Abstract
The anisotropic nanofluidic filter (nanofilter) array (ANA) is a unique molecular sieving structure for separating biomolecules. Here we describe fabrication of planar and vertical ANA chips and how to perform continuous-flow bioseparation using them. This protocol is most useful for bioengineers that are interested in developing automated multistep chip-based bioanalysis systems and assumes prior cleanroom microfabrication knowledge. The ANA consists of a two-dimensional periodic nanofilter array, and the designed structural anisotropy of the ANA causes different sized- or charged-biomolecules to follow distinct trajectories under applied electric fields, leading to efficient continuous-flow separation. Using microfluidic channels surrounding the ANA, the fractionated biomolecule streams are collected and routed to different fluid channels or reservoirs for convenient sample recovery and downstream bioanalysis. The ANA is physically robust and can be reused repeatedly. Compared to conventional gel-based separation techniques, the ANA offers the potential for faster separation, higher throughput, and more convenient sample recovery.
Keywords: Bioseparation, lab-on-chip, BioMEMS, nanofluidics, molecular sieve
INTRODUCTION
Compared to conventional bioanalysis and diagnostic methods, direct analysis of biologically-relevant molecules (e.g., proteins and nucleic acids) can potentially enhance speed, accuracy, and sensitivity1,2. Moreover, direct biomolecule observations and manipulations help investigators probe fundamental molecular processes in biochemistry and biophysics that are often obscured in ensemble assays3–5. Therefore, nanofluidic systems with characteristic dimensions comparable to molecular scale can provide new opportunities for direct observation, manipulation, and analysis of biomolecules, and these nanofluidic systems can potentially provide innovative platforms to achieve ultra-sensitive and high-resolution biosensing and detection6,7. Inspired by this concept, over the past ten years, there has been a surge of research effort from different scientific disciplines to design and fabricate different nanofluidic systems (with characteristic size scales between 10 to 100 nm) for biological and biomedical applications6,7. Among this surge, one notable research thrust is to design efficient, regular artificial molecular sieving structures that can be a potential alternative to the conventional gel-based separation methods to improve the speed and resolution of biomolecule separation8–10.
Development of efficient nanofluidic sieving structures is essential for optimizing biomolecule separation methods in a chip format within complete microanalysis environments8–10,11. These nanofluidic structures can provide flexible designs and offer precise control over the constraining geometries ideal for molecular sieving and separation. Moreover, the current research thrust in microfluidic based scaled-down analytical processes is stimulating development of different chip-based methods where bioanalysis can be carried out in a faster and lower cost fashion7,12,13. The random nanoporous gel materials that are conventionally used in routine bioseparation applications unfortunately have intrinsic difficulties being integrated into automated multistep chip-based bioanalysis systems. Therefore, the development of a versatile regular nanofluidic sieving structure that can be monolithographically integrated within highly complex bioanalysis microsystems will have profound implications for different biological and biomedical applications.
Over the past decade, there have been exciting developments in the field of artificial regular sieving structures due to the advance of micro- and nanotechnology8–10. So far, a myriad of regular sieve designs have been demonstrated with varying degrees of success in biomolecule separation14–18. These reported artificial sieving structures have been primarily successful for the separation of large biomolecules such as viral DNA (as reviewed in Ref. [10]). Recently, we have introduced a unique regular molecular sieving structure called the anisotropic nanofluidic filter (nanofilter) array (ANA)19,20, and have demonstrated its implementation for high-resolution continuous-flow separation of a wide range of DNA fragments (between 50 to 23,000 base pairs (bp)) and proteins (between 11 to 400 kDa) within a few minutes. In this protocol we describe the detailed fabrication of the ANA chips and how to perform continuous-flow bioseparation using these chips.
COMPARISON WITH GEL-BASED SEPARATION METHODS
Separation of biomolecules in a biology laboratory is currently routinely achieved with gel-based separation techniques, such as gel-exclusion chromatography and slab-gel electrophoresis21,22. These gel-based separation techniques use gelatinous materials that consist of cross-linked three-dimensional nanometer-sized pore networks. Despite the popularity of both the gel-based separation methods, they still exhibit several limitations that make neither method optimal in separating complex mixtures for downstream analysis21. For example, the chief limitations of gel-exclusion chromatography are that the separation can be slow and that the resolution of the emerging peaks is limited, and additionally, relatively large amounts of samples are necessary to obtain visible and well-resolved bands. In slab-gel electrophoresis, recovery of the separated sample is often problematic and inefficient: the location of the band in the gel must be physically cut out, and additional washing steps are necessary to extract the desired sample from the gel, which often lead to significant sample loss21.
Gel-based sieving structures have been recently successfully incorporated into different chip-based bioanalysis systems to separate both nucleic acids and proteins23,24. These gel-based microsystems have provided convincing evidences for separation speed and resolution. However, as mentioned earlier, these gel-based microsystems still pose the intrinsic difficulties for integration with other bioanalysis components and packaging as a whole lab-on-chip system. Most of these gel-based microanalysis systems are not reusable, economically not an ideal choice for large-scale screen for different systems-biology applications. In contrast, the ANA can be monolithographically integrated within bioanalysis microsystems as an upstream sample preparation component to separate and purify complex biological samples, and the ANA is physically robust and can be reused repeatedly19,20.
Compared to the gel-based sieving structures, two major limitations currently exist for the ANA: the separation resolution and the sample throughput. The ANA structure needs to be further optimized for separation resolution to fully realize its promise for on-chip based proteomic research and biomarker discovery. The current size selectivity of the ANA structure is about 2–3 nm, which corresponds to the end-to-end distance of 10 bp DNA; however, it is still not optimal for protein separation. In the future, an improved ANA structure should have size selectivity comparable to the gel-based techniques and should discriminate and separate proteins (either native or denatured) with a molecular weight difference of about 5 kDa. Two approaches might be undertaken to further improve size selectivity and therefore separation resolution of the ANA structure. First, the size selectivity of the ANA should be improved by scaling down the nanofilter structures (period, gap size, etc.) with advanced sub-100 nm resolution lithography techniques25,26. We have proved both theoretically and experimentally that the size selectivity of the ANA is inversely proportional to both the nanofilter period (pitch size) and the nanofilter gap size27,28. Second, it is possible to extend the separation functionality of the ANA structure by utilizing the Debye layer, electro-osmosis, and surface chemistries, together with the geometrical constraints of the ANA, to achieve biomolecule separation based on a suite of molecular properties (e.g. size, charge, or hydrophobicity)10. For example, we have recently demonstrated that by actively controlling the buffer ionic strength and therefore the Debye length, switchable size- or charge-based separation of proteins can be achieved within the ANA19.
The sample throughput of the ANA needs also to be improved for its future implementations in different biological and biomedical applications, even though this concern is alleviated greatly by the ever increasing number of on-chip biosensing and detection mechanisms and their much improved detection limits. For the current ANA design, the maximum volume throughputs for protein samples are in the range of μL/h. In the future, large-scale bioseparations in a chip format with high sample throughput (in the range of μL/min) would be desired. A key for this function goal would be to devise a convenient and inexpensive method to fabricate robust molecular sieves with highly parallel nanopores.
COMPARISON WITH OTHER MICRO/NANOFLUIDIC SIEVING STRUCTURES
Different regular micro/nanofluidic sieving structures have been reported recently in the literature to separate biomolecules such as viral DNA with fast speed and great resolution14–18. One notable example was the “DNA Prism” devised by Huang et al. to continuously separate long DNA fragments (61–209 kilo-base pairs (kbp)) within a few minutes15, a speed much faster than conventional pulsed-field gel electrophoresis (PFGE) and pulsed-field capillary electrophoresis. Han and Craighead recently designed an entropy trap array device to separate long DNA ladder samples (5–50 kbp) in about 30 min14. The ANA structure described in this protocol is a more recent development, and it offers several distinct advantages when compared to these previously reported micro/nanofluidic sieving structures, rendering it a promising generic molecular sieving structure for an integrated bioanalysis microsystem29–31. These unique advantages of the ANA include: (a) the capability to continuously separate physiologically-relevant molecules including proteins in a few minutes; (b) the versatile separation mechanisms (e.g., Ogston sieving, entropic trapping, and electrostatic sieving) that can take effect in the ANA to separate biomolecules covering broad biological size ranges and based on different molecular properties; (c) the continuous-flow operation of the ANA that facilitates the integration of the separation step with upstream or downstream analysis steps; thus, allowing the bioseparation to be performed “in-line” with other continuous flow processes; (d) the continuous-flow operation of the ANA allows downstream sampling (either of a detection signal or of the separated biomolecules themselves) to be time-integrated to improve the detection limit. In our opinion, these advantages of the ANA can have profound implications for proteomic research and biomarker discovery on a chip format32,33.
This Protocol is most useful for bioengineers and bioanalytical chemists that are interested in developing automated multistep chip-based bioanalysis systems7,12,13. Since the fabrication process of the ANA structure involves standard microfabrication techniques in a cleanroom environment, the researchers implementing this protocol need to receive necessary training from the cleanroom staffs about how to operate the required cleanroom equipments. Ideally, the researchers should either assume previous knowledge and experience in cleanroom microfabrication or have direct input of a cleanroom technologist in the research team. It is our goal that with the guidance of this protocol, the ANA structures can be fabricated in a fully-equipped and well-staffed cleanroom. It is also worth mentioning that, now that many different well-equipped microfabrication foundries are available worldwide for fabricating micro/nanoscale devices, even if the necessary microfabrication facility is not available locally or on campus, the designs of the ANA structure can still be out-sourced to fabrication foundries.
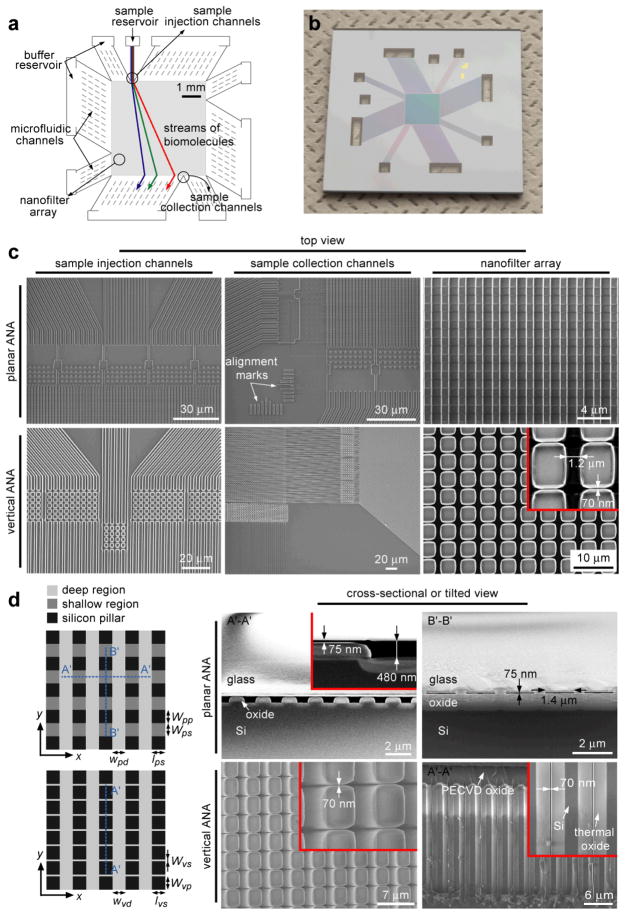
In the Protocol, the design and fabrication methods for two different types of the ANA-the planar ANA19 (Fig. 1a, as discussed in Procedure) and the vertical ANA20 (Fig. 1b, as discussed in Box 1)-will be discussed in detail. The design of both the ANA structures consists of a two-dimensional periodic nanofilter array, and the designed structural anisotropy of the ANA causes different sized- or charged-biomolecules to follow distinct trajectories, leading to efficient continuous-flow separation. The ANA structures are batch fabricated using conventional semiconductor microfabrication techniques on a silicon wafer. Using standard microfabrication techniques such as photolithography, reactive ion etching (RIE) or deep reactive ion etching (DRIE), and anisotropic potassium hydroxide (KOH) etching, the ANA structures with a nanofilter gap size down to about 10 nm can be fabricated27,34. Three different separation mechanisms have been demonstrated successfully with the ANA to separate both DNA and proteins based on either size (Ogston sieving or entropic trapping) or charge (electrostatic sieving)19. In the planar ANA structure, the relevant nanoscale constriction dimension of the planar nanofilter is etched by the RIE technique into the thickness direction of the silicon substrate (Fig. 1a); while in the vertical ANA, the nanoscale constriction of the vertical nanofilter is fabricated by taking advantage of the highly selective anisotropic KOH wet etching of the (110) silicon planes for high-aspect-ratio silicon structures with smooth and vertical sidewalls (Fig. 1b).
Figure 1.
Schematic of the fabrication process for both the planar and vertical ANA devices. In brief, for the planar ANA device (a), the shallow and deep regions of the ANA are defined and etched into a Si wafer using photolithography and RIE. KOH etching is performed to etch through the wafer for creating buffer access holes (not shown). A thick thermal oxide layer is grown to provide an electrical isolation between the conductive Si substrate and buffer solution. Finally, the ANA device is sealed by bonding a Pyrex wafer on the front surface of the silicon wafer. For the vertical ANA device (b), the narrow and wide regions of the ANA are defined and etched into a Si wafer using photolithography, DRIE, and wet anisotropic KOH etching. Then a thermal oxide layer is grown to further decrease the nanofilter gap size to a desired value. Finally, a uniform PECVD oxide layer is deposited to seal the ANA structure.
In the Experimental Design section, we first discuss the different separation mechanisms applicable for the ANA to separate biomolecules covering different size ranges and based on different molecular properties, and then we describe in detail the design guidelines of the ANA structure and the different microfabrication techniques involved in the ANA fabrication. Finally we describe the implementations of the ANA to separate different biomolecules such as DNA and proteins. In the Procedure section, a step by step description of the fabrication process is provided, followed by a troubleshooting table with information on how to troubleshoot the most likely problems encountered with the protocol. In the Anticipated Results section, we briefly describe the likely outcome of using the ANA to separate biomolecules for different applications.
EXPERIMENTAL DESIGN
SIEVING MECHANISMS IN ANA
Three different separation mechanisms (i.e., Ogston sieving, entropic trapping, and electrostatic sieving) have been applied in the ANA to separate biomolecules covering different size ranges and based on different molecular properties (see Ref. [10] for a detailed discussion of these separation mechanisms). For both Ogston sieving and entropic trapping, bioseparation should be conducted at high ionic strength where the Debye length becomes negligible compared to the nanofilter constriction size. Ogston sieving is effective for biomolecules with diameters smaller than the nanofilter constriction. Smaller molecules have a higher tendency to jump across the nanofilter and therefore assume a larger stream deflection angle in the ANA. In entropic trapping, diameters of biomolecules are greater than the nanofilter constriction size, and passage requires the molecules to deform to sneak through the nanofilter constriction. Since longer molecules have a greater probability to jump across the nanofilter constriction, they will assume a greater deflection angle in the ANA. For low ionic strength solutions where the Debye length becomes comparable to the nanofilter constriction size, electrostatic interactions (either repulsive or attractive) between charged biomolecules and charged nanofilter walls become prominent and start to dictate jump dynamics across the nanofilter. Therefore, similar sized biomolecules bearing different net charges are energetically favored to different degrees for passage through the nanofilter, resulting in efficient separation in the ANA. In practice, Ogston sieving and entropic trapping are most suitable for size separation of linear flexible biomolecules such as DNA and denatured proteins, while electrostatic sieving is most suitable for separation of native biomolecules such as globular proteins by both size and charge19. In addition, electrostatic sieving under the low ionic strength buffer can result in markedly increased size selectivity and therefore higher separation resolution for negatively charged biomolecules in the ANA35. This enhancement in size selectivity is likely due to an effective decrease in the nanofilter constriction size caused by electrostatic repulsion between negatively charged biomolecules and like-charged nanofilter walls.
DESIGN OF ANA STRUCTURE
The design of the ANA consists of a two-dimensional periodic nanofilter array. Nanofilters with a constriction size between 10–100 nm are arranged in rows and are separated by deep channels (Fig. 2). When injected into the ANA, the biomolecule stream is fractionated into different streams that are collected at intervals along the ANA opposite edge. Microfluidic channels are designed to surround the ANA, and they connect the ANA to fluid reservoirs where voltages are applied. The microfluidic channels provide sample loading and collection ports, and they also serve as electric-current injectors to generate uniform electric fields over the entire ANA structure (for more discussion, please refer to Ref. [15, 36]).
Figure 2.
Structure of the microfabricated devices incorporating the ANA structure. a shows the schematic of the ANA device, which includes the 5 mm × 5 mm rectangular ANA, sample and buffer reservoirs, microfluidic channels surrounding the ANA. b shows a high-resolution photograph of the ANA device. c–d show the scanning electron microscopy images of different regions of the ANA device (c: top view; d: cross-sectional or titled view). The planar ANA shown in c & d (top rows) have a period of 2 μm along both the x- and y-directions (x-direction: A′…A′; y-direction: B′…B′), a width of 1.4 μm (wps=1.4 μm), a length of 1 μm (lps=1 μm) and a depth of 75 nm (dps=75 nm). Deep microfluidic channels are 1 μm wide (wpd) and 480 nm deep (dpd). The vertical ANA shown in c & d (bottom rows) have a period of 3.5 μm along both the x- and y-directions, a nanofilter gap width of 70 nm (wvs=70 nm), and a wide region width of 1.2 μm (wvd=1.2 μm). All the channels have the same depth of about 15 μm. As indicated in a, after separation, the fractionated biomolecule streams are collected and routed to different fluid channels or reservoirs for convenient sample recovery and downstream bioanalysis.
The planar ANA contains planar nanofilters whose nanoscale constrictions are defined by the nanofilter shallow region and the top glass ceiling. The vertical ANA contains vertical nanofilters whose nanoscale constrictions are defined by the narrow gaps formed between adjacent silicon pillars. The key structural parameters of the planar ANA include the planar nanofilter width (wps), length (lps) and depth (dps), deep channel width (wpd) and depth (dpd), and rectangular pillar width (wpp) and length (lps) (Fig. 2d, top). The key structural parameters of the vertical ANA include the silicon pillar gap width (wvs), deep channel width (wvd) and depth (dvd), and rectangular pillar width (wvp) and length (lvs) (Fig. 2d, bottom). The structural parameters of the nanofilter depth dps and the deep channel depth dpd of the planar ANA and the silicon pillar gap width wvs and deep channel depth dvd of the vertical ANA are defined during the cleanroom fabrication process (for example, by controlling the RIE etching time), while the others are all defined during the photomask design. The size selectivity of the ANA is largely determined by the nanofilter constriction size and the nanofilter row pitch size (for the planar ANA: dps and wpd+ lps; for the vertical ANA: wvs and wvd+lvs). In principle, smaller nanofilter constriction size and nanofilter row pitch size will result in enhanced size selectivity for bioseparation. The sample throughput of the ANA is largely determined by the dimensions of the sample injection channels in the ANA. These key structural parameters can be optimized for different bioseparation applications.
MICROFABRICATION OF ANA STRUCTURE
The following techniques are involved in the ANA fabrication, and they need to be considered carefully: photomask manufacture, photolithography, RIE and DRIE, plasma-enhanced chemical vapor deposition (PECVD) and low-pressure chemical vapor deposition (LPCVD), KOH wet etching, and anodic bonding.
Photomask manufacture. Multiple photomasks are needed for fabrication of both the planar and vertical ANA structures. These photomasks are first in-house designed using computer-aided software, such as AutoCAD (Autodesk, Inc., CA) or L-Edit (Tanner Research, Inc., CA) (Note: the photomask files designed for both the planar and vertical ANA discussed here are available upon request. The mask design is machine-dependent). For proper registration and alignment between different layers of photomask patterning, designs of alignment marks need to be included into the photomasks. These photomasks are fabricated by commercial shops (e.g., Microtronics Inc., PA) using direct electron-beam writing on a sensitized, chrome-coated soda-lime glass or quartz plate. The chrome film is sputtered or evaporated onto the plate, and serves as an opaque layer to block ultraviolet (UV) light exposure. After resist development, the pattern transfer into the chrome is accomplished by wet etching to remove the exposed chrome area (see below for details of photolithography). The minimum feature size on the chrome photomask depends largely on the electron-beam spot size. For the ANA structures fabricated in this Protocol, the photomasks were fabricated with an electron beam spot size of 0.25 μm. The minimum feature size on the photomasks was about 1 μm.
Photolithography (including contact lithography and projection lithography). Different photolithography techniques can be used to transfer patterns of the ANA structures from the photomasks to the silicon wafers. During photolithography, layers of photoresist materials are first spin-coated onto the wafer substrates. Next, the resist layer is selectively exposed to UV light with an exposure tool (or ‘mask aligner’) and photomasks. The patterns in the resist are formed when the wafer undergoes the subsequent “development” step. The areas of resist remaining after development mask and protect the substrate regions. Pattern transfer onto the substrate surface is achieved by different additive or subtractive processes at the locations from which resist has been removed. If during UV light exposure the masks are placed into direct physical contact with the resist coated substrate, then this photolithography method is called “contact lithography”. The procedure of contact lithography results in a 1:1 image of the entire photomask transferred onto the silicon wafer. In projection lithography, a sophisticated refractive lens system is used to focus the mask image on the wafer surface, which is separated from the mask by a large distance. The mask pattern can be reduced by the imaging lens 1:5 or 1:10. Here it is worth noting the environments necessary for operating high performance photolithography and for performing the majority of photoresist processing steps. The light in the cleanroom needs to be filtered (i.e., yellow light) to minimize the resist exposure to UV wavelengths presented in unfiltered, ambient light. The cleanroom should be maintained at Class 100 or Class 10 cleanliness standards, and the temperature and humidity should be set at about 21°C and 50%, respectively. The mask aligners must be highly isolated from vibration by use of vibration isolation tables, as well as through appropriate vibration-isolation building design practices. For fabrication of the ANA, all the photomasks were designed for use in a 1:5 reduction projection lithography tool due to its high resolution (~0.3 μm) and high positional and overlay accuracy (<50 nm), except for the first two photomasks of the vertical ANA that were designed for use in a contact mask aligner.
Reactive ion etching (RIE) and deep-reactive ion etching (DRIE). The RIE process is a physical-chemical dry etching technique that can reproduce the features on the mask with great fidelity. The RIE technique uses chemically reactive plasma to etch the exposed wafer surface directly. Bombardment by energetic ions generated from the plasma can also disrupt an un-reactive wafer surface and cause damages such as dangling bonds and dislocations, resulting in a substrate more reactive towards etchant species. Vacuum pumping removes the volatile reaction products. In the RIE process, different degrees of anisotropic etching can be achieved. A modified version of RIE is the DRIE process (also called “the Bosch process”), which alternates repeatedly between two phases (an isotropic plasma etch mode and a passivation mode for conformal deposition of a chemically inert Teflon-like layer) to achieve nearly vertical deep structures. Each phase lasts for several seconds. The passivation layer can protect the entire wafer from further chemical etching. However, during the plasma etching phase, the directional bombardment by energetic ions attacks and sputters off the passivation layer at the bottom of the trench but not along the sides, therefore exposing the trench bottom areas to the chemical etchant. Photoresists can serve as the mask materials for both RIE and DRIE. For very deep silicon etching with DRIE, thermal dioxide can be used as mask material with a Si:SiO2 etch selectivity of more than 150:1. For fabrication of the ANA structures, RIE is the standard dry etching technique because of its precise control of the etch rate and its applicability to different semiconductor materials. DRIE was used only for the vertical ANA to achieve the desired vertical deep structures of the narrow and wide regions in the ANA.
Plasma-enhanced chemical vapor deposition (PECVD) and low-pressure chemical vapor deposition (LPCVD). In the chemical vapor deposition (CVD) process, the wafer is exposed to volatile precursors that react and/or decompose to produce the desired deposit on the wafer surface. The film materials commonly deposited include silicon, silicon dioxide, and silicon nitride in various forms such as monocrystalline and polycrystalline. A number of forms of CVD have been developed and they differ in the means by which chemical reactions are initiated (e.g., activation process) and process conditions. PECVD is a CVD process that utilizes plasma to enhance chemical reaction rates of the precursors. PECVD processing allows deposition at a relative low temperature (~ 400°C) and with a great deposition rate. LPCVD processes at sub-atmospheric pressures, leading to a growth limited by the surface reaction rate rather than by the rate of mass transfer to the substrates. Because of the low pressure and therefore greater mean free path, LPCVD allows producing uniform, high-quality films with excellent step coverage. In fabrication of both the planar and vertical ANA structures, LPCVD was used for deposition of a low-stress silicon nitride layer as the mask material for subsequent KOH etching (see below for KOH wet etching). PECVD was used to deposit a thick oxide layer to seal the vertical ANA structure.
KOH wet etching. The etch rate of silicon in KOH depends greatly on the crystallographic orientation of the etched Si surface as well as the temperature and concentration of the KOH solution. KOH etching of silicon (111) crystallographic planes is much slower (about hundreds of times slower) when compared with other planes such as (100) and (110) planes, where the etch rate is on the order of 1 μm/min. Photoresist cannot serve as mask material for KOH etching. Instead, LPCVD-deposited low-stress nitride is the desired mask material to minimize undercuts during KOH etching.
Anodic bonding. Anodic bonding is a commonly used technique to permanently bond glass to silicon. Before anodic bonding, both the glass and silicon wafers are pre-cleaned with a Piranha solution to render both glass and silicon surfaces highly hydrophilic. The glass wafer is then placed on the top surface of the Si wafer, and the two wafers can bond to each other spontaneously. During anodic bonding, a pin-point electrical contact is made to the uppermost surface of the glass which is held at a constant negative bias with respect to the electrically grounded silicon. Because of the applied electric field, mobile sodium ions in the glass migrate away from the interface region between the glass and silicon, which leaves the less mobile oxygen ions at the sodium depletion zone near the bonding interface. Because of the high bonding temperature, oxygen ions can diffuse into silicon surface and react with silicon to form an amorphous oxide layer. Anodic bonding can be accomplished on a hot plate in atmosphere between 300 to 500°C; typical voltages, depending on the glass thickness and the bonding temperature, range from 200 to 1000 V.
BIOSEPARATION THROUGH ANA
Continuous-flow bioseparation in the ANA is achieved by applying two orthogonal electric fields across the ANA to drive the biomolecules to be analyzed to migrate across the nanofilter array. The horizontal electric field, Ex, drives the biomolecules to jump across the nanofilter constrictions along the x-direction (Fig. 2d; also see Fig. 1 in Ref. [19]), and the vertical electric field, Ey, causes drifting of biomolecules in the deep (for planar ANA) or wide (for vertical ANA) microchannels (Fig. 2d). Separation speed and resolution of the ANA are mainly modulated by these two independent electric fields Ex and Ey, respectively: higher Ex leads to a greater separation resolution while higher Ey leads to a faster separation speed. Careful regulation of both Ex and Ey simultaneously will be necessary for a rapid separation with high resolution.
The ANA can be used to separate different physiologically-relevant biomolecules such as DNA, proteins, and carbohydrates. In our experiments, DNA molecules shorter than about 1 kbp and longer than about 2 kbp can be rapidly separated by the ANA based on Ogston sieving and entropic trapping, respectively, both with Tris-Borate-EDTA (TBE) 5× buffer (see Materials section)19. The size selectivity achieved so far for DNA in the Ogston sieving and entropic trapping regimes is about 10 bp and 1 kbp, respectively. For proteins (either native or denatured), either Ogston sieving or electrostatic sieving can be used for separation, depending on the ionic strength conditions chosen for different applications. TBE 5× and TBE 0.05× buffers have been used for separation of proteins by Ogston sieving and electrostatic sieving, respectively19. Additional 0.1% wt/vol sodium dodecyl sulfate (SDS) needs to be added to TBE 5× buffer for separation of denatured proteins by Ogston sieving. Non-specific adsorption of negatively charged native proteins in the ANA was not found to be significant, possibly due to electrostatic repulsion from the like-charged hydrophilic ANA walls. As a proof of principle, we have also recently achieved in the ANA separation of proteins with small molecules such as fluorescence dyes and different sized carbohydrates (collaboration with Dr. Ram Sasisekharan at the Massachusetts Institute of Technology, data not shown) in TBE 5× buffer based on Ogston sieving.
To visualize in-situ separation of biomolecules in the ANA, the biomolecules can be fluorescent-labeled with different dyes and then detected with the fluorescence microscopy method. To further detect and indentify the fractionated biomolecules after purification and separation in the ANA, the biomolecules can be routed by the microfluidic channels to different on-chip downstream bioanalysis components such as the microfluidic enzyme-linked immunosorbent assay (ELISA) to detect the presence of an antibody or an antigen in a complex biological sample.
MATERIALS
REAGENTS
Deionized and distilled water (DI water)
Hexamethyldisiliazane (HMDS, Cat. NO. H4875, Sigma-Aldrich, St. Louis, MO) ! CAUTION inflammable. Avoid long exposure time or inhalation.
Shipley SPR700 photoresist (Shipley Company, L.L.C., Marlborough, MA) ! CAUTION inflammable. Avoid long exposure time or inhalation.
95% vol/vol Sulfuric acid (Cat. NO. 320501, Sigma-Aldrich, St. Louis, MO) ! CAUTION Corrosive. Goggles and gloves must be used during operation.
30% vol/vol Hydrogen peroxide (Cat. NO. 216763, Sigma-Aldrich, St. Louis, MO) ! CAUTION Oxidizer and corrosive. Goggles and gloves must be used during operation.
49% vol/vol Hydrofluoric acid (HF, Cat. NO. 244279, Sigma-Aldrich, St. Louis, MO) ! CAUTION Highly corrosive and toxic. Goggles, aprons, and gloves must be used during operation.
27% vol/vol Ammonium hydroxide (Cat. NO. 221228, Sigma-Aldrich, St. Louis, MO) ! CAUTION Corrosive. Goggles and gloves must be used during operation.
27% vol/vol Hydrochloric acid (Cat. NO. 320331, Sigma-Aldrich, St. Louis, MO) ! CAUTION Corrosive. Goggles and gloves must be used during operation.
Potassium hydroxide (KOH, Cat. NO. 221473, Sigma-Aldrich, St. Louis, MO) ! CAUTION Corrosive. Goggles and gloves must be used during operation.
Tris-Borate-EDTA (TBE) powder (Cat. NO. T7527, Sigma-Aldrich, St. Louis, MO)
λ DNA–Hind III digest (Cat. NO. N3012L, New England BioLabs, Beverly, MA)
YOYO-1 dye (Cat. NO. Y3601, Molecular Probes, Eugene, OR)
Alexa Fluor 488-conjugated cholera toxin subunit B (Cat. NO. C22841, Molecular Probes, Eugene, OR)
Alexa Fluor 488-conjugated β-galactosidase (Cat. NO. G3153, Molecular Probes, Eugene, OR)
Fluorescent R-phycoerythrin (Cat. NO. P801, Alexis Biochemicals, San Diego, CA)
Fluorescein isothiocyanate (FITC, Cat. NO. F7250, Sigma-Aldrich, St. Louis, MO)
Sodium dodecyl sulfate (SDS, Cat. NO. L3771, Sigma-Aldrich, St. Louis, MO)
Dithiothreitol (DTT, Cat. NO. 43815, Sigma-Aldrich, St. Louis, MO)
EQUIPMENT (please refer to Suppl. Fig. S1 for photos of the relevant equipments)
Single-side polished silicon wafers (WaferNet Inc., CA; website: http://www.wafernet.com/)
Prime (110) silicon wafers (El-CAT, Inc., NJ; website: http://www.el-cat.com/)
Double-side polished Pyrex wafers (Sensor Prep Services, Inc., IL; website: http://www.sensorprepservices.com/)
High-resolution chrome mask (Microtronics Inc., PA; website: http://www.microtronicsinc.com/)
Pressurized nitrogen gas
Glass, quartz and Teflon containers and tanks
Wet processing station (with Piranha solution tank, diluted HF solution tank, and wafer dump rinser) (Model WPS-400 & 800, Semifab Inc., CA; website: http://www.semifab.com/)
RCA wet processing station (with RCA-1 clean tank, RCA-2 clean tank, diluted HF solution tank, and wafer dump rinser) (Model WPS-400 & 800, Semifab Inc., CA; website: http://www.semifab.com/)
KOH wet etching station
Wafer spin dryer
Automated photoresist coat and develop system (SSI 150 Dual Track Spinner System)
Projection stepper (Nikon NSR-2005i9, Nikon Precision Inc., CA; website: http://www.nikonprecision.com/)
Mask contact aligner (Electronic Visions EV620, E V Group Inc., AZ; website: http://www.evgroup.com/en/)
Oxygen photoresist stripper (Matrix 106 plasma asher)
Reactive-ion etcher (AME Model P5000, Applied Materials Inc., CA; website: http://www.appliedmaterials.com/)
Deep reactive-ion etcher (ICP Deep Trench Etching Systems, Surface Technology Systems plc, UK; website: http://www.stsystems.com/)
Vertical Thermal Reactor (VTR) Low Pressure Chemical Vapor Deposition (LPCVD) Furnace (SVG/Thermco 7000 Series, Silicon Valley Group, CA)
Atmospheric thermal oxide furnace (Thermco 10K Furnace Systems, Tetreon Technologies Limited, UK; website: http://thermcosystems.eu/)
Plasma enhanced chemical vapor deposition (PECVD) system (Novellus Concept One PECVD system, CA; website: http://www.novellus.com/default.aspx)
Optical upright microscope (Zeiss Axiotron Microscope, Carl Zeiss MicroImaging Inc.; website: http://www.zeiss.com/micro)
Surface profilometer (KLA-Tencor-Prometrix P10, KLA-Tencor Corp., CA; website: http://www.kla-tencor.com/)
Spectroscopic ellipsometer (KLA-Tencor-Prometrix UV-1280, KLA-Tencor Corp., CA; website: http://www.kla-tencor.com/)
Scanning electron microscopy (SEM, Zeiss SUPRA 40 High Throughput FESEM, Carl Zeiss NTS GmbH; website: http://www.smt.zeiss.com/nts)
Wafer aligner and bonder (Electronic Visions EV501-620, EV Group Inc., AZ; website: http://www.evgroup.com/en/)
Wafer die saw (Disco Abrasive System DAD-2H/6T, Disco Abrasive Systems K.K., Japan; website: http://www.disco.co.jp/das/eg/cp/index.html)
Inverted epi-fluorescence microscope (IX-71 Inverted Microscope, Olympus Imaging America Inc., PA; website: http://www.olympusamerica.com/)
Power supply
Custom-made voltage divider box
REAGENT SETUP
Piranha solution. Add 1 part of the 30% vol/vol hydrogen peroxide solution to 3 parts of the 95% vol/vol sulfuric acid solution in the quartz tank. Prepare the solution immediately before use.
Diluted HF solution. Dilute the 49% vol/vol hydrofluoric acid solution with DI water to the desired concentration in the Teflon tank. This diluted HF solution can be stored at room temperature (20–23°C) for at least 6 months.
RCA-1. Add 1 part of the 27% vol/vol ammonium hydroxide and 1 part of the 30% vol/vol hydrogen peroxide to 5 parts of DI water in the quartz tank. Heat the solution with hotplate to about 80°C and maintain the temperature of the solution. Prepare the solution immediately before use.
RCA-2. Add 1 part of the 27% vol/vol hydrochloric acid and 1 part of the 30% vol/vol hydrogen peroxide to 6 parts of DI water in the quartz tank. Heat the solution with hotplate to about 80°C and maintain the temperature of the solution. Prepare the solution immediately before use.
KOH solution. Add KOH pellets to DI water in the quartz tank to the desired working concentration. Heat the solution with hotplate to different temperatures and maintain the temperature of the solution. Prepare the solution immediately before use.
TBE 5× buffer. Dissolve TBE powder with DI water in a glass container to the desired working concentration. TBE 5× buffer contains 0.445 M Tris-Borate and 10 mM EDTA, and the pH is about 8.3. TBE 5× buffer can be stored at room temperature for about 2 months.
Labeled DNA ladder sample. λ DNA–Hind III digest is labeled with the intercalating fluorescence dye YOYO-1 in TBE 5× buffer. The dye to DNA base pair ratio is about 1:2 and the final DNA concentration is about 104 μg/ml. Prepare the sample immediately before use.
Labeled protein mixture. Alexa Fluor 488 conjugated cholera toxin subunit B and β-galactosidase and B-phycoerythrin are all denatured by adding SDS and DTT. The SDS-DTT protein mixture contains 2% wt/vol SDS and 0.1M DDT and is treated in an 80°C water bath for 10 min. The resultant SDS-protein complex solutions are mixed and further diluted in TBE 5× buffer. The final SDS-protein complex sample solution contains 15.1 μg/ml cholera toxin subunit B, 90.9 μg/ml β-galactosidase, 10 μg/ml R-phycoerythrin, 0.1% wt/vol SDS, and 5 μM DTT. Prepare the protein sample immediately before use.
EQUIPMENT SETUP
CUSTOM-MADE POLYOXYMETHYLENE (DELRIN) GADGET
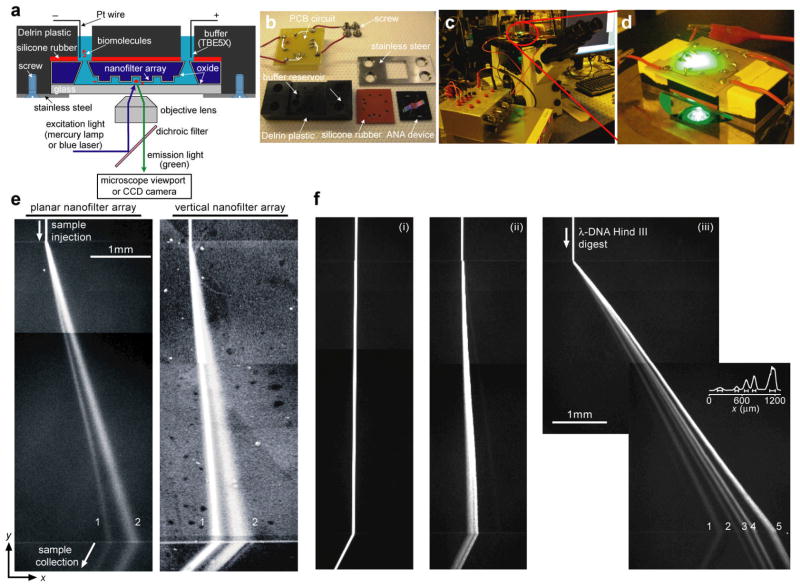
The Delrin gadget is designed to hold the ANA device during bioseparation experiments. The Delrin gadget contains four different machined parts: one Delrin rectangular cuboid, one stainless steel plate, one silicone rubber gasket, and one printed circuit board (PCB) with soldered Pt wires (Panel b of Fig 3). The Delrin cuboid contains ten drilled through-holes that can connect to the buffer access holes in the ANA device. During assembly, the stainless steel plate and the Delrin cuboid are screwed together with the ANA devices and the silicone rubber gasket to completely seal the ANA devices. Both the stainless steel plate and the silicone gasket have a square opening at the center to allow for observing bioseparation in the ANA using an inverted epi-fluorescence microscope. The Pt wires on the PCB serves as electrodes to provide an electric connection between the buffer solution and the external power supply.
Figure 3.
Experimental setup for separation of biomolecules through the ANA. a shows the schematic of the home-made Delrin plastic gadget holding the ANA. The biomolecules are labeled as red, and they are driven by the applied electric field in the buffer solution (blue) to migrate across the nanofilter array. (b) The Delrin gadget contains pre-drilled holes as buffer reservoirs that connect to the buffer access holes in the ANA device, and the gadget also includes a silicone rubber for sealing the ANA device. The Delrin gadget is connected to the external power supply through a custom-designed PCB and is mounted on the inverted epi-fluorescence microscope (c–d). e–f show composite fluorescent photographs of separation of proteins (e) and DNA molecules (f) through the ANA under different electric field conditions. Band assignments: (e, left, Ex=75 V/cm and Ey= 50 V/cm) (1) Alexa Fluor 488-conjugated cholera toxin subunit B (MW~11.4 kDa); (2) Alexa Fluor 488-conjugated β-galactosidase (MW~116.3 kDa); (e, right, Ex=250 V/cm and Ey= 40 V/cm) (1) R-phycoerythrin (MW~240 kDa); (2) FITC dye molecule (MW~389 Da); (f, λ DNA–Hind III digest) (1) 2,322 bp, (2) 4,361 bp, (3) 6,557 bp, (4) 9,416 bp, (5) 23,130 bp. f–i: Ex=15 V/cm and Ey=25 V/cm. f–ii: Ex=50 V/cm and Ey=25 V/cm. f–iii: Ex=185 V/cm and Ey=100 V/cm. The fluorescence intensity profile shown in f–iii is measured at the ANA bottom edge. The bars underneath the peaks are centered at the means and label the stream widths. Part of e is reprinted by permission from the Royal Society of Chemistry: Lab on a Chip, copyright (2009). Part of f is reprinted by permission from Macmillan Publishers Ltd: Nature Nanotechnology, copyright (2007).
PROCEDURE
CRITICAL The operational conditions of the microfabrication techniques used below depend strongly on the many process parameters (such as pressure, temperature, gas flows, and radio frequency (RF) power) and the specific machine used; therefore, the parameters listed in the Procedure are given only for guidance. These parameters should be optimized empirically for each type of application.
Patterning alignment marks (timing: 3.5 h)
-
1 | Start with a new 6 inch silicon wafer, immerse the wafer in the Piranha solution for 10 min to remove organic residue, rinse the wafer in DI water with a dump rinser (or dump rinse the wafer) for 10 min, and dry the wafer with a spin rinse dryer (or spin dry the wafer).
! CAUTION The Piranha solution is hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
2 | Bake the wafer at 200°C for 10 min on a contact hotplate to dehydrate the wafer, let the wafer cool down to room temperature, and condition (or prime) the wafer with HMDS vapor in a vacuum desiccator for 10 min.
! CAUTION HMDS is a flammable liquid and vapor. It is harmful if inhaled or absorbed through skin. Handle HMDS in a vented chemical hood with care.
-
3 | Dispense the Shipley SPR700-1.0 photoresist (~5 ml) on the center of the wafer, spin at 500 revolutions per minute (rpm, in the wafer spin dryer) for 8 s to spread a uniform photoresist layer across the entire wafer, accelerate the wafer as quickly as is practical to a final spin speed of 4000 rpm, and spin for 30 sec.
▴ CRITICAL STEP The wafer should be coated with photoresist as quickly as possible after HMDS priming, and it is recommended that coating be performed no later than 60 min after completing the priming step.
? TROUBLESHOOTING
4 | Soft bake the wafer at 95°C for 60 s on a contact hotplate to drive off solvent from the spun-on photoresist.
5 | Expose the wafer with the first photomask using either a mask contact aligner (e.g., the EV620 mask aligner) or a stepper (e.g., the Nikon projection stepper). The exposure dose is about 170 mJ/cm2, and this exposure dose might be machine-dependent.
6 | Post-exposure bake the wafer at 115°C for 60 s on a contact hotplate.
-
7 | Develop the wafer with Shipley CD-26 developer for 30 s, dump rinse the wafer with DI water for 2 min, and spin dry the wafer.
? TROUBLESHOOTING
-
8 | Inspect the wafer under an optical microscope to determine if the desired patterns on the mask have been successfully transferred to the photoresist. If not, strip the photoresist with the Piranha solution, and restart the fabrication process from Step 1.
▴ CRITICAL STEP The after-develop-inspection step is critical for monitoring if a) the correct mask has been used; b) the quality of the photoresist film is acceptable; c) the critical dimensions are within the specified tolerances; d) the registration or mask alignment is within specified limits.
9 | Hard bake the wafer at 130°C for 60 s on a contact hotplate.
-
10 | Measure the thickness of patterned photoresist film after the hard bake step with the surface profilometer.
▪ PAUSE POINT The wafer patterned with photoresist can be stored in a dry and clean environment at room temperature for at least one week.
- 11 | Use gentle O2 plasma with the RIE machine to remove photoresist residue (scum) left over after photoresist development (see the Descum recipe listed below).
Gas O2 (10 sccm) Pressure 150 mTorr RF Power 100 W Time 40 s -
12 | Dip the wafer into the diluted HF solution (DI:HF=50:1) for 10 s to remove native oxide on the wafer, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
▴ CRITICAL STEP Both Step 11 and Step 12 are essential for an accurate control of the Si etch depth in Step 14.
! CAUTION HF is an extremely hazardous liquid and vapor, and is highly corrosive to eyes and skin. Proper goggles, aprons, and gloves must be used during operation.
-
13 | Run O2 plasma for 10 min to clean the internal surface of the RIE machine chamber (see the Cleaning recipe listed below). This cleaning helps eliminate the buildup organic residue in the RIE chamber. Let the RIE chamber cool down to room temperature.
Gas O2 (40 sccm) Pressure 150 mTorr RF Power 300 W Time 10 min ▴ CRITICAL STEP Step 13 is necessary to ensure no black silicon etching to occur in Step 14. Ion bombardment during RIE etching can cause re-deposition of chemically inert residue from the RIE chamber on the silicon wafer, which can cause the black silicon phenomenon.
-
14 | Etch the wafer with the RIE machine through the opened photoresist area using Cl2 and HBr plasma, the etch rate of which can be tightly controlled to be about 2.7 nm/s using the Si-etch recipe listed below.
Gas Cl2 (20 sccm), HBr (20 sccm) Pressure 200 mTorr RF Power 300 W Time 2.7 nm/s ? TROUBLESHOOTING
-
15 | Strip photoresist with the Piranha solution for 10 min, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
! CAUTION The Piranha solution is hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
16 | Measure the silicon etch depth with the surface profilometer. The uniformity of the silicon etch profile can be monitored with the surface profilometer to be less than 8% across the entire wafer.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
Fabrication of nanofilter shallow regions (timing: 3.5 h)
-
17 | Repeat from Steps 1–16 to fabricate the nanofilter shallow regions with the second photomask.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
? TROUBLESHOOTING
Fabrication of nanofilter deep regions (timing: 3.5 h)
-
18 | Repeat from Steps 1–16 to fabricate the nanofilter deep regions as well as the microfluidic channels surrounding the ANA with the third photomask.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
? TROUBLESHOOTING
Fabrication of buffer access holes (timing: 20 h)
-
19 | Immerse the wafer in the RCA-1 solution at 80°C for 15 min, and dump rinse the wafer with DI water for 10 min.
! CAUTION RCA-1 is extremely corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
20 | Transfer the still-wet wafer to the diluted HF solution (DI:HF=10:1) for 15 s to remove hydrous oxide film formed during Step 19, dump rinse the wafer with DI water for 20 s.
▴ CRITICAL STEP The short DI rinse minimizes re-growth of the oxide.
! CAUTION HF is an extremely hazardous liquid and vapor, and is highly corrosive to eyes and skin. Proper goggles, aprons, and gloves must be used during operation.
-
21 | Immerse the still-wet wafer from Step 20 into the RCA-2 solution at 80°C for 15 min, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
! CAUTION RCA-2 is extremely corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
22 | Deposit a thin layer of low-stress silicon nitride on both sides of the wafer using the LPCVD furnace. The thickness of the nitride film is about 100 nm.
Gas NH3 (25 sccm), SiH2Cl2 (250 sccm) Pressure 250 mTorr Temperature 775 °C Nitride deposition rate 3 nm/min ▴ CRITICAL STEP The nitride film should be deposited as quickly as possible after Step 21 to minimize re-growth of the oxide. Only low-stress nitride film can serve as a KOH etch mask, since the conventional high-stress nitride film tends to possess pinholes and cracks during KOH etching.
-
23 | Measure the nitride film thickness with the spectroscopic ellipsometer.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
24 | Repeat from Steps 2–11 to pattern the photoresist for the buffer access holes with the fourth photomask.
- 25 | Etch the nitride layer with the RIE machine through the opened photoresist area using CF4 and O2 plasma, the etch rate of which can be tightly controlled to be about 3.4 nm/s using the SiN-etch recipe listed below.
Gas CF4 (8 sccm), O2 (6 sccm) Pressure 50 mTorr RF Power 250 W Nitride etch rate 3.4 nm/s -
26 | Strip photoresist with the Piranha solution for 10 min, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
! CAUTION The Piranha solution is hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
27 | Measure the nitride etch depth with the surface profilometer to ensure the nitride layer etched through. The uniformity of the nitride etch profile can be monitored with the surface profilometer to be less than 8% across the entire wafer.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
-
28 | Etch the buffer access holes through the silicon wafer with a KOH solution (20% wt/vol) at 80°C for about 8 h. The Si etch rate with the 20% wt/vol KOH solution at 80°C is about 1.33 μm/min.
! CAUTION The KOH solution is extremely hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
? TROUBLESHOOTING
29 | Dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
30 | Immerse the wafer in the Piranha solution for 10 min, and dump rinse the wafer with DI water for 10 min.
31 | Immerse the still-wet wafer from Step 30 into the second Piranha solution for 10 min, and dump rinse the wafer with DI water for 10 min.
-
32 | Immerse the still-wet wafer from Step 31 into the diluted HF solution (DI:HF=50:1) for 30 s, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
▴ CRITICAL STEP Steps 30–32 are necessary to remove KOH contaminants before growing thermal oxidation on the wafer.
! CAUTION HF is an extremely hazardous liquid and vapor, and is highly corrosive to eyes and skin. Proper goggles, aprons, and gloves must be used during operation.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
-
33 | Immerse the wafer in a concentrated HF solution (49% vol/vol HF) for 20 min (the etch rate of nitride in 49% vol/vol HF is about 5 nm/min), dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
! CAUTION HF is an extremely hazardous liquid and vapor, and is highly corrosive to eyes and skin. Proper goggles, aprons, and gloves must be used during operation.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
Thermal oxidation and anodic bonding (timing: 12 h)
34 | Repeat from Steps 19–21 to clean the wafer before thermal oxidation in Step 35.
-
35 | Thermally grow a thick layer of oxide (300 nm to 500 nm) with an atmosphere furnace to provide an electrical isolation between the conductive Si substrate and buffer solution. Let the wafer cool down to room temperature.
? TROUBLESHOOTING
-
36 | Measure the oxide film thickness with the spectroscopic ellipsometer.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
-
37 | Immerse both the silicon wafer and another double-side polished Pyrex wafer in the Piranha solution for 10 min, dump rinse both the wafers with DI water for 10 min, and spin dry the wafers.
! CAUTION The Piranha solution is hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
-
38 | Carefully place the Pyrex wafer on the front surface of the silicon wafer, and anodically bond the two wafers with a wafer bonder (e.g., Electronic Visions EV501-620 wafer aligner and bonder) at about 350°C and 800 V for about 30 min.
! CAUTION Anodic bonding is conducted under high temperature and high voltage.
? TROUBLESHOOTING
-
39 | Cut the bonded wafers with a wafer die saw into individual ANA devices. The final ANA device has a dimension of about 26 mm by 26 mm.
▴ CRITICAL STEP During die saw cutting, the buffer access holes of the ANA device should be properly sealed with a wafer dicing tape to prevent water leak into the ANA device.
40 | Assemble the ANA device with the custom-made Delrin gadget, as described in the Experimental Design section (Fig. 3a–d)
-
41 | Fill the ANA device with TBE buffer through the buffer reservoirs on the Delrin gadget (for separation of denatured proteins, additional 0.1% wt/vol SDS should be added to the TBE buffer). Since both thermal oxide and glass surface of the ANA device are hydrophilic, the TBE buffer can fill the ANA device spontaneously.
▴ CRITICAL STEP The ANA device should be filled with the buffer solution immediately after the die saw cutting, since the hydrophilicity of both the thermal oxide and the glass surface in the ANA device deteriorates over time.
? TROUBLESHOOTING
-
42 | Dip platinum wires into the buffer reservoirs on the gadget and connect them to an external power supply through the custom-designed PCB (Fig. 3a–d). Run electro-osmosis through the ANA device to remove air bubbles (if any) for a few hours. The ANA device is now ready for bioseparation experiments.
▪ PAUSE POINT The ANA device filled with the buffer solution can be stored at 4°C for at least half a year. If stored, the gadget holding the ANA should be rapped with parafilm completely to prevent buffer evaporation.
Bioseparation with ANA (timing: 3 h)
43 | Load the biomolecule mixture to be analyzed into the sample reservoir of the Delrin gadget.
44 | Mount the Delrin gadget holding the ANA on an inverted epi-fluorescence microscope. The external power supply is connected to the gadget through a custom-made voltage divider box. The voltages at the different buffer reservoirs of the ANA can be adjusted continuously to generate different electric fields in the ANA.
45 | During separation, the CCD camera attached to the microscope can be used to visualize and record the fluorescence images of the fractionated biomolecule streams in the ANA. The voltages at the different buffer reservoirs of the ANA can be fine tuned to achieve a desired separation.
46 | After separation, the fractionated biomolecules can be collected using a micropipette in the different ANA reservoirs for downstream analysis.
47 | Replace the buffer in the ANA device with fresh TBE buffer, and run electro-osmosis through the ANA device for one hour to make sure no biomolecule left in the ANA device.
48 | Replace the buffer in the ANA device with fresh TBE buffer. The ANA device is now ready for a next bioseparation experiment or can be stored at 4°C for at least half a year.
TIMING
(Planar ANA discussed in the Procedure)
Patterning alignment marks (timing: 3.5 h): Step 1–10: 2 h; Step 11–16: 1.5 h.
Fabrication of nanofilter shallow regions (timing: 3.5 h): Step 17: 3.5 h.
Fabrication of nanofilter deep regions (timing: 3.5 h): Step 18: 3.5 h.
Fabrication of buffer access holes (timing: 20 h): Step 19–23: 5 h; Step 24–27: 2 h; Step 28–32: 12 h; Step 33: 1 h.
Thermal oxidation and anodic bonding (timing: 12 h): Step 34–36: 5 h; Step 37–42: 7 h.
Bioseparation with ANA (timing: 3 h): Step 43–48: 3 h.
(Vertical ANA discussed in the Box 1)
Patterning alignment marks to find (111) planes of (110) wafer (timing: 10 h): Step i–iii: 5 h; Step iv–vi: 1 h; Step vii: 1 h; Step viii–x: 3 h.
Patterning alignment marks (timing: 2 h): Step xi: 2 h.
Fabrication of ANA narrow channels (timing: 4 h): Step xii: 4 h.
Fabrication of ANA wide channels and other microfluidic channels (timing: 17.5 h): Step xiii: 0.5 h; Step xiv–xxii: 11 h; Step xxiii–xxv: 6 h.
Fabrication of buffer access holes (timing: 8.5 h): Step xxvi: 0.5 h; Step xxvii–xxxi: 8 h.
Bioseparation with ANA (timing: 3 h): Step xxxii: 3 h.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| STEP | PROBLEM | POSSIBLE REASON | SOLUTION |
|---|---|---|---|
| 3 | Spin-coated photoresist is not uniform and has traces of particulates and particles. |
|
|
| 7 | Photoresist pattern on the wafer doesn’t reproduce the photomask features with good fidelity. |
|
|
| 14, 17, 18 | After RIE process, some black silicon appears on the etched area. |
|
|
| 28 | KOH etch rate for silicon is slow and not uniform across the entire wafer. |
|
|
| 35 | Silicon wafer breaks during thermal oxidation process. |
|
|
| 38 | Bonding strength between Pyrex and silicon wafers is not strong after anodic bonding, and the wafers can easily become delaminated. |
|
|
| 41 | TBE buffer is not spontaneously filling the entire ANA device. |
|
|
| 45 | After application of the electric fields, the initial biomolecule stream is not injected from the sample injection channels. |
|
|
| x | It is hard to determine the (111) planes. |
|
|
| xii | Irregular trenches (e.g., non-uniform width or depth) of the ANA narrow regions are generated after KOH etching. |
|
|
| xiv | Spin-coated photoresist can not uniformly cover the wafer. |
|
|
| xviii | DRIE etching doesn’t reproduce the features on the mask with good fidelity. |
|
|
| xxv | The final gap size of the ANA narrow regions is either too small or too large. |
|
|
ANTICIPATED RESULTS
The stream deflection pattern during separation depends on the separation mechanisms applied in the ANA. For Ogston sieving, smaller molecules will assume a larger steam deflection angle in the ANA and will be collected by the microfluidic channels on the right-hand side of the ANA opposite edge. In contrast, for entropic trapping, since longer molecules assume a greater deflection angle in the ANA, they will be deflected towards the right-hand side of the ANA opposite edge and collected by the microfluidic channels. For electrostatic sieving, the separation pattern would be more difficult to predict. In principle, smaller biomolecules bearing less negative charges will be deflected more in the ANA, towards the right-hand side of the ANA opposite edge.
During separation, we can use the CCD camera attached to the microscope to visualize and record the migration trajectories of fluorescence-labeled biomolecules in the ANA. These fluorescence images can be further analyzed with the image processing software (e.g., IPLab, BD Biosciences Bioimaging, Rockville, MD) to generate fluorescence intensity profiles. Gaussian functions can be used for fitting these fluorescence intensity profiles to determine the means (the maximum intensity) as well as the widths of the fractionated biomolecule streams19. The separation efficiency of the ANA can then be further quantified by calculating both the size selectivity and the effective peak capacity under different electric field conditions22. Figure 3e–f shows examples of composite fluorescent photographs of continuous-flow separation of proteins (Fig. 3e) and DNA (Fig. 3f) through both the planar and the vertical ANA, respectively. Upon application of the electric field Ey along the y-axis of the ANA, the initial biomolecule stream is continuously injected into the deep (for the planar ANA) or wide (for the vertical ANA) channels on the top left of the ANA structure (Fig. 3fi). After the orthogonal electric field Ex is superimposed along the x-axis across the nanofilters, the drifting biomolecules in the deep channels start to jump across the nanofilters and separate from each other (Fig. 3fii&iii). With properly adjusted values of both Ex and Ey, both the protein mixtures and the long DNA digest can be based-line separated into distinct streams within a few minutes (Fig. 3e–f). The sample volume throughput of the ANA structure can be estimated based on the migration speed of biomolecules in the ANA and the dimensions of the ANA sample injection channels. For the protein separation shown in Fig. 3e, the sample volume throughputs for the planar and vertical ANA are in the range of nL/h and μL/h, respectively. The sample throughput of the ANA can be scaled up by parallelism with multi-device processing. For the vertical ANA, the sample throughput can be further enhanced by increasing the depth of the ANA structure to a maximum value of the silicon wafer’s thickness, which is about 500 μm for a 6″ wafer.
Box 1. FABRICATION OF VERTICAL ANA DEVICE
Patterning alignment marks to find (111) planes of (110) wafer (timing: 10 h)
i | Start with a new 6 inch (110) wafer, and clean the wafer as described in Steps 19–21 in the Procedure before the nitride deposition in Step ii.
-
ii | Deposit a thin layer of low-stress silicon nitride on both sides of the wafer with the LPCVD furnace. The thickness of the nitride film is about 300 nm.
Gas NH3 (25 sccm), SiH2Cl2 (250 sccm) Pressure 250 mTorr Temperature 775 °C Nitride deposition rate 3 nm/min ▴ CRITICAL STEP The nitride film should be deposited as quickly as possible after Step i to minimize re-growth of the oxide. Only the low-stress silicon nitride film can serve as a etch mask for the KOH etching, since the conventional high-stress silicon nitride film tends to possess pinholes and cracks during the KOH etching step.
-
iii | Measure the nitride film thickness with the spectroscopic ellipsometer.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
iv | Spin-coat the wafer with photoresist as described in Steps 2–4 of the Procedure.
v | Expose the wafer using a mask contact aligner (e.g., the EV620 mask aligner) for the first photomask. The exposure dose is about 170 mJ/cm2, and this dose value might be machine-dependent.
-
vi | Perform Steps 6–11 of the Procedure to post-exposure bake the wafer, develop the photoresist, inspect the wafer under microscope, hard bake the wafer, measure the photoresist thickness using the surface profilometer, and use gentle O2 plasma to remove photoresist residue.
▪ PAUSE POINT The wafer patterned with photoresist can be stored in a dry and clean environment at room temperature for at least one week.
-
vii | Etch the nitride layer with the RIE machine using CF4 and O2 plasma, strip photoresist with the Piranha solution, dump rinse the wafer with DI water, spin dry the wafer, and measure the nitride etch depth with the surface profilometer as described in Steps 25–27 of the Procedure.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
-
viii | Etch the wafer with the KOH solution (44% wt/vol) at 73°C for 30 min to find the (111) planes. The Si etch rate by the 73°C KOH solution is about 1 μm/min.
! CAUTION The KOH solution is extremely hot and corrosive. Proper goggles, aprons, and gloves must be used during operation.
ix | Remove the KOH contaminants before growing thermal oxidation on the wafer as described in Steps 29–32 of the Procedure.
-
x | Inspect the fan-shaped alignment marks under the optical microscope to locate the silicon trench with the minimum undercut during the KOH etching.
▴ CRITICAL STEP This step is critical to find the (111) planes on the (110) Si wafer. The Si etch should be deep enough for sharp contrast between adjacent Si trenches with different levels of undercuts.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
? TROUBLESHOOTING
Patterning alignment marks (timing: 2 h)
-
xi | Repeat from Steps iv to vii to pattern the photoresist layer with the second photomask for the alignment marks, etch the nitride layer, strip photoresist with the Piranha solution, dump rinse the wafer with DI water, spin dry the wafer, and measure the nitride etch depth with the surface profilometer.
▴ CRITICAL STEP During the exposure step, the features on the mask must be aligned to the (111) planes as determined in Step x, not to the wafer flat. The etch depth of the nitride layer should be around 100 to 200 nm, less than the total thickness of the nitride layer. Exposing the underlying silicon surface (if the nitride layer is etched through) will cause damage to the alignment marks during the following KOH etching. The alignment marks patterned in this step can be used for either contact lithography or projection lithography.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
Fabrication of ANA narrow channels (timing: 4 h)
-
xii | Repeat from Steps iv to ix to pattern the photoresist layer with the third photomask for the ANA narrow channels, etch the nitride layer, strip the photoresist with the Piranha solution, dump rinse the wafer with DI water, spin dry the wafer, measure the nitride etch depth with the surface profilometer, etch the wafer with the KOH solution, and clean the wafer to remove KOH contaminants.
▴ CRITICAL STEP Please note that the vertical ANA narrow channels are etched in the KOH solution at room temperature. The Si etch rate of the KOH solution at room temperature is about 75 nm/min. Si trenches with uniform depths across the entire wafer can be fabricated under this condition.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
? TROUBLESHOOTING
Fabrication of ANA wide channels and other microfluidic channels (timing: 17.5 h)
-
xiii | Deposit a thick layer of oxide (~ 3 μm) on the wafer using the PECVD system to seal the ANA narrow channels.
Gas SiH4 (50 sccm), N2O (800 sccm) Pressure 2.7 Torr Temperature 400 °C RF Power 270 W Oxide deposition rate 10 nm/sec ▴ CRITICAL STEP This step is critical to cover deep trenches of the ANA narrow channels and therefore ensure uniform photoresist coating on the wafer. The thickness of this PECVD oxide layer varies with different trench dimensions.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
-
xiv | Perform steps 2–10 of the Procedure to pattern the ANA wide channels and other surrounding microfluidic channels with the fourth photomask.
? TROUBLESHOOTING
xv | Use O2 plasma to remove photoresist residue and clean the internal surface of the RIE machine chamber as described in Step 11 and Step 13 of the Procedure.
- xvi | Etch PECVD oxide through the opened photoresist area with the RIE machine using CHF3/CF/Ar plasma. The oxide etch rate is about 3.5 nm/s using the Oxide-etch recipe listed below.
Gas CHF3 (45 sccm), CF4 (15 sccm), Ar (100 sccm) Pressure 100 mTorr RF Power 200 W Etch rate 3.5 nm/s xvii | Perform Steps 25–27 of the Procedure to etch the nitride layer, strip the photoresist, and measure the etch depth with the surface profilometer to ensure both the oxide and nitride layers etched through.
-
xviii | Etch the wafer with the DRIE machine for the ANA wide channels. The etch rate is about 1.47 μm/min using the SiDRIE-etch recipe listed below. Measure the silicon etch depth with the surface profilometer.
Etch mode Passivation mode Process time: 6 s 4.5 s Overrun: 0.5 s 0 s Platen generator power: 80 W 60 W Coil generator power: 600 W 600 W Gas: SF6 (70 sccm) C4F8 (35 sccm) Etch rate: 1.47 μm/min N/A ▴ CRITICAL STEP The Bosch process used in the DRIE etching should end with the SF6 etching cycle. The SiDRIE-etch recipe listed above is characterized with a minimum side-wall roughness and a near-vertical etch profiles. The DRIE Si etch depth should equal to the ANA narrow channel depth.
? TROUBLESHOOTING
-
xix | Run O2 plasma with the O2 plasma asher to clean the wafer for at least 4 h.
Gas O2 (45 sccm) Pressure 100 mTorr RF Power 200 W Time 240 min ▴ CRITICAL STEP This step is critical to remove the Teflon-like polymer layer left after the DRIE Bosch process.
xx | Strip photoresist with the Piranha solution for 10 min, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
xxi | Immerse the wafer in the concentrated HF solution (49% vol/vol HF) for 2 hours to strip both the oxide and the nitride layer, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
-
xxii | Inspect the wafer in the SEM to measure the width of the ANA narrow and wide channels.
▴ CRITICAL STEP Besides checking the uniformity of the DRIE etching profile, this step is critical in determining the desired thickness of the thermal oxide grown in Step xxiv.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
xxiii | Clean the wafer before thermal oxidation as described in Steps 19–21 of the Procedure.
- xxiv | Thermally grow an oxide layer with an appropriate thickness in an atmosphere furnace to provide an electrical isolation between the conductive Si substrate and buffer solution. Let the wafer cool down to room temperature.
Gas H2 (5 sccm), O2 (10 sccm) Pressure 1 atm Temperature 1000°C -
xxv | Repeat Step xxii to determine the final gap sizes of the ANA narrow and wide channels.
? TROUBLESHOOTING
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
Fabrication of buffer access holes (timing: 8.5 h)
-
xxvi | Repeat Step xiii to deposit a thick layer of oxide (~ 6 μm) on the wafer using the PECVD system.
▪ PAUSE POINT The wafer can be stored in a dry and clean environment at room temperature for a long period of time.
xxvii | Perform Steps 2–9 of the Procedure to pattern the photoresist with the fifth photomask for buffer access holes. Spin coat a thick photoresist (~ 10 μm) instead of a thin one.
-
xxviii | Perform Step 15 and Step 16 of the Procedure to use O2 plasma to remove photoresist residue and to clean the internal surface of the RIE machine chamber, and to etch the PECVD oxide. The etch time is controlled to ensure the buffer access holes are open.
▴ CRITICAL STEP The etch time should be well controlled so that the holes are completely open and have connection with other channels. In addition, if the wafer is over etched, silicon surface in some areas will be exposed, which will cause current leakage in the separation experiment.
xxix | Strip photoresist with the Piranha solution for 10 min, dump rinse the wafer with DI water for 10 min, and spin dry the wafer.
xxx | Cut the wafer with diesaw into the individual ANA devices. The final ANA device has a dimension of about 26 mm by 26 mm.
-
xxxi | Perform Steps 40–42 of the Procedure to assemble the ANA device with the Delrin gadget, and fill the ANA device with TBE buffer, run electro-osmosis through the ANA device to remove air bubbles (if any) for a few hours. The ANA device is ready for bioseparation experiments.
▪ PAUSE POINT The ANA device filled with the buffer solution can be stored at 4°C for at least half a year. If stored, the gadget holding the ANA should be rapped with parafilm completely to prevent buffer evaporation.
Bioseparation with ANA (timing: 3 h)
xxxii | Separate biomolecules for different applications as described in Steps 43–48 of the Procedure.
Acknowledgments
The authors acknowledge financial support from the National Institute of Health (EB005743), Korea Institute of Science and Technology–Intelligent Microsystems Center (KIST-IMC), and the Singapore-MIT Alliance (SMA-II, CE program). We also thank J. Yoo for his contribution in the experimental setup, H. Bow and S. Reto for helpful discussions. The MIT Microsystems Technology Laboratories is acknowledged for support in microfabrication.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS
J. Fu, P. Mao, and J. Han conceived and designed the ANA chips. J. Fu and P. Mao fabricated the ANA chips. J. Fu and P. Mao designed and performed experiments, and analyzed data. J. Fu and P. Mao wrote manuscript.
Contributor Information
Jianping Fu, Email: jpfu@umich.edu.
Pan Mao, Email: panmao@mit.edu.
Jongyoon Han, Email: jyhan@mit.edu.
References
- 1.Goodsell DS. Bionanotechnology, Lessons from Nature. Wiley-Liss; New Jersey: 2004. [Google Scholar]
- 2.Jain KK. Nanobiotechnology in Molecular Diagnostics, Current Techniques and Applications. Horizontal Bioscience; Norfolk, U.K: 2006. [Google Scholar]
- 3.Chu S. Biology and polymer physics at the single-molecule level. Philos Trans R Soc London Ser A-Math Phys Eng Sci. 2003;361:689–693. doi: 10.1098/rsta.2002.1157. [DOI] [PubMed] [Google Scholar]
- 4.Fortina P, Kricka LJ, Surrey S, Grodzinski P. Nanobiotechnology: the promise and reality of new approaches to molecular recognition. Trends Biotechnol. 2005;23:168–173. doi: 10.1016/j.tibtech.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Hinterdorfer P, Dufrêne YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nature Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 6.Eijkel JCT, van den Berg A. Nanofluidics: what is it and what can we expect from it? Microfluid Nanofluid. 2005;1:249–267. [Google Scholar]
- 7.Craighead H. Future lab-on-a-chip technologies for interrogating individual molecules. Nature. 2006;442:387–393. doi: 10.1038/nature05061. [DOI] [PubMed] [Google Scholar]
- 8.Tegenfeldt JO, et al. Micro- and nanofluidics for DNA analysis. Anal Bioanal Chem. 2004;378:1678–1692. doi: 10.1007/s00216-004-2526-0. [DOI] [PubMed] [Google Scholar]
- 9.Han J, Fu J, Schoch RB. Molecular sieving using nanofilters: past, present and future. Lab Chip. 2008;8:23–33. doi: 10.1039//b714128a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu J, Mao P, Han J. Artificial molecular sieves and filters: a new paradigm for biomolecule separation. Trends Biotech. 2008;26:311–320. doi: 10.1016/j.tibtech.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Slater GW. Theory of DNA electrophoresis: A look at some current challenges. Electrophoresis. 2000;21:3873–3887. doi: 10.1002/1522-2683(200012)21:18<3873::AID-ELPS3873>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 13.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Craighead HG. Separation of long DNA molecules in a microfabricated entropic trap array. Science. 2000;288:1026–1029. doi: 10.1126/science.288.5468.1026. [DOI] [PubMed] [Google Scholar]
- 15.Huang LR, et al. A DNA prism: high speed continuous fractionation of large DNA molecules. Nature Biotech. 2002;20:1048–1051. doi: 10.1038/nbt733. [DOI] [PubMed] [Google Scholar]
- 16.Baba M, et al. DNA size separation using artificially nanostructured matrix. Appl Phys Lett. 2003;83:1468–1470. [Google Scholar]
- 17.Kaji N, et al. Separation of long DNA molecules by quartz nanopillar chips under a direct current electric field. Anal Chem. 2004;76:15–22. doi: 10.1021/ac030303m. [DOI] [PubMed] [Google Scholar]
- 18.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 19.Fu J, Schoch RB, Bow H, Stevens AL, Tannenbaum SR, Han J. A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins. Nature Nanotech. 2007;2:121–128. doi: 10.1038/nnano.2006.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao P, Han J. Massively-parallel ultra-high-aspect-ratio nanochannels as mesoporous membranes. Lab Chip. 2009;9:586–591. doi: 10.1039/b809370a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scopes RK. Protein Purification, Principles and Practice. 3. Springer-Verlag; New York: 1993. [Google Scholar]
- 22.Giddings JC. Principles and Theory. Part 1. Marcel Dekker; New York: 1965. Dynamics of Chromatography. [Google Scholar]
- 23.Yao G, et al. SDS capillary gel electrophoresis of proteins in microfabricated channels. Proc Natl Acad Sci U S A. 1999;96:5372–5377. doi: 10.1073/pnas.96.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callewaert N, et al. Total serum protein N-glycome profiling on a capillary electrophoresis-microfluidics platform. Electrophoresis. 2004;25:3128–3131. doi: 10.1002/elps.200406020. [DOI] [PubMed] [Google Scholar]
- 25.Turner SW, Perez AM, Lopez A, Craighead HG. Monolithic nanofluid sieving structures for DNA manipulation. J Vac Sci Technol B. 1998;16:3835–3840. [Google Scholar]
- 26.Cao H, et al. Fabrication of 10 nm enclosed nanofluidic channels. Appl Phys Lett. 2002;81:174–176. [Google Scholar]
- 27.Fu J, Mao P, Han J. Nanofilter array chip for fast gel-free biomolecule separation. Appl Phys Lett. 2005;87:263902. doi: 10.1063/1.2149979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu J, Yoo J, Han J. Molecular sieving in periodic free-energy landscapes created by patterned nanofilter arrays. Phys Rev Lett. 2006;97:018103. doi: 10.1103/PhysRevLett.97.018103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eijkel JCT, van den Berg A. Nanotechnology for membranes, filters and sieves. Lab Chip. 2006;6:19–23. doi: 10.1039/b516903h. [DOI] [PubMed] [Google Scholar]
- 30.Austin R. Nanofluidics: a fork in the nano-road. Nature Nanotech. 2007;2:121–128. doi: 10.1038/nnano.2007.18. [DOI] [PubMed] [Google Scholar]
- 31.Pamme N. Continuous flow separations in microfluidic devices. Lab Chip. 2007;7:1644–1659. doi: 10.1039/b712784g. [DOI] [PubMed] [Google Scholar]
- 32.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 33.Righetti PG, Castagna A, Herbert B, Reymond F, Rossier JS. Prefractionation techniques in proteome analysis. Proteomics. 2003;3:1397–1407. doi: 10.1002/pmic.200300472. [DOI] [PubMed] [Google Scholar]
- 34.Mao P, Han J. Fabrication and characterization of 20 nm nanofluidic channels by glass-glass and glass-silicon bonding. Lab Chip. 2005;5:837–844. doi: 10.1039/b502809d. [DOI] [PubMed] [Google Scholar]
- 35.Bow H, Fu J, Han J. Decreasing effective nanofluidic filter size by modulating electrical double layers: Separation enhancement in microfabricated nanofluidic filters. Electrophoresis. 2008;29:4646–4651. doi: 10.1002/elps.200800256. [DOI] [PubMed] [Google Scholar]
- 36.Huang LR, et al. Generation of large-area tunable uniform electric fields in microfluid arrays for rapid DNA separation. Tech Dig Int Elect Dev Mtg. 2002:363–366. [Google Scholar]