Abstract
The use of luciferase reporter genes together with luminescence detection has enabled high frequency monitoring of molecular circadian clock function in living tissues. With the help of an intensified CCD camera combined with an inverted epifluorescence microscope, the authors have established a new imaging strategy that makes use of transgenic cell type-specific expression of fluorescent proteins to identify cells of interest for subsequent circadian luminescence recording at single-cell resolution.
Keywords: circadian clock, Drosophila, imaging, luciferase, luminescence, fluorescence, gene expression
Circadian clocks are internal daily time-keeping mechanisms employed by a wide range of organisms to both predict daily environmental rhythms and organize many aspects of their physiology and behavior in a coherent daily schedule. All known circadian clocks rhythmically control biological functions by controlling gene expression and in most known cases the central time-keeping mechanism itself is a gene expression feedback circuit (e.g., Wijnen and Young, 2006). Circadian clock function can, therefore, be studied directly by monitoring circadian gene expression rhythms in tissues and cells from transgenic animals.
Traditionally, clock-controlled gene expression has been assayed by sampling from a synchronized population of cells, large heterogenous tissue explants, or whole organisms over the course of multiple days. This approach has at least two important limitations: 1) individual cellular clocks cannot be followed over time, making it difficult to separate defects in synchrony from defects in cell-autonomous clock function, and 2) it is relatively labor intensive, making this approach poorly suited to achieving high temporal resolution of circadian gene expression or high-throughput screening of molecular circadian pheno-types. The development of transgenic reporter constructs that make use of the firefly luciferase gene has provided an alternative method of assaying circadian gene expression that does not suffer from these limitations (Stanewsky, 2007; Welsh et al., 2005; Yamazaki and Takahashi, 2005; Yu and Hardin, 2007). Luciferase enzyme activity is reliably linked to gene expression rhythms and can be monitored noninvasively by assaying luminescence via luminometry or imaging, provided that the enzymatic substrate luciferin is made available to the cells of interest. In comparison with most fluorescent signals, luciferase-generated luminescence is much weaker, but it offers the advantage that background luminescence is close to zero and phototoxicity associated with long-term excitatory illumination can be avoided. Moreover, the luciferase protein is sufficiently unstable to allow for the detection of circadian rhythms generated at the transcript level.
The most recent technological developments in circadian bioluminescence imaging include the use of cooled charge-coupled device (CCD) cameras allowing highly sensitive imaging of live tissues and cultures with extremely low background. Some of these camera systems take advantage of ultra-cooled intensified CCD technology to achieve a single-photon threshold of detection. As a result, circadian bioluminescent signals have been detected from single cultured cells or tissue slices, allowing analysis of oscillator properties at the single-cell level as well as synchrony between cells in a population.
Here, we describe a new application of in vivo circadian luciferase imaging in combination with fluorescent microscopy aimed at selective monitoring of molecular clock function in individual cells. The introduction of fluorescent marker proteins enables the association of circadian luciferase reporter rhythms with spatiotemporal expression patterns of interest. This allows the selection of relevant subsets of native or functionally manipulated cells for bioluminescent imaging as well as verification of assumed or predicted spatiotemporal luciferase expression patterns.
In the examples shown in Figures 1 and 2 and Supplemental Material S2 and S3, cell type-specific expression of the membrane- tethered green fluorescent protein CD8::GFP (Lee and Luo, 1999) allowed in vivo monitoring of individual clock neurons for circadian gene expression rhythms in preparations of dissected whole Drosophila brains. Due to the relative weakness of the luminescent signal and small number of clock-bearing neurons in the adult Drosophila brain (~150; Hall, 2005) the ability to preselect cells for luciferase imaging based on GFP expression greatly enhanced the efficiency of recording circadian gene expression rhythms for individual cells in these preparations. Although visualization of Drosophila clock neurons with luciferase reporters has recently been reported (Sehadova et al., 2009; Yoshii et al., 2009), the present study is, to our knowledge, the first to demonstrate successful circadian bioluminescence imaging of these cells. Single-cell circadian bioluminescence rhythms were recorded for various types of Drosophila clock neurons including the peptidergic large ventral lateral neurons (l-LNVs) as well as dorsal lateral neurons (LNds).
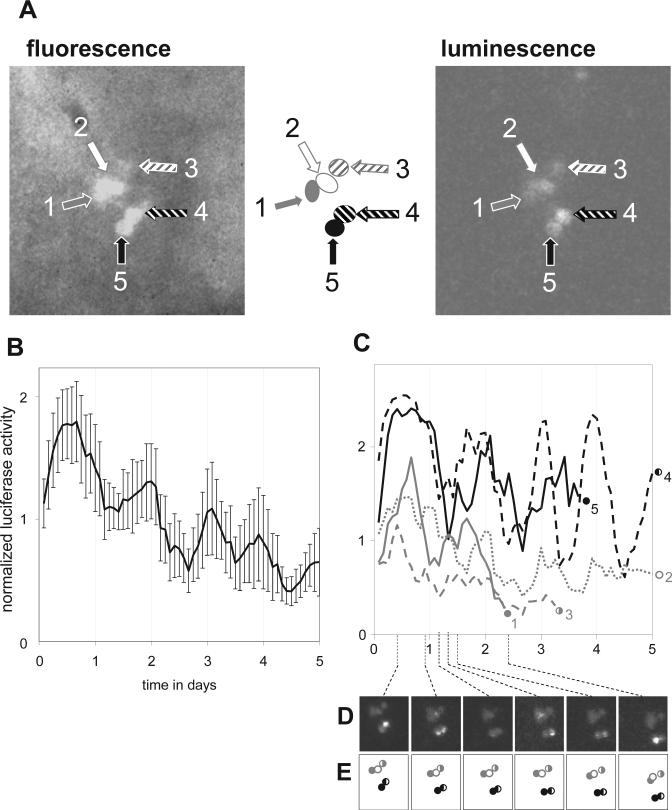
Figure 1.
Imaging of single-cell circadian gene expression in cultured Drosophila brains (see also Supplemental Material S2). A cluster of 5 ventral lateral clock neurons was identified based on cell type-specific fluorescence (Pdf>GFP) and imaged for luminescence from a circadian luciferase reporter (tim-luc) in the dark. Fluorescence and luminescence images of these cells at the start of the time course are shown in A. Mean single-cell bioluminescence (± SEM) time course data (normalized to the experimental average) is graphed in B, whereas separate analyses for individual cells are graphed in C. Data corresponding to poor quality signals for 3 of the cells has been omitted causing 3 of the line graphs in C to end prematurely. Individual images from select time points are shown in D to illustrate the phase differences between different neurons. The diagrams in E indicate the positions of the imaged neurons (same symbols as in C) in each of the images in D.
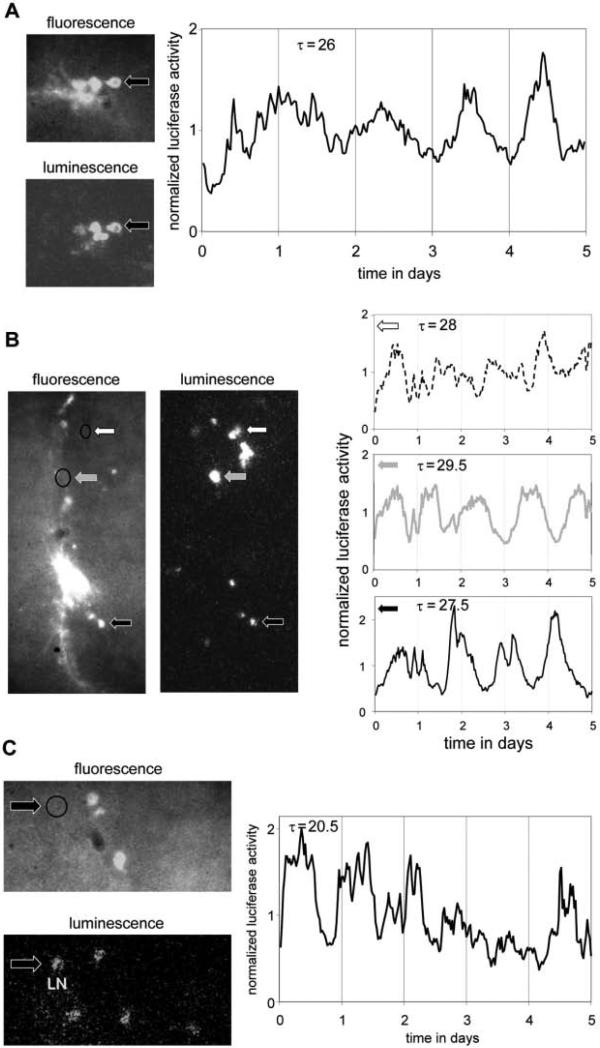
Figure 2.
Circadian imaging of individual clock neurons across different cell types and genotypes (see also Supplemental Material S3). Using the same fluorescent and bioluminescent reporters as in Figure 1 (Pdf>GFP and tim-luc) significant circadian luminescence rhythms were detected in individual ventral and dorsal lateral neurons (LNvs and LNds) in cultured brains from wild-type (A, B) or short period mutant clock mutant (dbts/+) flies (C). Period lengths (τ) of significant circadian luminescence rhythms during days 2 to 5 (p < 0.001; A, B) and 1 to 5 (p < 0.025; C) are indicated. Arrows and graphs are patterned in C to identify the cells whose luciferase rhythms are plotted. The absence of fluorescence in clock neurons imaged in B and C (circled areas) indicates that these are not PDF-expressing cells. Based on their location and morphology these cells were identified as LNd (B) or LN (either 5th sLNv or LNd; C).
In the experiment represented in Figure 1 and Supplemental Material S2, a cluster of five l-LNVs was identified based on their cell size and relative strength of expression of CD8::GFP under control of Pigment dispersing factor (Pdf) promoter sequences. These 5 cells were then imaged during 5 days for bioluminescence produced by a reporter for the timeless promotor and 5′ untranslated region (tim-luc) (Stanewsky et al., 1998). The luminescence signal for each of the 5 cells appeared to be rhythmic with an approximate circadian period length during the portion of the time course where it could be reliably detected (see Fig. 1C). Indeed, χ2 periodogram analyses for cells 2 and 4 across days 2 to 5 indicated significant periodicity at period lengths of 23 h (p < 0.05) and 23.5 h (p < 0.001), respectively. Circadian rhythmicity was also suggested by the average signal among the 5 cells (see Fig. 1B). However, because the rhythms of the individual cells showed considerable variation in phase, rhythms were actually more robust at the single-cell level. Similar observations were made in independent imaging experiments for l-LNv and LNd neurons in brains of the same genotype (see Fig. 2A, B; Supp. Material S3). Robust single-cell rhythms were identified with considerable phase differences even among cells in the same clusters. The rhythms observed, here, under constant conditions for the transcriptional tim-luc reporter in individual l-LNvs are consistent with previous studies that reported tim transcript rhythms (Peng et al., 2003; Stoleru et al., 2005) despite greatly reduced PER and TIM protein oscillations (Helfrich-Forster et al., 2007).
Next, we introduced a dominant short period mutation at the double-time locus (dbts) (Price et al., 1998) in the tim-luc; Pdf>CD8::GFP genetic background. As expected, single-cell bioluminescence recordings from LNs in the brains heterozygous for dbts showed robust rhythms with a shortened period length (see Fig. 2C). Similar manipulations with combinations of other classical mutations and transgenic constructs potentially offer a plethora of phenotypic contexts for studying circadian bioluminescence.
Although the experimental applications of circa-dian fluorescence/bioluminescence imaging presented here are limited to preparations of Drosophila brains, practically any tissue can be imaged in this way as long as in vivo circadian bioluminescence reporters and fluorescent markers are available. Finally, we would like to point out the potential for employing fluorescent fusion proteins to add another functional dimension to the circadian fluorescence/luminescence dual mode imaging experiment. Core circadian clock components show rhythms in protein abundance, and subcellular localization that can be readily visualized with the help of fluorescent fusion proteins (Meyer et al., 2006). As long as the light associated with fluorophore excitation and emission does not interfere with the circadian parameters of interest, parallel fluorescence/luminescence imaging can be used to describe the relationship between the clock-controlled gene expression rhythms reported by luciferase and rhythms in subcellular localization or protein abundance reported by fluorescently tagged clock components.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Ralf Stanewsky, Michael Rosbash, Michael Young, and the Bloomington Stock Center for fly stocks. Funding for this research was provided by NIH grants GM78339 to HW, and MH56647 to MM.
APPENDIX
Drosophila genetic crosses were conducted to combine clock neuron-specific expression of membrane-bound GFP with circadian expression of luciferase. Specifically, an X-chromosomal tim-luc transgene (Stanewsky et al., 1998) was combined with Pdf-Gal4 (2nd chromosome) (Park et al., 2000) and UAS-CD8::GFP (3rd chromosome; Lee and Luo 1999) transgenes (together abbreviated as Pdf>CD8::GFP), which produce Gal4-mediated expression of CD8::GFP in the ventral lateral clock neurons. In a separate genetic cross, flies were generated that contained the same transgenes, but also carried one copy of the short period mutation dbts (Price et al., 1998). Adult Drosophila brains of both experimental genotypes were dissected on a chilled metal surface and kept in insect tissue culture media without luciferin [85.9% Shields and Sang M3 insect tissue culture media, 12% fetal bovine serum (heat inactivated for 30 min at 60 °C), 1% penicillin-streptomycin mixture, and 0.1% insulin (1 mg/mL) solution] until mounting. At least 1 day prior to imaging, the brains were mounted on a sterile filter insert (Millicell-CM; Millipore Inc.) and immobilized under a 13-mm coverglass with the help of sterile vacuum grease (see Supp. Fig. S1B). The insert was then placed in a sterile glass-bottom dish (FluoroDish FD35PDL) containing 1.2 mL of insect tissue culture media with 0.1 mM luciferin. Sterile vacuum grease was then applied to seal the dish. Because we generally observed considerable movement in the brain tissue during the 1st day after mounting, imaging time courses were started with a delay of at least 1 day. Fluorescence/luminescence circadian imaging was conducted with an inverted epifluorescence microscope (Olympus CKX-41 equipped with an X-CITE 120 microscope light source system and mineralogic objective Olympus LUCPLNFL 40X LWD, NA 0.6, WD 2.7-4.0 mm with correction collar) and cooled intensified CCD camera (Mega10Z; Stanford Photonics Inc., Palo Alto, CA) housed in a light-tight wooden dark box. Sample temperature was controlled during recordings by a forced air heater (WPI Inc., Sarasota, FL). Following an initial fluorescent reference image, luminescence images (66.6 msec/ frame × 900 frames = 1 min; TIFF format) were collected (1440 images/24 h) and processed offline using Piper image analysis software (Stanford Photonics). Dark noise (read noise) was eliminated by setting the detection threshold in the Piper software just above the level of noise generated by simply operating the camera in total darkness. The use of a software integrated “discrimination filter” allowed for real-time online removal of any frames containing cosmic ray events (approximately 5-7% of frames), therefore significantly improving signal-noise ratio of the images. At the end of the recording, raw 1-min images were further integrated into 3-min images and a 3-h moving average (over 60 images) was applied to smooth the data. For data analysis, 1 of every 10 smoothed 3-min images (1 image every 30 min) was selected and combined to create an image stack (48 images/24 h). Image stacks were then used to identify regions of interest (single cells) and luminescent pixel intensity was quantified with ImageJ (NIH) and plotted using Excel (Microsoft). Finally, circadian rhythmicity of cellular luminescence was evaluated using the χ2 periodogram feature of the ClockLab software package. At least 4 independent experiments with 1 to 5 informative individual cells each were carried out for each genotype. Representative results are shown.
Footnotes
Supplementary material for this article is available on the Journal of Biological Rhythms Web site at http://jbr .sagepub.com/supplemental.
REFERENCES
- Hall JC. Systems approaches to biological rhythms in Drosophila. Methods Enzymol. 2005;393:61–185. doi: 10.1016/S0076-6879(05)93004-8. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Yoshii T, Wulbeck C, Grieshaber E, Rieger D, Bachleitner W, Cusamano P, Rouyer F. The lateral and dorsal neurons of Drosophila melanogaster: New insights about their morphology and function. Cold Spring Harb Symp Quant Biol. 2007;72:517–525. doi: 10.1101/sqb.2007.72.063. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R. Temperature entrainment of Drosophila's circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron. 2009;64:251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Stanewsky R. Analysis of rhythmic gene expression in adult Drosophila using the firefly luciferase reporter gene. Methods Mol Biol. 2007;362:131–142. doi: 10.1007/978-1-59745-257-1_9. [DOI] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuro-peptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Use of firefly luciferase activity assays to monitor circadian molecular rhythms in vivo and in vitro. Methods Mol Biol. 2007;362:465–480. doi: 10.1007/978-1-59745-257-1_38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.