incubated in an Applied Biosystems GeneAmp PCR system 9700 for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and held at 4°C. Quantitative real time PCR was performed as previously described [28] with the following exception: all reactions were run in duplicate, on an ABI 7500 Fast Real Time PCR Detection System. All quantitative real time PCR data were analyzed using the comparative CT method, normalizing against expression of RNU48.
2.5. miR-146a overexpression construct transfection
Single stranded oligonucleotides were designed containing the mature miR-146a sequence, represented in bold (top strand: 5’ TGCTGTGAGAACTGAATTCCATGGGTGTTTTGGCCACTGACTGACGACTACACATCAGCGATTT 3′; bottom strand: 5’ CCTGTGAGAACTGAACCATGGGTGTCAGTCAGTGGCCAAAACACCCATGGAATTCAGTTCTCAC 3’). The oligos were annealed and the now double-stranded oligos were ligated into Block-iT™ Pol II miR RNAi Expression Vectors according to the manufacturer’s instruction (Invitrogen). Using One Shot TOP10 Transformation Protocol, TOP10 Competent E. coli (Invitrogen) were transformed with the miR-146a plasmid or a negative control plasmid (included in kit) containing a sequence that is processed into a mature miRNA but does not target any known vertebrate genes. Transformants were analyzed by sequencing to ensure proper incorporation of oligos into plasmids. Transformants were expanded and plasmids purified by maxi-prep (Qiagen). Cells were seeded to 90% confluency for transfection in 6-well plates. The vectors containing either miR-146a or negative control plasmid were transfected into 3A cells using Lipofectamine-2000 (Invitrogen) per manufacturer’s guidelines. Stable transfectants were selected using 4 µg/ml blasticidin in medium, and stable expression followed using fluorescence microscopy to determine GFP expression. Expression of the miR-146a or negative control miRNA was confirmed using RT-PCR in all stable lines.
2.6. Proliferation Assay
Ninety-six well plates were seeded with 7000 cells per well in replicates of 12. At indicated time-points, cells were stained with 20 µL per well of CellTiter 96 Aqueous One Solution (Promega) for one hour. A SpectraMax M2 and SoftMax Pro software (Molecular Devices) were used for measurement and analysis of absorbance. Absorbance was measured at 6 hours after seeding, once cells adhered, followed by readings at 24, 48, and 72 hours.
2.7. Colony formation assay
Ten centimeter dishes were seeded with 3×103 cells and allowed to adhere for 24 hours, after which they were treated with individual chemicals at the following doses: 0, 2.5 or 25 µg/ml BPA in DMSO; or 0, 2.5, or 5 µg/ml bleomycin in DMSO. For BPA exposure, cells were treated with exposure medium for 6 days, and exposure medium was refreshed on days 3 and 5. Due to the toxicity of bleomycin, cells were exposed to the bleomycin in serum-free medium for 1 hour, and then returned to regular medium conditions for the remaining 6 days. All experimental and control conditions were performed in triplicate.
3. Results
3.1. miRNA expression profiles of placental cell lines following Bisphenol A exposure
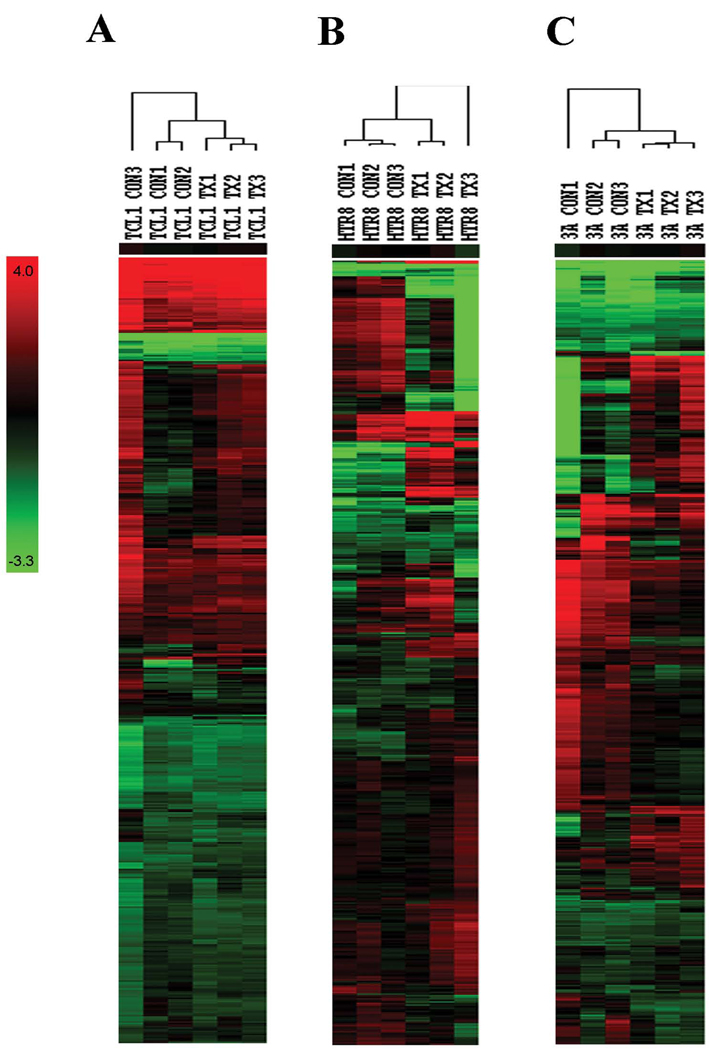
To examine the effects of BPA exposure on miRNA expression in placental cell lines, 3A, TCL-1, and HTR-8 cells were treated over a six-day period with 25 ng/µL of BPA and a microarray platform was used to determine microRNA expression of all the known miRNA sequences based on the Sanger Institutes microRNA database Release 10.0 as well novel proprietary miRNAs in multiple species. Data from the microarray analysis were visualized using heat maps generated using unsupervised hierarchical clustering based on all the normalized signal data for each cell line and both treatment groups. These heat maps and their associated dendrograms demonstrate that the three cell lines each show disparate patterns of miRNA expression. While BPA treatment did not result in obvious segregation of treatment and control groups in TCL-1 cells (Figure 1A), BPA exposed HTR-8 and 3A cells formed distinct clusters away from their control counterparts (Figure 1B and C).
Figure 1.
Unsupervised hierarchical clustering of BPA-treated (TX) and control (CON) based on all miRNA spotted on the microarray in (B) TCL-1 cells, (C) HTR-8 cells, and (D) 3A cells. Columns represent samples and rows represent individual miRNAs. Colors represent fold change intensity.
Overall, a total of 25 miRNAs were considered differentially expressed at a false discovery rate of <0.2 in 3A cells, and 60 miRNA were considered significantly differentially expressed in HTR-8 cells, including proprietary novel miRNAs (Supplementary Table 1). Interestingly, the majority of miRNAs that were differentially expressed in BPA-treated 3A cells were also differentially expressed in BPA-treated HTR-8 cells, with 21 miRNAs in common between the two. Additionally, this analysis revealed that following FDR correction, no miRNAs were identified as significantly altered by BPA exposure in TCL-1 cells. Table 1 lists the annotated human miRNAs found to be most significantly differentially expressed in 3A and HTR-8 cell lines.
Table 1.
Significantly differentially expressed annotated human miRNA in BPA treated cells vs. Control
| 3A | ||
| miRNA | Fold change | q-value |
| hsa-let-7f | 2.80 | 0.14 |
| hsa-let-7g | 2.85 | 0.14 |
| hsa-miR-146a | 2.77 | 0.17 |
| hsa-miR-21 | 3.50 | 0.13 |
| HTR-8 | ||
| miRNA | Fold Change | q-value |
| hsa-let-7f | 4.89 | 0.07 |
| hsa-let-7g | 4.26 | 0.07 |
| hsa-let-7i | 3.16 | 0.11 |
| hsa-miR-106a | 2.67 | 0.18 |
| hsa-miR-106b | 3.04 | 0.10 |
| hsa-miR-146a | 7.87 | 0.07 |
| hsa-miR-155 | 2.92 | 0.08 |
| hsa-miR-16 | 3.9 | 0.10 |
| hsa-miR-19b | 2.68 | 0.19 |
| hsa-miR-20a | 4.57 | 0.07 |
| hsa-miR-21 | 9.22 | 0.07 |
| hsa-miR-26b | 3.43 | 0.07 |
| hsa-miR-29a | 3.21 | 0.07 |
| hsa-miR-335 | 4.31 | 0.07 |
| hsa-miR-376c | 2.74 | 0.10 |
3.2. Quantitative real time PCR validation of miRNA expression
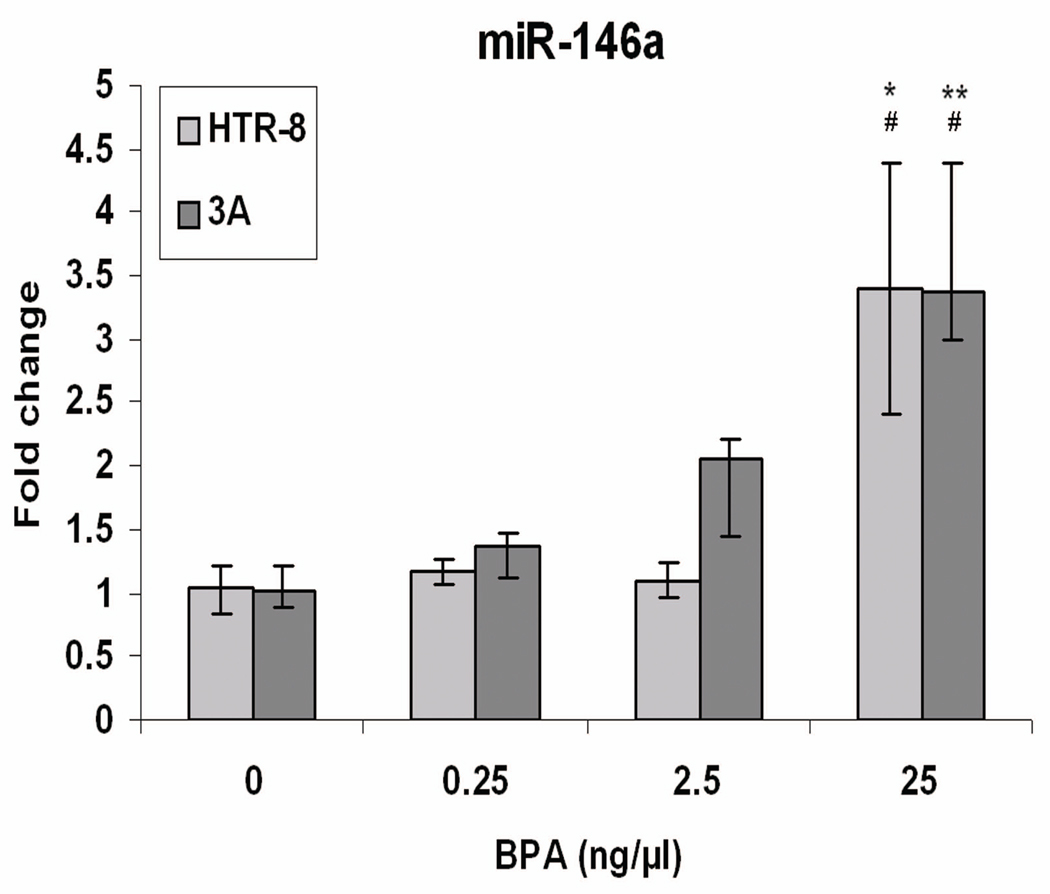
The expression of the four annotated human miRNAs that were identified from the microarray analysis as differentially expressed in both 3A and HTR-8 cell lines (let-7f, let-7g, mir-21, and mir-146a) was assayed by quantitative real-time PCR after repeating the BPA exposure experiment in all three cell lines. Figure 2 illustrates the change in miR-146a expression with increasing doses of BPA. miR-146a was confirmed as significantly upregulated in both 3A and HTR-8 cells (P < 0.01 and P < 0.05, respectively) and it was shown to vary significantly across treatments in both cell types (ANOVA; P < 0.05). TCL-1 cells showed no detectable miR-146a expression, consistent with the results of the microarray. Results of validation of the three other miRNAs are shown in Supplementary Figure 1.
Figure 2.
Quantitative real time PCR analysis of miR-146a expression in cells treated with increasing doses of BPA for 6-days. Fold change refers to the fold change in expression of selected miRNAs in BPA treated triplicates compared to the average of control triplicates. Values represent the mean ±SE of three individual experimental samples run in triplicate.
* P < 0.05, for t-test between control (0) and 25 ng/µl dose
** P < 0.01, for t-test between control (0) and 25 ng/µl dose
# P < 0.05, for ANOVA across all treatments
3.3. Effects of miR-146a overexpression in 3A placental cell lines
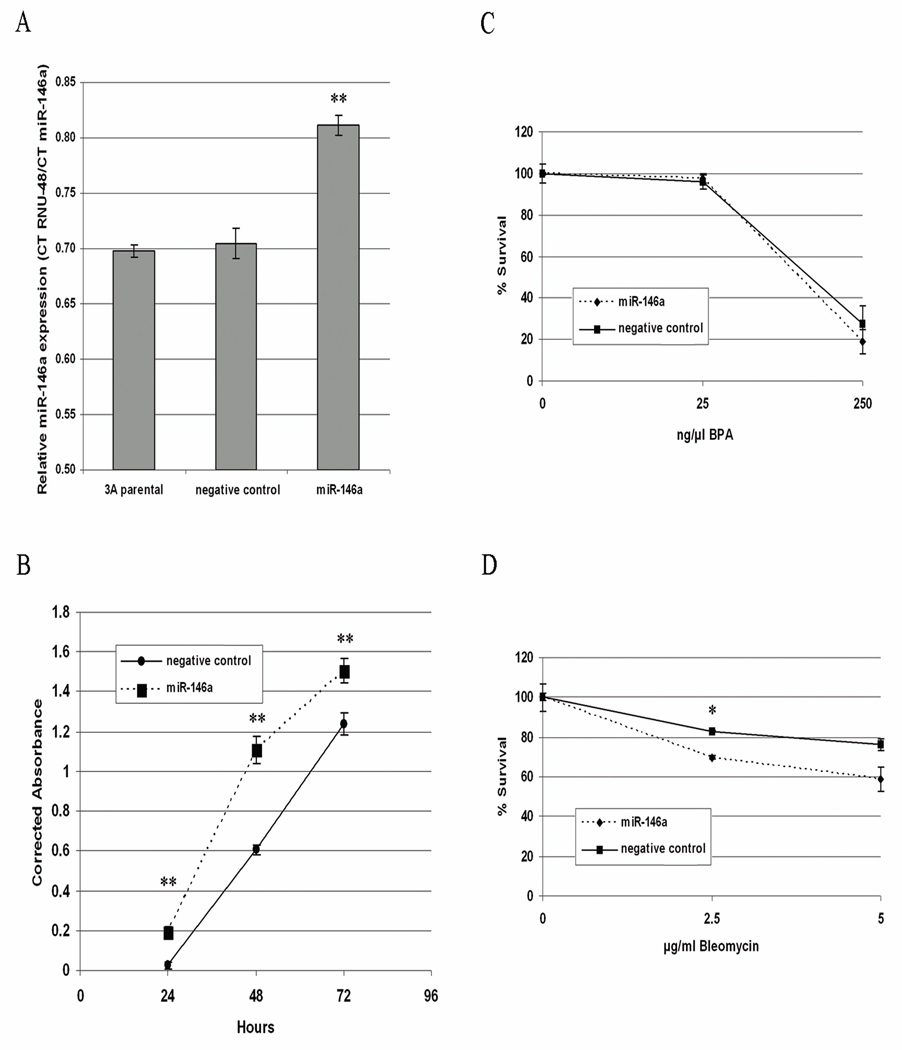
miR-146a was found to be upregulated in both 3A and HTR-8 cells in response to BPA treatment, and was previously found to be upregulated in response to cellular stress including folate deficiency in human lymphoblast cells [18]. This suggests that miR-146a may be commonly induced by stress and toxicant exposure, and thus we sought to examine if there are functional consequences of its overexpression. Quantitative real-time PCR results showed that miR-146a was significantly upregulated in cells transfected with the miRNA compared with control-transfected cells (P < 0.001; Figure 3A), corresponding to a 32-fold increase in expression. In order to examine the effects of miR-146a overexpression on cell growth, an MTS proliferation assay was used to measure normal proliferation of transfected cells. Results showed that cells which overexpress miR-146a had a significantly decreased rate of cell proliferation compared to control-transfected cells at 24, 48 and 72-hour time-points (P < 0.0001 for all comparisons; Figure 4B). We used a clonogenic survival assay to determine whether upregulation of miR-146a alters cells’ responses to various doses of BPA or bleomycin. Though no significant differences were observed in BPA-treated cells, cells over-expressing miR-146a showed significantly increased sensitivity to bleomycin at the 2.5 µg/ml doses (P = 0.002; Figure 4C).
Figure 3.
Experiments with 3A cells transfected with miR-146a overexpression construct or scrambled control miRNA. (A) Quantitative real time PCR showing expression of miR-146a transfected cells and non-transfected parental 3A cells. (B) Cell proliferation assay showing corrected absorbance at 24, 48 and 72 hours after cell attachment to culture dish. (C) Clonogenic survival of transfected cells following exposure to increasing doses of BPA or (D) bleomycin.
* P < 0.01
** P < 0.001
4. Discussion
The commercial use of BPA is increasing in frequency and research has shown that exposure to BPA, for example through plastic beverage containers, can significantly increase urinary BPA concentrations [29, 30]. The increase in maternal exposure during critical windows of fetal development is of particular concern as even low-dose exposure to BPA can lead to adverse effects in laboratory animals including delayed developmental outcomes in offspring of exposed mothers [31–33]. Importantly, a recent study demonstrated that low dose exposure to BPA also leads to apoptosis and necrosis in primary human placental cytotrophoblast cells [24]. Though the mechanism of BPA toxicity is still unclear, previous studies with BPA have demonstrated its ability to alter DNA methylation patterns [13–15], suggesting an epigenetic component to its toxicity. This work has explored an alternative epigenetic mechanism by determining the effects of the toxicant on miRNA expression following exposure of three different placental cell lines to BPA.
A miRNA microarray showed a total of 85 miRNAs to be significantly differentially expressed in BPA-treated 3A and HTR-8 cells compared to mock-treated controls, twenty one of which were altered in both cell lines. A heat map from hierarchical clustering also showed the overall miRNA profiles of the two cell lines to be similar. This overlap may be accounted for by the fact that both 3A and HTR8 cells are derived from the first trimester placenta, although they play distinct roles in the tissue. 3A cell lines are derived from first trimester villous cells, which ultimately give rise to the syncytiotrophoblast, a layer of cells which has important metabolic and endocrine functions in the placenta. Meanwhile, HTR-8 cells are derived from first trimester extravillous cells, which are involved in invasion and anchoring of the placenta into the maternal decidua. Though these data suggest similar responses to BPA exposure at the level of miRNA expression, it is reasonable to predict that these alterations may have different implications for these tissues based on their disparate functions. Similarly, the lack of significantly differentially expressed miRNAs observed in TCL-1 cells may reflect the origin of these cells. As they are third trimester extravillous cells, they are essentially terminally differentiated and may not represent the critical window for exposure to BPA.
miR-146a was the only miRNA validated by qRT-PCR as significantly upregulated in both 3A and HTR-8 cells with BPA treatment. This miRNA also showed the highest fold-change in up-regulation in these validation experiments in both cell lines, though its expression was not detectable in TCL-1 cells. Past studies have demonstrated that miR-146a can have both tumor suppressive and oncogenic roles in different cancers [34–36] and that its expression can be altered by arsenic treatment and folate deficiency in the human lymphoblastoid cell line TK6 [18]. Additionally, miR-146a has been shown to be involved in immunity, potentially regulating Toll-like receptor and cytokine signaling [37]. Together with our observations, these studies demonstrate the importance of this miR-146a in response to environmental stress and suggest that it may be environmentally labile, and have important roles in inflammatory response and cell growth.
Our work also demonstrates that stable overexpression of miR-146a in 3A placental cell lines leads to a significant decrease in proliferation, a result that is consistent with several other studies (Bhaumik et al., 2008; Lin et al., 2008). Additionally, we show that miR-146a overexpression increased sensitivity to the DNA damaging agent bleomycin. Bleomycin is a glycopeptide known to induce DNA double strand breaks, inhibit DNA synthesis and repair and, to some extent, protein and RNA synthesis as well [38]. As a test mutagen, bleomycin exposure can reveal cells that have compromised DNA repair mechanisms therefore suggesting that targets of miR-146a may be involved in the DNA damage response. Exposure to BPA, through its alterations of cellular miRNA, may interfere with DNA repair thus increasing the cells’ sensitivity to various other agents, both exogenous and endogenous.
The increasing presence of BPA in the environment indicates a need for a thorough examination of human responses to BPA exposure, including a consideration of molecular effects. The importance of studying BPA’s effects on the placenta is underscored by BPA’s tendency to accumulate in that tissue [3] and the general importance of placenta in fetal development [39]. The ability of BPA to alter miRNA expression in the placenta might reflect a mechanism of BPA toxicity which may carry detrimental consequences for the developing fetus. Given the growth-inhibiting and growth-promoting functions of several human miRNAs in the placenta, normal miRNA regulation in placental tissue is extremely important. This study serves to demonstrate the utility of miRNA expression profiles in bettering our understanding of the action of environmental toxicants and may aid in improved prevention, diagnosis, and treatment of exposure-related effects.
Supplementary Material
Acknowledgments
Funding Information
This work was supported by [National Institutes of Health NCRR P20 RR018728, P20 RR016457]; [National Institutes of Health – National Institute of Environmental Health Sciences P42 ES013660] and [National Science Foundation- The Experimental Program to Stimulate Competitive Research 0554548].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
References
- 1.vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environ Health Perspect. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang JH, Kondo F, Katayama Y. Human exposure to bisphenol A. Toxicology. 2006;226:79–89. doi: 10.1016/j.tox.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110:A703–A707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi O, Oishi S. Disposition of orally administered 2,2-Bis(4-hydroxyphenyl)propane (Bisphenol A) in pregnant rats and the placental transfer to fetuses. Environ Health Perspect. 2000;108:931–935. doi: 10.1289/ehp.00108931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabbe SG, Niebyl JR, Simpson JL. Obstetrics : normal and problem pregnancies. 5th ed. Philadelphia, PA: Churchill Livingstone/Elsevier; 2007. [Google Scholar]
- 6.Rubin BS, Murray MK, Damassa DA, King JC, Soto AM. Perinatal exposure to low doses of bisphenol A affects body weight, patterns of estrous cyclicity, and plasma LH levels. Environ Health Perspect. 2001;109:675–680. doi: 10.1289/ehp.01109675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacLusky NJ, Hajszan T, Leranth C. The environmental estrogen bisphenol a inhibits estradiol-induced hippocampal synaptogenesis. Environ Health Perspect. 2005;113:675–679. doi: 10.1289/ehp.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriyama K, Tagami T, Akamizu T, Usui T, Saijo M, Kanamoto N, et al. Thyroid hormone action is disrupted by bisphenol A as an antagonist. J Clin Endocrinol Metab. 2002;87:5185–5190. doi: 10.1210/jc.2002-020209. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed SA. The immune system as a potential target for environmental estrogens (endocrine disrupters): a new emerging field. Toxicology. 2000;150:191–206. doi: 10.1016/s0300-483x(00)00259-6. [DOI] [PubMed] [Google Scholar]
- 10.Palanza P, Gioiosa L, vom Saal FS, Parmigiani S. Effects of developmental exposure to bisphenol A on brain and behavior in mice. Environ Res. 2008;108:150–157. doi: 10.1016/j.envres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Wetherill YB, Akingbemi BT, Kanno J, McLachlan JA, Nadal A, Sonnenschein C, et al. In vitro molecular mechanisms of bisphenol A action. Reprod Toxicol. 2007;24:178–198. doi: 10.1016/j.reprotox.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Keri RA, Ho SM, Hunt PA, Knudsen KE, Soto AM, Prins GS. An evaluation of evidence for the carcinogenic activity of bisphenol A. Reproductive toxicology (Elmsford, N.Y. 2007;24:240–252. doi: 10.1016/j.reprotox.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 17.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 18.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 19.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2008 doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect. 2009;117:1745–1751. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–298. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 24.Benachour N, Aris A. Toxic effects of low doses of Bisphenol-A on human placental cells. Toxicol Appl Pharmacol. 2009;241:322–328. doi: 10.1016/j.taap.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 27.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carwile JL, Luu HT, Bassett LS, Driscoll DA, Yuan C, Chang JY, et al. Use of Polycarbonate Bottles and Urinary Bisphenol A Concentrations. Environmental Health Perspectives. 2009 doi: 10.1289/ehp.0900604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Miao M, Herrinton LJ, Wu C, Yuan W, Zhou Z, et al. Bisphenol A levels in blood and urine in a Chinese population and the personal factors affecting the levels. Environ Res. 2009;109:629–633. doi: 10.1016/j.envres.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Vandenberg LN, Maffini MV, Wadia PR, Sonnenschein C, Rubin BS, Soto AM. Exposure to environmentally relevant doses of the xenoestrogen bisphenol-A alters development of the fetal mouse mammary gland. Endocrinology. 2007;148:116–127. doi: 10.1210/en.2006-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howdeshell KL, Hotchkiss AK, Thayer KA, Vandenbergh JG, vom Saal FS. Environmental toxins: Exposure to bisphenol A advances puberty. Nature. 1999;401:763–764. doi: 10.1038/44517. [DOI] [PubMed] [Google Scholar]
- 33.Zhou R, Zhang Z, Zhu Y, Chen L, Sokabe M. Deficits in development of synaptic plasticity in rat dorsal striatum following prenatal and neonatal exposure to low-dose bisphenol A. Neuroscience. 2009;159:161–171. doi: 10.1016/j.neuroscience.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 35.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barranco SC, Humphrey RM. The effects of bleomycin on survival and cell progression in Chinese hamster cells in vitro. Cancer Res. 1971;31:1218–1223. [PubMed] [Google Scholar]
- 39.Iyengar GV, Rapp A. Human placenta as a 'dual' biomarker for monitoring fetal and maternal environment with special reference to potentially toxic trace elements. Part 3: toxic trace elements in placenta and placenta as a biomarker for these elements. Sci Total Environ. 2001;280:221–238. doi: 10.1016/s0048-9697(01)00827-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.