Abstract
Mesenchymal stem cells (MSCs) hold a great promise for application in several therapies due to their unique biological characteristics. In order to harness their full potential in cell-or gene-based therapies it might be advantageous to enhance some of their features through gene delivery strategies. Accordingly, we are interested in developing an efficient and safe methodology to genetically engineer human bone marrow MSC (BM MSC), enhancing their therapeutic efficacy in Regenerative Medicine. The plasmid DNA delivery was optimized using a cationic liposome-based reagent. Transfection efficiencies ranged from ~2% to ~35%, resulting from using a Lipid/DNA ratio of 1.25 with a transgene expression of 7 days. Importantly, the number of plasmid copies in different cell passages was quantified for the first time and ~20,000 plasmid copies/cell were obtained independently of cell passage. As transfected MSC have shown high viabilities (>90%) and recoveries (>52%) while maintaining their multipotency, this might be an advantageous transfection strategy when the goal is to express a therapeutic gene in a safe and transient way.
1. Introduction
Mesenchymal stem cells (MSCs) transplantation has been proven to be an efficient method to treat a large spectrum of diseases. It is noteworthy that both autologous and allogeneic MSCs have not induced host immunoreactivity upon local transplantation or systemic administrations. Therefore, MSCs are an ideal carrier to deliver genes into the tissues of interest for gene therapy applications [1]. Genetically manipulated MSC can be used in different therapeutic strategies, either as immunosuppressive agents or as engineered cells to secrete a variety of different proteins in vitro and in vivo that could potentially treat a variety of serum protein deficiencies and other genetic or acquired diseases, such as bone, cartilage, and bone marrow (BM) disorders. In addition, the ability to genetically modify these MSCs would further contribute to Tissue Engineering settings enabling the selective enhancement of specific differentiation pathways [2]. As MSCs are not immunologically rejected and possibly home to damaged tissues, they represent an opportunity to deliver therapeutic proteins. The advantages of MSC gene therapy over pharmaceutical agents are the potential of long-term effects after a single intervention and the local expression of the desired gene [3]. Gene therapy can increase survival of engrafted stem cells when transgenes are inserted into the cell to prevent or reduce apoptosis and inflammatory injury.
Despite the promise of stem cell-based gene therapy to have an impact on human health, technical challenges remain to be solved in order to harness the full potential of stem cells. Presently, the widely used method to transfer genes to MSC is performed through defective viruses, such as adenovirus, lentivirus, and retrovirus [4]. When MSCs are used to compensate or correct a genetic pathology and must express the therapeutic gene for the duration of a patient's life (permanent expression), integrating viruses, such as lentivirus or retrovirus, are preferred because of their well-known capacity for long-term expression. On the contrary, when MSCs are used to treat noninherited diseases and are only required to express the therapeutic gene for a short period of time (transient expression), nonintegrating vectors including adenoviruses and nonviral gene delivery systems are preferred [5]. Although these cells can be more efficiently modified using viral methods, safety issues including mutagenesis, toxicity, and the immunogenicity of the virus itself remain considerable concerns. Alternatively, and despite its less efficiency compared to viral methods, the advantage of using nonviral methods resides on its safety, demonstrating no immunogenicity, negligible toxicity, and easier preparation, and having the ability to carry larger therapeutic genes [6]. Overall, by using plasmids it is possible to modify genes or introduce new ones to make the cell undergo apoptosis or survive longer, secrete proteins or switch off genes, differentiate or not differentiate, and even proliferate [7]. For these reasons, there is an increased interest in the development of a safe and efficient nonviral gene delivery system that can overtake the limitations associated to the viral approach. Importantly, for in vitro analysis and subsequent use for transplantation, the selected system should not affect MSC proliferation and differentiation after transfection. Among the current nonviral methods, liposome carriers and electroporation-based gene transfer techniques were determined most efficient for transfecting MSC [8]. Electroporation, while effective in transfecting stem cells, is rather harsh and leads to excessive cell death [5, 9–11].
In a few reports, some lipofection reagents were described to successfully introduce transgenes and small interfering RNAs (siRNAs) into MSC, while these cells have maintained their proliferation capacity and ability to differentiate into different mesodermal lineages (bone, cartilage and fat) without loss of transgene expression [12]. The main reason why cationic liposomes have demonstrated lower transfection efficiencies compared to viral vectors is that these nonviral vectors are not provided with specific devices for controlling intracellular gene trafficking, turning its optimization essential [13, 14]. So far, the development of liposomal vectors has been an empirical process taking into account the measurement of liposomal transfection by the percentage of cells expressing a reporter protein encoded by a plasmid determined either by microscopy or flow cytometry. Nevertheless, although gene expression is the main goal of transfection, it depends on several factors including plasmid uptake, intracellular plasmid stability, plasmid access to nucleus, and finally transcription and translation efficiency. In this context, more promising enhanced vectors might be developed considering that all barriers that delivered DNA must traverse in its journey from the outside to the nucleus of target cells. Furthermore, the mechanism of nuclear transport of the delivered gene, as well as the relationship between the amounts of gene delivered to the nucleus and gene expression, is unclear at the present. Thus, a quantitative understanding of the intracellular trafficking of plasmids delivered by these vectors is required to understand the factors governing the efficiency of gene expression [14]. To achieve this goal, the quantification of the number of plasmid molecules that enter the cells can not only contribute to understanding the underlying mechanisms of liposomal gene delivery but it can also be a helpful tool to optimize the liposomal vectors increasing their transfection efficiencies. Herein, by using this approach we were able to determine the optimal amount of delivered DNA required for the best transgene expression and compare delivery efficiency in different cell passages of human adult BM-derived MSC. Overall, we believe our findings are extremely useful towards the maximization of gene delivery to human MSC, without compromising cell function and viability, when the final goal is (i) to express a therapeutic gene in vivo in a safe and transient way (ii) or to improve their in vitro expansion or controlled differentiation by overexpression of stemness- or specific lineage commitment-related genes/proteins, respectively.
2. Materials and Methods
2.1. Plasmid Construction, Production, and Purification
pVAX-GFP (3697 bp) plasmid was obtained by modification of the commercial plasmid pVAX1lacZ (6050 bp, Invitrogen), by replacement of the β-galactosidase reporter gene by the enhanced Green Fluorescent Protein (eGFP, referred to as GFP thereafter) gene. The details of the construction are described elsewhere [15].
The plasmid contains the human cytomegalovirus (CMV) immediate-early promoter, a ColE1 type origin of replication and the kanamycin resistance gene for bacterial selection. Plasmid DNA (pDNA) was obtained by growing E.coli cultures (harbouring pVAX-GFP) overnight, in 2 L shake-flasks containing 250 mL LB medium and antibiotics (30 μg/mL of kanamycin). Plasmid purification was performed according to the Endotoxin-free Plasmid DNA Purification Kit protocol (Macherey-Nager). The concentration of purified pDNA solutions was assayed by spectrophotometry at 260 nm (Nanodrop, Thermo Scientific), and DNA integrity was confirmed by DNA agarose gels stained with ethidium bromide.
2.2. Real-Time PCR for Determination of MSC Plasmid Content
Quantitative Real-Time PCR was performed by amplification of a 108 bp sequence within the GFP gene (forward primer: 5′-TCG AGC TGG ACG GCG ACG TAA A-3′; reverse primer: 5′-TGC CGG TGG TGC AGA TGA AC-3′). PCR reactions were carried out in a Roche LightCycler detection system using the FastStart DNA Master SYBR Green I kit (Roche Diagnostics) as recommended by the manufacturer. Each 20 μL of final reaction volume contained 2.0 μL of the 10x SYBR Green mixture, 0.4 μL of each primer (0.4 μM final concentration), 1.6 μL of MgCl2 solution (3.0 mM final concentration), and 2–7 μL of our sample (corresponding volume to 10,000 MSC) and the remaining volume was fulfilled by PCR grade water. Reactions were incubated at 95°C for 10 minutes to activate FastStart DNA polymerase and lyse cells, followed by 40 cycles of 10 seconds at 95°C, 5 seconds at 55°C, and 7 seconds at 72°C (adapted from [16]).
Calibration curves were constructed by adding serial dilutions of pDNA standards (pVAX-GFP) to a suspension of nontransfected MSC cells (10,000 cells per reaction). These samples were then mixed with the other PCR reagents as described above. Two negative controls were always included; one containing the same amount of nontransfected cells, exposed to the pDNA, but not to Lipofectamine, and in the other, PCR grade water was used instead of control cells to detect undesired contamination.
2.3. Mesenchymal Stem Cells Isolation, Thawing, and Expansion
Bone marrow (BM) aspirates were obtained from adult volunteer donors, after informed consent at Instituto Português de Oncologia de Lisboa Francisco Gentil. Low density BM mononuclear cells (MNCs) were separated by a Ficoll density gradient (1.077 g/ml) (GE Healthcare) and then washed twice in Dulbecco's Modified Essential Medium (DMEM, Gibco) with 10% Fetal Bovine Serum (FBS) MSC qualified (GibcoBRL), 1% (v/v) penicillin (10,000 U/mL)/streptomycin (10,000 g/ml), and 0.1% (v/v) Fungizone (Gibco). BM MNCs were then plated at a density of 2 × 105 cells/cm2 on plastic culture flasks in DMEM with 10% FBS at 37°C and 5% CO2 in a humidified atmosphere. Medium was changed twice a week. BM MSCs were isolated based on adherence to plastic, and near cell confluence (70%–80%) exhausted medium was removed from the flasks, cells were washed with phosphate buffered saline (PBS; Gibco) and detached from the flask by adding Accutase solution (Sigma) for 7 minutes at 37°C. Upon isolation, MSCs were expanded for 2–4 passages and kept frozen in liquid nitrogen. Isolated BM MSC, expressed their characteristic immunophenotype being CD73, CD90, and CD105 positive and negative for CD31, CD34, CD45, and CD80 [17]. When necessary, cells were thawed in a 37°C water bath during approximately 1 minute and resuspended in 5 mL of prewarmed Iscove's modified Dulbecco's medium (IMDM, Gibco) supplemented with 20% FBS. Then the suspension was centrifuged at 1250 rpm for 7 minutes, and the pellet was resuspended in prewarmed (37°C) DMEM-10% FBS. After determination of the cell number and cell viability using the Trypan Blue (Gibco) dye exclusion test, the cells were plated on 75 cm2 T-flasks at a density of 3000–6000 cells/cm2 and kept at 37°C in a humidified atmosphere containing 5% CO2. The medium was changed every 3-4 days. When cultures reached approximately 80%–90% confluence, cells were washed with PBS and harvested by enzymatic treatment (Accutase; Sigma). After discarding the supernatant, the cells were resuspended in 1 mL DMEM. Finally, cells were counted and seeded on T-flasks or 24 well-plates for the transfection protocol.
2.4. Lipofection
A total number of 50,000 cells were plated per well in a 24-well plate. After 24 hours, Lipofectamine2000-mediated (LF2000) (1 mg/mL) (Invitrogen) transient transfection was performed according to the protocol given by the supplier (Invitrogen), varying transfection reagent volumes and the amount of DNA (pVAX-GFP) in ratios LF2000/pDNA from 1 to 2.5. Dilutions of pVAX-GFP and LF2000 were carried out in OPTIMEM 1 (Gibco), without serum or antibiotics. Before the transfection, mixture has been added to the adherent MSC, and the culture medium was changed to serum- and antibiotics-free DMEM. Five hours after transfection the medium was replaced with fresh 500 μL DMEM supplemented with serum and antibiotics. Cell viability was determined by trypan blue exclusion method. For each transfected sample (t), Cell Recovery (CR) was determined using the equation
| (1) |
where CAt is the number of transfected cells alive, and CAc is the number of non-transfected cells alive (control).
2.5. Detection of GFP Expression and Cell Sample Preparation for RT-PCR
Determination of transfection efficiency was performed 24 hours after transfection through fluorescence microscopy and flow cytometry analysis. Culture medium was removed and 500 μL of PBS were added to each well. Cells were observed immediately under a fluorescence optical microscope Leica DMI 3000B (Leica Microsystems GMbH), and digital images were taken with a digital camera Nikon DXM 1200 F.
Cells were then harvested by enzymatic treatment, and cells suspension was then collected into a conical tube and centrifuged at 1250 rpm for 7 minutes, and after discarding the supernatant, the cells were resuspended in 1 mL PBS. Thereafter, 500 μL of this cell resuspension were collected into FACS tubes for GFP expression analysis using a FACScalibur equipment and the CellQuest software (BD Biosciences). The other 500 μL were used to determine the cell number and cell viability as previously described. Finally, cells were centrifuged again at 2500 rpm for 6 minutes, the supernatant solution was removed, and pellets were stored at −80°C for subsequent Real-Time PCR analysis.
2.6. Immunophenotyping and Evaluation of Multilineage Differentiative Potential
Approximately 50,000 transfected and non-transfected (control) MSCs per FACS tube were incubated in the dark, with phycoerythrin- (PE-) conjugated monoclonal antibodies anti-CD73PE (BD Biosciences), CD105PE (Invitrogen), and CD90PE (RD Systems), for 15 minutes at room temperature. Then the cells were washed in PBS and fixed with 2% (w/v) paraformaldehyde (PFA; Sigma). Appropriate isotype controls IgG γ1 and IgG γ2 (BD Biosciences) were also considered.
When cultured MSC reached total confluence, osteogenic and adipogenic differentiation was induced through replacement of expansion medium by Stem PRO(R) Osteogenesis Differentiation kit and Stem PRO (R) Adipogenesis Differentiation kit (Gibco), respectively. The medium was changed every 4 days and after 14 days on culture the differentiative potential of MSC was checked by observation of the standard staining tests.
Oil red-O staining (adipogenesis) —
Cells in culture were washed with PBS, fixed with 2% (w/v) PFA for 45 minutes, washed again with distilled water and incubated with 0.3% Oil Red-O solution (Sigma) for 1 hour. After a second wash with distilled water, cells were observed under the microscope.
Alkaline Phosphatase and Von Kossa Stainings (osteogenesis) —
Cells in culture were washed with PBS, fixed with 2% PFA for 45 minutes, and washed again with distilled water. Then cells were incubated in a solution of 1 : 3 Naphtol AS-MX phosphate and Fast Violet (Sigma) for 45 minutes, washed three times with distilled water, and observed under the microscope. Cells were then stained for Von Kossa by incubation with 2.5% (w/v) Silver Nitrate solution (Fluka) for 30 minutes and were finally observed.
2.7. Colony-Forming Units (CFUs) Assay
MSC were plated on 25 cm2 T-flasks with 5 mL DMEM containing 10% FBS (MSC qualified) at a density of 10 cells/cm2 (250 cells per T-flask). After 14 days in culture, without medium replacement, the cells were washed with PBS, a solution of 0.5% (w/v) crystal violet (Sigma) was added and the cells were kept at room temperature for 30 minutes. After crystal violet solution removal, the cells were washed 4 times with PBS and 1 time with distilled water. Finally, all the freestanding water was pipetted off and when the T-flasks were dry, the number of colony-forming units (CFUs) was counted.
3. Results
3.1. Lipofection Optimization
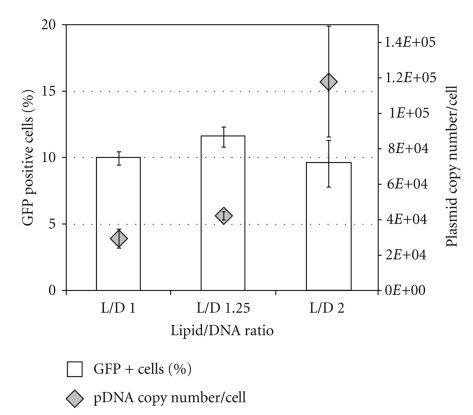
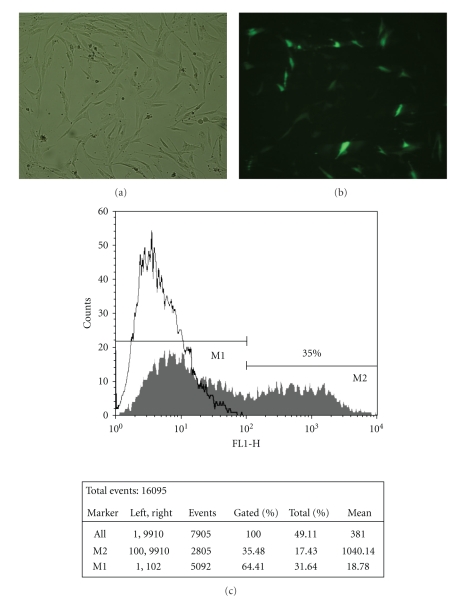
Different strategies were tested to enhance and optimize the MSC transfection protocol using Lipofectamine2000 (LF2000). The evaluation of the transfection efficiency was based on the percentage of cells expressing GFP assessed by cytometry analysis and on the determination of plasmid copy number by quantitative real-time PCR. To obtain the highest transfection efficiency and low cytotoxicity, transfection conditions were optimized by varying pDNA (D) and LF2000 (L) amount. Lipoplexes were firstly prepared using three different L/D ratios using the same amount of pDNA (2 μg). Although it could be observed nearly the same number of GFP positive (GFP+) cells in all three L/D ratios (approximately 10%), the average number of plasmids per cell for L/D = 2 is 2.5-fold higher when compared to the other studied L/D ratios (Figure 1). By phase contrast microscopic images of transfected MSC (Figure 2(a)) it was possible to verify that no relevant changes have occurred in MSC morphology upon lipoplexes contact. Moreover, cells expressing GFP (Figure 2(b)) also maintained the characteristic spindle-shaped morphology of MSC. By flow cytometry we compared the number of GFP+ cells of transfected samples with a control (cells previously in contact with the same amount of lipid). In Figure 2(c) an overlay of two histograms of transfected and non-transfected cells is shown. Around 35% of the cells were GFP+ with mean fluorescence intensity (MFI) around 1000 a.u. (% Gated and Mean of M2, respectively, shown on the table).
Figure 1.
Differences between the percentage of GFP positive cells assessed by flow cytometry analysis (white bars) and intracellular plasmid copy number determined by RT-PCR (grey ⋄). MSCs were transfected with three different Lipofectamine/pDNA (L/D) ratios: 1, 1.25, and 2. Results are the mean of four independent experiments with standard deviation.
Figure 2.
Bright field (a) and fluorescence (b) microscopic images of lipofectamine-mediated pDNA-GFP transfer in MSC, 24 hours after transfection. Images obtained by fluorescence optical microscopy (100x). Flow cytometry profiles of nontransfected hMSC (white) and hMSC transfected with pDNA-GFP using Lipofectamine2000 L/D = 1.25 (grey) and respective statistical analysis (c).
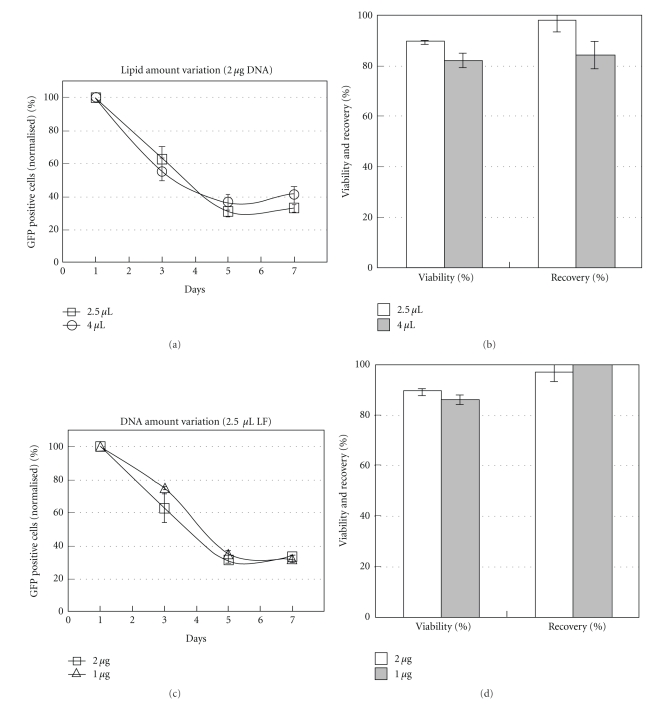
Transgene expression on MSC was evaluated up to 7 days after transfection (Figures 3(a) and 3(c)). This assay was performed using lipoplexes with L/D ratios 1.25, 2, and 2.5. Varying either lipid amount (2.5 μL or 4 μL) or DNA quantity (1 μg or 2 μg) a 70% decrease of GFP+ cells was observed along 7 days (Figures 3(a) and 3(c)). Cellular viabilities and cell recoveries of all studied conditions are shown in Figures 3(b) and 3(d). While higher lipid amounts decreased cell viability (from 90% to 80%) and cell recovery (from 95% to 85%), apparently DNA amount did not influence cell survival, when using 2.5 μL of LF2000.
Figure 3.
Evaluation of GFP expression along 7 days after transfection of MSC with Lipoplexes (a, c) and percentage of cell viability and cell Recovery of the different liposomal formulations measured 24 hours after transfection (b, d). Variation of liposome amount, maintaining DNA amount (2 μg) at charge ratios L/D 1.25 and 2, using 2.5 μL and 4 μL of LF2000, respectively (a, b). Variation of DNA amount, maintaining liposome volume (2.5 μL) at charge ratios L/D 1.25 and 2.5, using 2 μg and 1 μg of DNA, respectively (c, d). The data was obtained from the average of two independent experiments with duplicates ± SEM.
3.2. Influence of Cell Passage Number on Transfection Efficiency
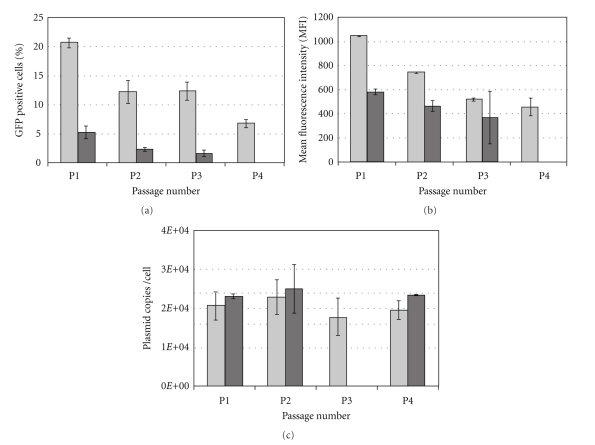
Typically, as MSCs are rare in their niches (as the BM), they have to be cultivated for several passages in order to reach clinically relevant cell numbers. Thus, the influence of MSC passage number on transfection efficiency was studied herein. To this end, MSCs from 2 different donors, from passage 1 to 4, were used. The cells were analysed by evaluating the number of cells expressing GFP, the MFI value (measure of GFP expression level), and the number of plasmids per cell (Figures 4(a), 4(b) and 4(c), resp.). We have found that the percentage of GFP+ cells and the MFI decreased with the passage number for both donors. This decrease is more evident between passages 1 and 2.
Figure 4.
Percentage of GFP positive cells (a) and mean fluorescence intensity (b) assessed by flow cytometry analysis and intracellular plasmid copy number (c) determined by RT-PCR after lipofection of MSC (L/D = 1.25) of two different donors, at different cell passages. Results from one experiment with duplicates ± standard deviation are displayed.
At early passages, the number of GFP+ cells or the MFI was donor dependent since the percentage of GFP+ cells is 4-fold higher for one of the donors. Interestingly, the average number of plasmids per cell appeared to be approximately constant (~20,000 plasmids/cell) and independent of the donor and passage number (Figure 4(c)). This seems to indicate that the decrease in the number of GFP+ cells observed at higher passages is not related to plasmid uptake by the cell but to the efficiency of GFP expression. This higher efficiency observed at lower passages may be the result of a higher nuclear uptake and/or a more efficient transcription/translation of the GFP gene.
Cell viability and cell recovery after transfection are critical parameters to consider when determining the efficacy of nonviral gene delivery methodology. By analysing Table 1, it is remarkable that, under optimal transfection conditions (L/D = 1.25 and plating cell density of 25,000 cells/cm2), LF2000 led to high cell viabilities at all cell passages (90%–95%). Moreover, cell recoveries from 50% to 98% were obtained, which is the real measure of the number of cells that remained adherent, and supposedly functional, after lipoplexes uptake.
Table 1.
Percentage of MSC viability and recovery at different passages (1–7), 24 hours after transfection. Controls are non-transfected cells. Results are presented as mean ± standard deviation of duplicates.
| Passage number | Cell Viability (%) | Cell Recovery (%) | |
| Control | Transfected cells | Transfected cells | |
| 1 | 90.1 (±3.3) | 93.6 (±3.8) | 52.5 (±11.2) |
| 2 | 91.5 (±3.1) | 89.7 (±8.0) | 80.4 (±33.6) |
| 4 | 85.5 (±4.1) | 89.7 (±1.7) | 98.0 (±11.5) |
| 5 | 86.6 (±6.1) | 92.4 (±0.6) | 79.1 (±6.3) |
| 6 | 94.1 (±2.9) | 94.7 (±1.9) | 74.2 (±5.5) |
| 7 | 91.4 (±0.4) | 91.1 (±9.2) | 88.0 (±4.6) |
3.3. Assessment of Lipofected MSC Multipotency
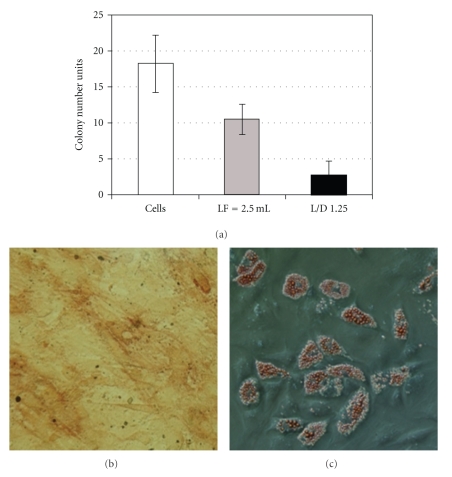
According to the high capacity of MSC to proliferate and differentiate in vitro into different mesodermal lineages, both clonogenic and differentiative potential assays were performed to verify to what extent lipofection could affect MSC characteristics. It was also analysed if MSC maintained their characteristic immunophenotypic profile. In order to verify if MSC maintained the capacity to proliferate and originate colonies, a CFU assay was carried out for transfected MSC and two controls. One control included cells incubated with 2.5 μL of LF2000 (Figure 5(a), white bar) and the other cells that were neither in contact with LF2000 nor pDNA (Figure 5(a), grey bar). The transfected cells (black bar), which had a transfection efficiency of nearly 20%, have considerably lost their clonogenic potential when compared to the control of cells only (an 85% decrease). Nevertheless, independently of pDNA transfection, LF2000 itself had a harmful effect on MSC clonogenic potential, since a decrease of 43% in colonies was observed. Considering the maintenance of MSC multilineage differentiative potential, transfected cells (with transfection efficiency near 20%) were induced to differentiate into the adipogenic and osteogenic lineages (Figure 5). Controls with non-transfected cells and cells incubated with LF2000 were also performed. As can be seen in Figures 5(b) and 5(c) transfected MSC were showed to preserve the capacity to differentiate into adipocytes (Figure 5(b)) and osteocytes (Figure 5(c)). After 15 days in culture, the adipogenic differentiation was confirmed through the Oil red-O test, where stained red lipid droplets inside vacuoles of mature adipocytes were visualized. On the other hand, osteogenic differentiation was confirmed with the appearance of nodules that stained positive (in brown) for calcified matrix through the Von Kossa staining and an increasing alkaline phosphatase activity was also observed, which is a distinctive characteristic of osteogenic commitment (in pink).
Figure 5.
(a) CFU assay of MSC after transfection (in black). Two different controls were assayed: cells which have not been in contact with lipofectamine and pDNA (white), and cells which have only been in contact with lipofectamine (L = 2.5 μL—grey). MSC were transfected at passage 4 with a transfection efficiency of ~20%. Results are presented as mean ± SEM of two independent experiments with duplicates. (b) Oil red-O staining and (c) Alkaline Phosphatase and Von Kossa stainings were tested in order to analyse the maintenance of adipogenic and osteogenic differentiation potential after transfection, respectively. The assay was performed in duplicate, and images were obtained through fluorescence optical microscopy (200x).
Through immunophenotype staining, it was verified whether transfected MSC (with a transfection efficiency of nearly 20%) maintained its characteristic phenotypic profile, namely over 90% positive for CD73, CD90, and CD105 [18]. As expected, more than 90% of the non-transfected cells (control) expressed the three markers. When analysing the cell population that underwent the transfection protocol, a slight decrease (<9%) on the percentage of cells expressing these markers compared to controls was observed (Table 2). However, when considering only GFP+ cells from the transfected population, a higher decrease was observed, namely, 83.0%, 74.2%, and 86.5% of cells expressed CD73, CD105, and CD90, respectively.
Table 2.
Immunophenotype evaluation of transfected MSC through flow cytometry analysis. The identification of MSC characteristic phenotype was assessed by fluorescent-conjugated monoclonal antibodies against CD73, CD105, and CD90 receptors. Controls with non-transfected cells were also performed as well as the appropriate isotype controls IgG γ1 and IgG γ2.
| Phenotype marker | CD73+ | CD105+ | CD90+ |
|---|---|---|---|
| MSC (control) | 96.5% | 95.3% | 92.1% |
| Transfected MSC | 89.3% | 87.0% | 89.8% |
| GFP+ MSC | 83.0% | 74.2% | 86.5% |
4. Discussion
BM MSCs have been widely investigated due to their regenerative potential; however, in many cases it might be useful to enhance some of their features through gene delivery strategies to harness their full potential in cell/gene-based therapies.
Previously, a method for plasmid delivery quantification was successfully established by us, fulfilling a lack in the study of MSC transfection, since an efficient quantification of the number of plasmids that enter into the cell was required. This new RT-PCR quantification method has been revealed to be an extremely useful tool for the optimization of MSC transfection using cationic liposomes, particularly to understand the relationship between expression and the plasmid uptake within a population of transfected cells.
Recently, an RT-PCR methodology was established to quantify the number of plasmids inside CHO cells and bacteria [16]. Taking advantage of the usefulness of this technique we have previously established a rapid quantitative RT-PCR assay to determine intracellular plasmid DNA in MSC (submitted for publication). In this previous work, the RT-PCR method showed good reproducibility, high sensitivity, and a wide linear range of 75 to 2.5 × 106 plasmid DNA copies per cell. The influence of cell number on the RT-PCR sensitivity was also evaluated. In the work described herein we have studied the effect of the initial plasmid amount, cationic lipid, and cells passage number on the plasmid uptake for bone marrow-derived MSC.
By varying cationic liposome amount, using the same quantity of pDNA, we have verified that major amounts of lipids did not correlate with higher amounts of pDNA inside the cells. In fact, it has been shown that for each cell type there is an optimal dose of plasmid, above which any additional plasmid will not contribute to transgene expression even though it may be delivered to the nucleus [13]. Moreover, higher amounts of transfection reagent were thought to increase pDNA protection against intracellular degradation by endonucleases, therefore increasing pDNA entrance into the nucleus. However, it was reported that the highest values of DNA protection do not correlate with higher values of transfection activity, even when using DOPE [19]. This neutral lipid at physiological pH is included in LF2000 formulation and reacts to the acidic pH of endosomes promoting pDNA release before their maturation into lysosomes [20]. It was verified by others that DNA complexes giving rise to effective transfection are released from endocytic vesicles at an early stage of endocytosis, that is, mainly before 1.5 hour after uptake, when the DNA is still mostly nondegraded [21, 22].
Moreover, recently, the existence of a nuclear clumping of extranuclear DNA was observed 24 hours after transfection of B16F10 mouse melanoma cells and A549 human lung carcinoma, with LF2000 [13].Contrarily, other authors did not detect extranuclear DNA 24 hours after lipofection of NIH3T3 cells [23]. Our methodology does not include any separation step before RT-PCR. This is advantageous because it is less time consuming but does not allow a distinction between pDNA inside the nucleus or in the perinuclear space. Assuming that different cells may lead to different rates of DNA nuclear uptake and knowing that NIH3T3 cells (mouse embryonic fibroblasts cell line) may show more physiological similarities with BM MSC, which are fibroblast precursors, we may suggest that plasmid copies detected by our method 24 hours after transfection were mainly from the nucleus. Nevertheless, higher amount of pDNA copies was obtained at higher charge ratios (L/D = 2) although not correlating with higher transgene expression. This lower expression could be either related to a higher level of DNA aggregation, within lipids or proteins, in the perinuclear space, or due to saturation of GFP expression [13, 23], or both. Indeed, with the increase of cationic reagent, apart from the occurrence of aggregation, higher number of plasmids may also enter into the nucleus because, in this case, more pDNA molecules may be complexed. However, as observed in our case, an increase in pDNA inside the nucleus could not lead to any increase in protein expression [13]. Furthermore, we verified that this formulation, L/D = 2, led to higher levels of cell death because a reduction of around 10% in cell viability and recovery was observed when the lipid amount was increased from 2.5 μL to 4 μL. This event may be a reflection of excessive foreign DNA molecules inside these cells. More fundamental studies on pDNA intracellular trafficking in BM MSC after lipofection are required in order to validate its kinetics, especially using different amounts of DNA and cationic liposomes.
Since the plasmid used in this study does not have the capability of replicating inside cell, it will be diluted after transfection and with consecutive cellular divisions. Thus it would be expected to observe a decrease in the number of cells expressing GFP over time as a consequence of the decrease in plasmid copy numbers inside the cells. As previously shown by others using CHO cells transfected with the same plasmid and using an RT-PCR quantification method, the plasmid copy number inside these cells decreases exponentially along seven days [16]. In contrast to the viral delivery systems, which are frequently used to long-term expression of a gene of interest, in many cases of non-inherited diseases it may be desirable to express the therapeutic gene transiently. In fact, since the transfection method used was only expressed for a short period of time (≈7 days) it is an ideal candidate for this kind of therapeutic approach, such as, MSC transient expression of anti-inflammatory genes to suppress an acute inflammation or expression of homing chemokines to move MSC towards the site of injury [24]. In addition, the expression profiles reported can also be useful in other Regenerative Medicine applications, including Tissue Engineering, where proliferation and/or differentiation into a specific lineage can be favoured by transient expression of a target gene involved in self-renewal or cell commitment, respectively [25].
Regarding transfection efficiencies using cationic liposomes, although the values obtained are still considerably lower than those obtained with the viral vectors and some nonviral vectors, namely, electroporation and nucleofection [25, 26], the results presented herein are very encouraging since a maximum value of 35% was reached using L/D = 1.25. In fact, from all the available commercial reagents LF2000 has been shown to be more efficient on MSC transfection, and efficiencies of 5%–20% have been reported when similar detection methods were used [24, 27].
It is worth mentioning that for some applications lower transfection efficiencies may be required, because in most cases, high transfection efficiencies are directly associated to a higher cell death [28]. In fact, it was recently shown that cationic liposomes overcome not only the known clinical safety problems related to viral vectors but also the toxicity problem, since lipofectamine toxicity is generally less than 20%, which is considered to be adequate for several applications [24, 29].
In this work it was verified that human MSC transgene expression is cell passage dependent, in accordance with the results reported by others using similar [27] and different transfection systems and cells [28, 29]. Indeed, only one report was found in the literature focusing on the effect of the cell passage number of MSC on the transfection efficiency [27]. These authors, however, only studied two different passages (5 and 7). Herein, regardless the decrease in GFP+ cells along cell passages, the same amount of plasmid entered the cells along the passages. According to our results around 20,000 pDNA copies were detected inside human MSC in the first four passages. The effects of MSC consecutive cell doublings on their genotypic and phenotypic characteristics were previously reported [30, 31], but to our best knowledge no adverse effect has been detected at passages 2, 3, and 4. We hypothesize that upon the first passages the transcriptional mechanism of MSC is being delayed which may explain the decrease of GFP+ cells and MFI whereas cells enclose similar amounts of plasmid DNA. Indeed, expression of nonnative proteins requires extra energy from the cells [32], and probably at early passages mammalian cells have more energy available to use in transcriptional activities of foreign genes. It is important to notice that our results were obtained with cells from donors above 60 years old, and cells from younger donors may lead to different results, since in few studies it has been reported that donors' age might influence for example the replicative lifespan of cells [33] or their capability to differentiation [34] which are directly related to endogenous protein expression along different cell passages. The variability observed for the different donors might be due to intrinsic donor-to-donor variability. To date, and to our best knowledge, there are no reports establishing any relationship between plasmid copy number inside cells after lipofection and passage number of human MSC.
In this work, cell viabilities above 85% surpass those recently reported by others using nucleofection method for gene delivery to human MSC. Using this nucleofection procedure, higher transfection efficiency was achieved (~50%) but over 50% cell death postnucleofection was induced [26]. Similarly, other authors reported less than 50% cell viability after MSC electroporation; in this case, a 90% gene delivery efficiency to MSC is reported using 60 μg pDNA to 1 × 106 cells, which in turn represents ~17,000 cells/μg DNA, a lower cell/DNA ratio than the one used in this work (25,000 cells/μg DNA) [25]. The cell recovery results obtained by lipofection (53%–100%) were better than those obtained by nucleofection (≈40%) [35] or other electroporation methods (35%–40%) [25]. Interestingly, no other reports on cell recovery (i.e., the real measure of cells that survived after lipofection) of MSC lipofection were found in the literature.
In what concerns the multipotency of MSC, it was possible to conclude that transfection does not affect MSC differentiative potential since transfected MSC could differentiate into cells from two different mesodermal lineages: adipocytes and osteocytes. This is especially interesting when considering MSC as one of the preferable starting cell source for Tissue Engineering applications.
On the other hand, it is believed that the decrease observed in the expression of the characteristic markers CD73, CD90, and CD105, as well as in the clonogenic potential, does not jeopardize MSC therapeutic potential especially if one considers that most applications with MSC in Cellular Therapy are based on their intrinsic trophic (secretory) activity which is believed to be immunomodulatory (i.e., treatment of graft-versus-host disease). Other potential applications of MSC would also take advantage from demonstrated features as being angiogenic, anti-inflammatory, antiapoptotic, and promitotic [1]. In this context, it would be interesting to determine how transfection can affect MSC secretory activity of different factors as for instance interleukin-10 (IL-10) or vascular endothelial factor (VEGF).
Although all transfection experiments performed herein were based on expressing a reporter protein encoded by a plasmid, our final goal is to develop efficient and safe methodologies to geneticallyengineer MSC to enhance their therapeutic efficacy in different Regenerative Medicine applications, namely, for tissue regeneration and cancer treatment. Indeed, genetic modification of MSC with genes encoding for tissue-specific growth factors and cytokines (e.g., bone morphogenetic proteins) can promote, both in vitro and in vivo, lineage-specific differentiation and improve tissue function [36]. On the other hand, several studies have shown that MSCs engineered to express antitumour factors (e.g., tumour necrosis factor apoptosis ligand (TRIAL), interferon (IFN)-β, -γ, and interleukin-2) (reviewed in [37]) represent an attractive tool as a cell-mediated gene therapy to counteract tumour growth.
5. Conclusions
In summary, this study showed that cationic liposomes are promising transfection agents to human MSC, especially for applications only requiring low and transient expression of proteins. We were able to achieve up to 35% of cells expressing GFP using L/D = 1.25 at early passages. Moreover, their differentiative potential and viability were not compromised. Furthermore, we found that human MSC lipofected at different passages, using the same amount of plasmid displayed, different levels of protein expression regardless similar amount of plasmids were detected inside the cells. These findings contributed to better understanding of MSC lipofection and also underlined the need to focus on more fundamental studies to comprehend the pDNA trafficking mechanism from the cytoplasm into the nucleus in these cells. Accordingly, this concept will be undoubtedly extremely useful in future developments of novel vectors for nonviral gene delivery to human MSC, contributing to an optimization of the gene delivery processes as well as to broadening the field of MSC applications.
Acknowledgments
C. Madeira (SFRH/BPD/41755/2007) and S. C. Ribeiro (SFRH/BPD/41824/2007) acknowledge Fundação para a Ciência e Tecnologia, Portugal for financial support. The authors ackowledge funding from the MIT-Portugal Program, Bioengineering Focus Area, Fundação para a Ciência e Tecnologia and from Associação Portuguesa contra a Leucemia (Portuguese Association against Leukemia). J. S. Boura is a recipient of a fellowship from Associação Portuguesa contra a Leucemia.
References
- 1.Baksh D, Song L, Tuan RS. Adult mesenchymal stem cells: characterization, differentiation, and application in cell and gene therapy. Journal of Cellular and Molecular Medicine. 2004;8(3):301–316. doi: 10.1111/j.1582-4934.2004.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guillot PV, Cui W, Fisk NM, Polak DJ. Stem cell differentiation and expansion for clinical applications of tissue engineering. Journal of Cellular and Molecular Medicine. 2007;11(5):935–944. doi: 10.1111/j.1582-4934.2007.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai Y, Drobinskaya I, Kolossov E, Chen C, Linn T. Genetic modification of cells for transplantation. Advanced Drug Delivery Reviews. 2008;60(2):146–159. doi: 10.1016/j.addr.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Godbey WT. Viral vectors for gene delivery in tissue engineering. Advanced Drug Delivery Reviews. 2006;58(4):515–534. doi: 10.1016/j.addr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Park J, Ries J, Gelse K, et al. Bone regeneration in critical size defects by cell-mediated BMP-2 gene transfer: a comparison of adenoviral vectors and liposomes. Gene Therapy. 2003;10(13):1089–1098. doi: 10.1038/sj.gt.3301960. [DOI] [PubMed] [Google Scholar]
- 6.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Therapy. 2008;15(16):1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 7.Phillips MI, Tang YL. Genetic modification of stem cells for transplantation. Advanced Drug Delivery Reviews. 2008;60(2):160–172. doi: 10.1016/j.addr.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira E, Potier E, Logeart-Avramoglou D, Salomskaite-Davalgiene S, Mir LM, Petite H. Optimization of a gene electrotransfer method for mesenchymal stem cell transfection. Gene Therapy. 2008;15(7):537–544. doi: 10.1038/gt.2008.9. [DOI] [PubMed] [Google Scholar]
- 9.Kim JA, Cho K, Shin MS, et al. A novel electroporation method using a capillary and wire-type electrode. Biosensors and Bioelectronics. 2008;23(9):1353–1360. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Hoelters J, Ciccarella M, Drechsel M, et al. Nonviral genetic modification mediates effective transgene expression and functional RNA interference in human mesenchymal stem cells. Journal of Gene Medicine. 2005;7(6):718–728. doi: 10.1002/jgm.731. [DOI] [PubMed] [Google Scholar]
- 11.Vanderbyl S, MacDonald GN, Sidhu S, et al. Transfer and stable transgene expression of a mammalian artificial chromosome into bone marrow-derived human mesenchymal stem cells. Stem Cells. 2004;22(3):324–333. doi: 10.1634/stemcells.22-3-324. [DOI] [PubMed] [Google Scholar]
- 12.Yuan B, Zhang Y, Zhao Z, et al. Treatment of chronical myocardial ischemia by adenovirus-mediated hepatocyte growth factor gene transfer in minipigs. Science in China C. 2008;51(6):537–543. doi: 10.1007/s11427-008-0073-1. [DOI] [PubMed] [Google Scholar]
- 13.Cohen RN, van der Aa MAEM, Macaraeg N, Lee AP, Szoka FC., Jr Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. Journal of Controlled Release. 2009;135(2):166–174. doi: 10.1016/j.jconrel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachibana R, Harashima H, Ide N, et al. Quantitative analysis of correlation between number of nuclear plasmids and gene expression activity after transfection with cationic liposomes. Pharmaceutical Research. 2002;19(4):377–381. doi: 10.1023/a:1015162722295. [DOI] [PubMed] [Google Scholar]
- 15.Azzoni AR, Ribeiro SC, Monteiro GA, Prazeres DMF. The impact of polyadenylation signals on plasmid nuclease-resistance and transgene expression. Journal of Gene Medicine. 2007;9(5):392–402. doi: 10.1002/jgm.1031. [DOI] [PubMed] [Google Scholar]
- 16.Carapuca E, Azzoni AR, Prazeres DMF, Monteiro GA, Mergulhao FJM. Time-course determination of plasmid content in eukaryotic and prokaryotic cells using Real-Time PCR. Molecular Biotechnology. 2007;37(2):120–126. doi: 10.1007/s12033-007-0007-3. [DOI] [PubMed] [Google Scholar]
- 17.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 18.Akita H, Ito R, Kamiya H, Kogure K, Harashima H. Cell cycle dependent transcription, a determinant factor of heterogeneity in cationic lipid-mediated transgene expression. Journal of Gene Medicine. 2007;9(3):197–207. doi: 10.1002/jgm.1010. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira AC, Ferraz MP, Monteiro FJ, Simoes S. Cationic liposome-DNA complexes as gene delivery vectors: development and behaviour towards bone-like cells. Acta Biomaterialia. 2009;5:2142–2151. doi: 10.1016/j.actbio.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Therapy. 2001;8(15):1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 21.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Molecular Therapy. 2004;9(3):443–451. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Hellum M, Hogset A, Engesaeter BO, et al. Photochemically enhanced gene delivery with cationic lipid formulations. Photochemical and Photobiological Sciences. 2003;2(4):407–411. doi: 10.1039/b211880g. [DOI] [PubMed] [Google Scholar]
- 23.Kamiya H, Fujimura Y, Matsuoka I, Harashima H. Visualization of intracellular trafficking of exogenous DNA delivered by cationic liposomes. Biochemical and Biophysical Research Communications. 2002;298(4):591–597. doi: 10.1016/s0006-291x(02)02485-3. [DOI] [PubMed] [Google Scholar]
- 24.Gheisari Y, Soleimani M, Azadmanesh K, Zeinali S. Multipotent mesenchymal stromal cells: optimization and comparison of five cationic polymer-based gene delivery methods. Cytotherapy. 2008;10(8):815–823. doi: 10.1080/14653240802474307. [DOI] [PubMed] [Google Scholar]
- 25.Helledie T, Nurcombe V, Cool SM. A simple and reliable electroporation method for human bone marrow mesenchymal stem cells. Stem Cells and Development. 2008;17(4):837–848. doi: 10.1089/scd.2007.0209. [DOI] [PubMed] [Google Scholar]
- 26.Aslan H, Zilberman Y, Arbeli V, et al. Nucleofection-based ex vivo nonviral gene delivery to human stem cells as a platform for tissue regeneration. Tissue Engineering. 2006;12(4):877–889. doi: 10.1089/ten.2006.12.877. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Green JJ, Dinio T, et al. Gene delivery to human adult and embryonic cell-derived stem cells using biodegradable nanoparticulate polymeric vectors. Gene Therapy. 2009;16:533–546. doi: 10.1038/gt.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye L, Haider HKh, Esa WB, et al. Nonviral vector-based gene transfection of primary human skeletal myoblasts. Experimental Biology and Medicine. 2007;232(11):1477–1487. doi: 10.3181/0706-RM-175. [DOI] [PubMed] [Google Scholar]
- 29.Santos JL, Oramas E, Pego AP, Granja PL, Tomas H. Osteogenic differentiation of mesenchymal stem cells using PAMAM dendrimers as gene delivery vectors. Journal of Controlled Release. 2009;134(2):141–148. doi: 10.1016/j.jconrel.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Crisostomo PR, Wang M, Wairiuko GM, et al. High passage number of stem cells adversely affects stem cell activation and myocardial protection. Shock. 2006;26(6):575–580. doi: 10.1097/01.shk.0000235087.45798.93. [DOI] [PubMed] [Google Scholar]
- 31.Derubeis AR, Cancedda R. Bone marrow stromal cells (BMSCs) in bone engineering: limitations and recent advances. Annals of Biomedical Engineering. 2004;32(1):160–165. doi: 10.1023/b:abme.0000007800.89194.95. [DOI] [PubMed] [Google Scholar]
- 32.Glick BR. Metabolic load and heterologous gene expression. Biotechnology Advances. 1995;13(2):247–261. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- 33.Klinger RY, Blum JL, Hearn B, Lebow B, Niklason LE. Relevance and safety of telomerase for human tissue engineering. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2500–2505. doi: 10.1073/pnas.0508184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretlow JD, Jin Y-Q, Liu W, et al. Donor age and cell passage affects differentiation potential of murine bone marrow-derived stem cells. BMC Cell Biology. 2008;9, article 60 doi: 10.1186/1471-2121-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aluigi M, Fogli M, Curti A, et al. Nucleofection is an efficient nonviral transfection technique for human bone marrow-derived mesenchymal stem cells. Stem Cells. 2006;24(2):454–461. doi: 10.1634/stemcells.2005-0198. [DOI] [PubMed] [Google Scholar]
- 36.Kofron MD, Laurencin CT. Bone tissue engineering by gene delivery. Advanced Drug Delivery Reviews. 2006;58(4):555–576. doi: 10.1016/j.addr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Lazennec G, Jorgensen C. Concise review: adult multipotent stromal cells and cancer: risk or benefit? Stem Cells. 2008;26(6):1387–1394. doi: 10.1634/stemcells.2007-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]