Abstract
It is now established that the transcription factors E2A, EBF1 and Foxo1 play critical roles in B cell development. Here we show that E2A and EBF1 bound regulatory elements present in the Foxo1 locus. E2A and EBF1 as well as E2A and Foxo1, in turn, were wired together by a vast spectrum of cis-regulatory codes. These associations were dynamic during developmental progression. Occupancy by the E2A isoform, E47, directly elevated the abundance as well as the pattern of histone H3K4 monomethylation across putative enhancer regions. Finally, the pro-B cell epigenome was divided into clusters of loci that show E2A, EBF and Foxo1 occupancy. From this analysis a global network consisting of transcriptional regulators, signaling and survival factors, was constructed that we propose orchestrates the B cell fate.
The basic unit of the chromatin fiber is the nucleosome. A nucleosome consists of a 146 bp DNA segment folded around an octamer of histones that are comprised of two copies each of H2A, H2B, H3 and H4. The N-terminal tails of H2A, H2B, H3 and H4 are epigenetically marked and can be post-translationally modified by acetylation of lysines, methylation of arginines and lysines, ubiquitination of lysines as well as phosphorylation of serines and threonines1. The acetylation of core histone residues generally correlates well with a chromatin fiber that is accessible to the transcriptional machinery2. Histone acetyltransferases catalyze acetylation of histone tails. In contrast, deacetylation of histone core tails is mediated by the histone deacetylases and is primarily associated with transcriptional repression. Methylation of histone residues promotes interactions of core histones with factors that play critical roles in chromatin remodeling (chromodomains as well as with plant homeodomain (PHD) domains). Methylation of core histone lysine residues is mediated by methyltransferases whereas histone demethylases act to remove methyl groups from the tails of core histones. Nucleosome depletion is generally associated with active promoters in yeast as well as in Drosophila2. Active promoters are epigenetically marked by the acetylation of multiple H3 and H4 residues as well as trimethylation of H3 residue lysine 4 (H3K4; ref. 3). In contrast, enhancer elements are characterized by the monomethylation of H3K4 (ref. 4).
Hematopoiesis is initiated from a small pool of hematopoietic stem cells (HSCs). HSCs are pluripotent and have the ability to differentiate into either lymphoid-primed multipotent progenitors (LMPPs) or alternatively into erythroid/egakaryocytic progenitors. LMPPs, in turn, differentiate into common lymphoid progenitors (CLPs) or granulocyte-macrophage progenitors (GMPs). B cell development requires a hierarchy involving multiple transcription factors, including E2A [http://www.signaling-gateway.org/molecule/query?afcsid=A000804]. In CLPs the helix-loop-helix protein, E47 [http://www.signaling-gateway.org/molecule/query?afcsid=A000806], an isoform encoded by the E2A gene, induces the expression of early B cell factor (EBF1 [http://www.signaling-gateway.org/molecule/query?afcsid=A000809])5–8. EBF1, in turn, activates the expression of Pax5 [http://www.signaling-gateway.org/molecule/query?afcsid=A000403] (ref. 9). Pax5 gene expression is also regulated by the concerted activities of the ETS-family transcription factor PU.1, and interferon regulatory factors IRF4 and IRF8 (refs 9–11).
In addition to E2A, EBF1 and Pax5, other transcriptional regulators modulating the developmental progression of B lineage cells have recently been identified. Prominent among these are Bcl11A and Foxo1 [http://www.signaling-gateway.org/molecule/query?afcsid=A000944]. B cell development in Bcl11A-deficient mice is arrested at a stage similar as described for E2A and EBF1-deficient mice12. Foxo1-deficient mice show a block at the pro-B cell stage and Foxo1 directly activates Rag1 and Rag2 gene expression13,14. Collectively, these factors form the core of the transcriptional machinery that promotes commitment to the B cell lineage, suppress the expression of genes associated with alternate cell fates and coordinate cellular expansion with developmental progression15,16.
As a first approach to determine how these factors are linked to promote the B cell fate, we performed a genome-wide analysis for E2A, EBF1 and Foxo1. We show that the DNA binding activities of E2A, EBF1 and Foxo1, were linked to global patterns of H3K4 methylation. Cistromes associated with E2A occupancy were primarily H3K4 monomethylated but underwent further epigenic alteration during the transition from the pre-pro-B to the pro-B cell stage. These altered cistromes frequently associated with EBF1 and with Foxo1, albeit to a lower extent. We show that in pro-B cells, E2A and EBF1 bound regulatory elements in the Foxo1 locus and that E2A, EBF and Foxo1, in turn, interact with putative enhancer elements in the Pax5 locus. We demonstrate that the coordinate DNA binding activities of E2A and Foxo1 have functional significance since mice heterozygous for both E2A and Foxo1 show a near complete absence of B cells. These data link E2A and the PI3K cascade into a common pathway. Finally, we describe the pro-B cell state in terms of a global network of transcription factors that involve E2A, EBF1 and Foxo1.
RESULTS
Cis-regulatory codes associated with E2A occupancy
Previous observations have established that the E2A proteins act to modulate lymphocyte differentiation, cell survival, cellular expansion and to suppress the development of lymphoma17. To determine how the E2A proteins mechanistically coordinate developmental progression and cell growth, E2A occupancy in pre-pro-B (EBF1-deficient) and pro-B cells (RAG1-deficient) was determined using chromatin immunoprecipitation combined with deep DNA sequencing (ChIP-Seq). More than 4531 E2A bound sites were identified in pre-pro-B cells, whereas in pro-B cells 11846 binding sites showed E2A occupancy. Approximately 44% of the E2A-bound sites in pre-pro-B occupied the same sites in pro-B cells (1993 out of 4531 sites). However, the majority of E2A occupancy was exclusively associated with pro-B cells, consistent with the critical roles that E2A proteins play at this developmental stage (9853 out of 11846 sites). Although DNA binding within close proximity from the transcription initiation sites was enriched, E2A occupancy was widespread and predominantly associated with either introns or intergenic regions (Supplemental Fig. 1a and Fig. 1a).
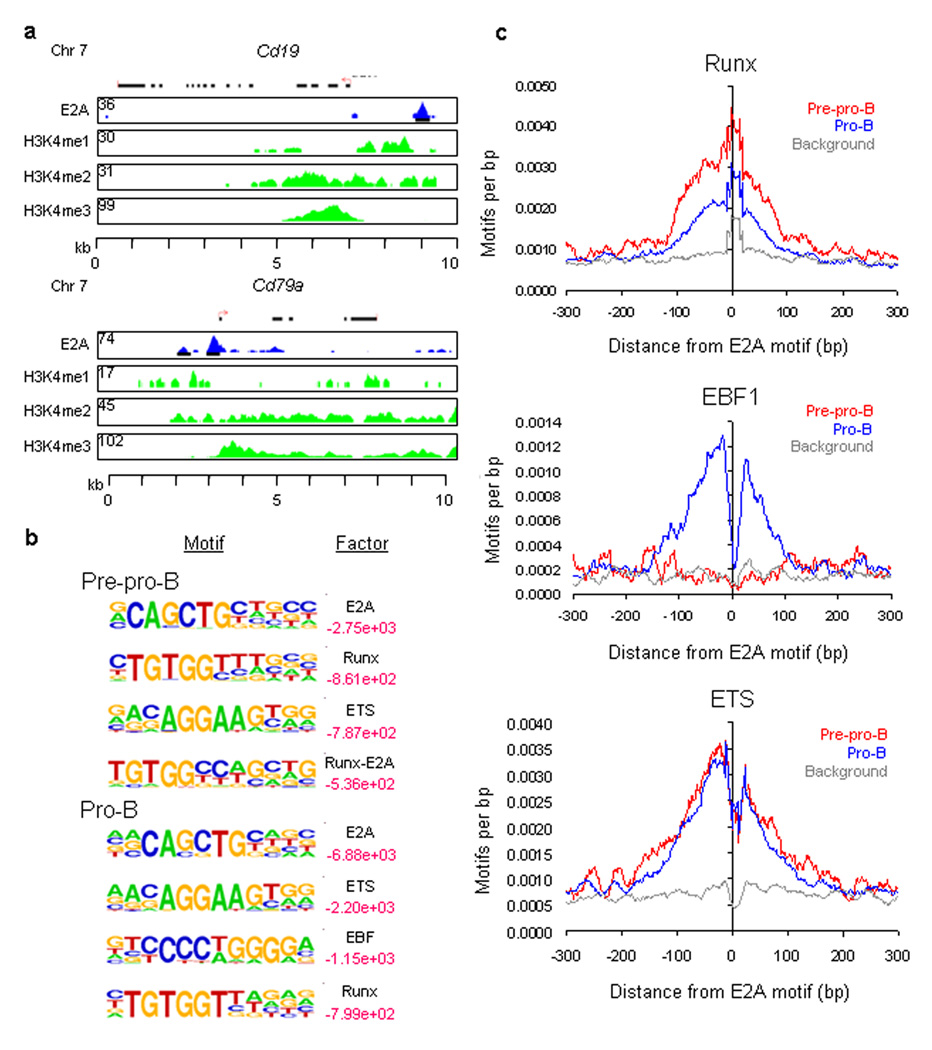
Figure 1.
E2A occupancy and epigenetic marking in cultured EBF-deficient pre-pro-B and RAG1-deficient pro-B cells. (a) E2A DNA binding sites in the Cd19 and Cd79a loci. Binding sites were identified using ChIP-Seq. UCSC Genome Browser was used to visualize binding patterns. Blue pattern refers to E2A occupancy. Green patterns refer to H3K4 methylation. Red arrow refers to transcription initiation sites. Numbers refer to the number of tags observed. Regions of H3K4me1, H3K4me2 and H3K4me3 are shown. (b) Cis-regulatory sequences associated with E2A occupancy in pre-pro-B and pro-B cells. Motifs that companion E2A occupancy were identified by comparing enriched peaks to random selected genomic DNA sequences. Log p-values are indicated in red. Smaller values represent relative high enrichment for a given motif as compared to randomly selected regions. (c) E2A occupancy is tightly associated with cis-regulatory elements containing EBF, ETS and RUNX consensus binding sites. Relative frequencies of EBF, ETS and RUNX consensus binding sites localized within close genomic proximity of E2A bound regions. Two independent ChIP-Seq experiments were performed in order to determine E2A occupancy for pre-pro-B and pro-B cells. A total of three independent ChIP-Seq experiments were performed in order to determine the pattern of H3K4me1, H3K4me2 and H3K4me3 in pro-B cells.
To determine whether E2A interacts collaboratively with alternative transcriptional regulators in EBF1-deficient pre-pro-B and RAG1-deficient pro-B cells, we examined E2A-associated sites for enriched sequence elements. A de novo motif finder algorithm, HOMER (Hypergeometric Optimization of Motif EnRichment) http://biowhat.ucsd.edu/homer/, was utilized to identify motifs localized ± 100 bp from E2A occupancy (Fig. 1c and Supplementary Table 1). The HOMER algorithm ranks motifs in transcription factor-associating DNA sequences based on their statistical enrichment. As expected, the E2A consensus DNA sequence (GCAGCTG) was ranked as the top-scoring motif in both pre-pro-B and pro-B cells (Fig. 1c). In pre-pro-B cells, E2A-bound regions were enriched for both Runx1 [http://www.signaling-gateway.org/molecule/query?afcsid=A000523] and ETS consensus motifs (Fig. 1c). In contrast to pre-pro-B cells, E2A-bound regions in pro-B cells were highly enriched for the EBF1 high-affinity binding sites (Fig. 1c,d and Supplementary Table 1).
To determine the relationship between E2A occupancy and its associated cis-regulatory elements, the genomic distances that separate EBF1, ETS and Runx consensus motifs from E2A binding sites were plotted (Fig. 1d). This analysis revealed that EBF1, Runx and ETS motifs were highly enriched within a 200 bp window centered across E2A-bound regions (Fig. 1d). Notably, the pattern of Runx occupancy was quite distinct from that observed for EBF1 and ETS DNA binding. Rather than showing a bimodal distribution, the Runx consensus motif appeared to overlap with E2A occupancy. These data indicate that the spacing separating E2A and Runx binding sites is under strict selective pressure and may reflect a special requirement for cooperative rather than collaborative DNA binding.
E2A occupancy is associated with H3K4 methylation
Previous studies have demonstrated a tight correlation between transcriptionally active promoters and H3K4 trimethylation, whereas H3K4 monomethylation has been associated with enhancer activity2–4,18–20. To determine whether E2A occupancy is allied with either promoter or enhancer regions, cell lysates derived from RAG1-deficient pro-B cells were immunoprecipitated with antibodies directed against H3K4me1, H3K4me2 and H3K4me3 and analyzed by ChIP-Seq. Consistent with previous observations, H3K4me3 was closely associated with the active promoter regions of the Cd19 and Cd79a loci (Fig. 1b). RAG1-deficient pro-B cells were also analyzed separately for global patterns of acetylated H3 (K9 and K14) (ref 21). The majority of E2A binding was associated with H3K4 monomethylation, whereas only a subset was associated with H3 acetylation (Supplementary Fig. 1b).
To determine whether epigenetic marks are globally aligned with E2A occupancy, cumulative sequence tag frequencies associated with E2A-bound sites were plotted for each of the histone marks (Fig. 2; Supplementary Fig. 1). To reveal the distribution of H3K4me1, the degree of H3K4me1 was plotted as a function of genomic separation from the E2A-bound sites (Fig. 2a, b). This analysis revealed a bimodal distribution with a substantial decrease in H3K4 methylation centered on the E2A-bound sites (Fig. 2a, b).
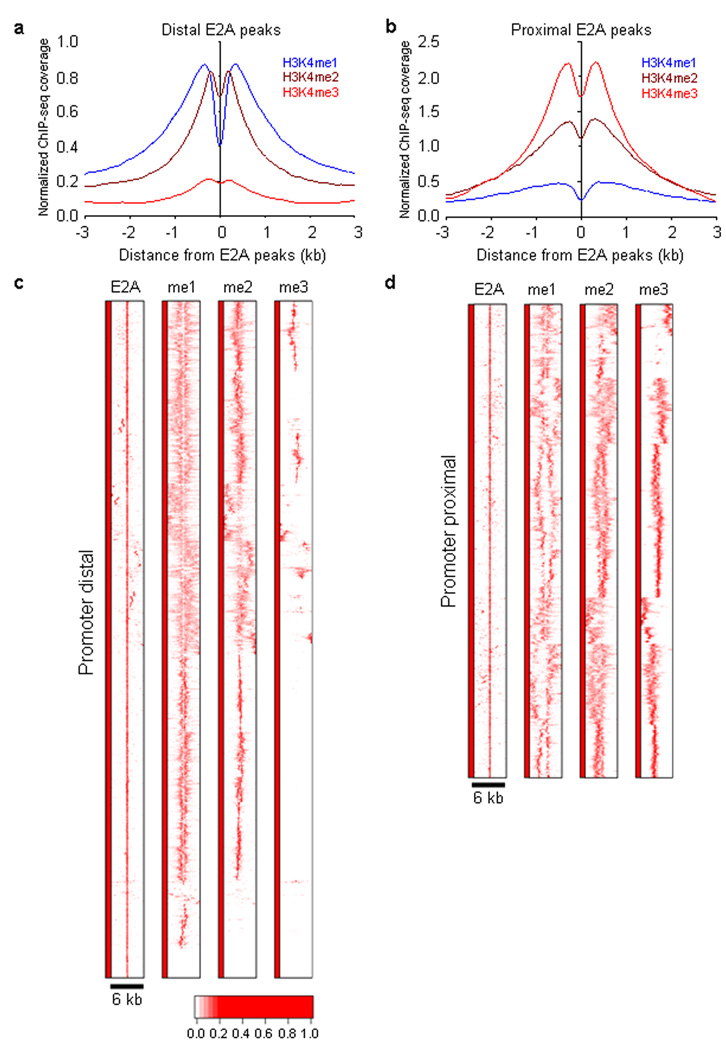
Figure 2.
E2A occupancy and H3K4 methylation in cultured RAG1-deficient pro-B cells. (a) Heat map of E2A occupancy and H3K4me1, H3K4me2 and H3K4me3 patterns for promoter-distal DNA sequences (> 3 kb separation from nearby transcription start sites). Each row represents a 6 kb genomic region showing E2A occupancy. The heat map reflects the enrichment of E2A binding and H3K4 methylation by color according a scale as indicated below. (b) Heat map of E2A occupancy and distributions of H3K4me1, H3K4me2 and H3K4me3 for promoter-proximal (< 3kb from nearby transcription initiation sites) region. (c) E2A occupancy correlates with H3K4 mono- and di-methylation in promoter distally located regions. The genomic distance distributions of H3K4 methylation (H3K4me1, H3K4me2 and H3K4me3) centered across E2A occupancy are shown across a 6 kb genomic region. (d) E2A occupancy correlates with H3K4 mono- and di-methylation in promoter-proximal located regions. The genomic distance distributions of H3K4 methylation (H3K4me1, H3K4me2 and H3K4me3) centered across E2A occupancy are shown across a 6 kb genomic region. A total of four independent ChIP-Seq experiments were performed in order to determine genome-wide E2A occupancy as well as patterns of H3K4me1, H3K4me2 and H3K4me3.
—To examine the shape of the distributions centered across individual E2A bound sites, heat maps of 10,000 bound regions were generated and analyzed genome-wide. We examined E2A occupancy in promoter-distal (>3 kb separated from the transcription start site (TSS)) as well as promoter-proximal (<3 kb separated from the TSS) regions (Fig. 2c,d). Normalized tag densities for either promoter-distal or promoter-proximal regions were plotted for E2A occupancy as well as H3K4me1, H3K4me2 and H3K4me3 methylation. E2A binding sites were aligned at the centers and designated as position zero. Genomic regions located within a 3 kb window upstream and downstream of E2A occupancy were identified and visualized (Fig. 2c,d). E2A occupancy was primarily associated with H3K4me1 and H3K4me2 in distal regions (Fig. 2c). Only a minor proportion of E2A occupancy was accompanied by H3K4me3 in promoter-distally located regions (Fig. 2c). As expected, however, in proximal elements the majority of E2A DNA binding was linked with H3K4 trimethylation (Fig. 2c).
Two basic patterns emerged: a focused and narrow distribution of H3K4 monomethylation as well as a diffuse and asymmetric pattern (Fig. 2c,d). The pattern of H3K4 monomethylation in promoter regions was diffuse for the majority of the segments that were analyzed (Fig. 2d)22. Focusing on H3K4 monomethylated sites present in pro-B cells, 22% were associated with E2A occupancy (data not shown). In sum, this analysis indicates that a substantial proportion of putative enhancer elements in pro-B cells are bound by E2A. Furthermore, the data show that the patterns of H3K4 methylation linked with E2A occupancy are distinct in promoter-proximal versus promoter-distal regions.
E2A occupancy and H3K4 methylation
To determine whether E2A occupancy changes during early B lineage development, we examined E2A DNA binding in both EBF1-deficient pre-pro-B and RAG1-deficient pro-B cells. The binding sites were gated on promoter-distal located genomic regions. Ten thousand randomly chosen E2A binding sites in pre-pro-B as well as pro-B cells were aligned at centers. The promoter-distal subset of these sites (7296) were plotted and directly compared to the patterns of H3K4me1, H3K4me2 and H3K4me3.
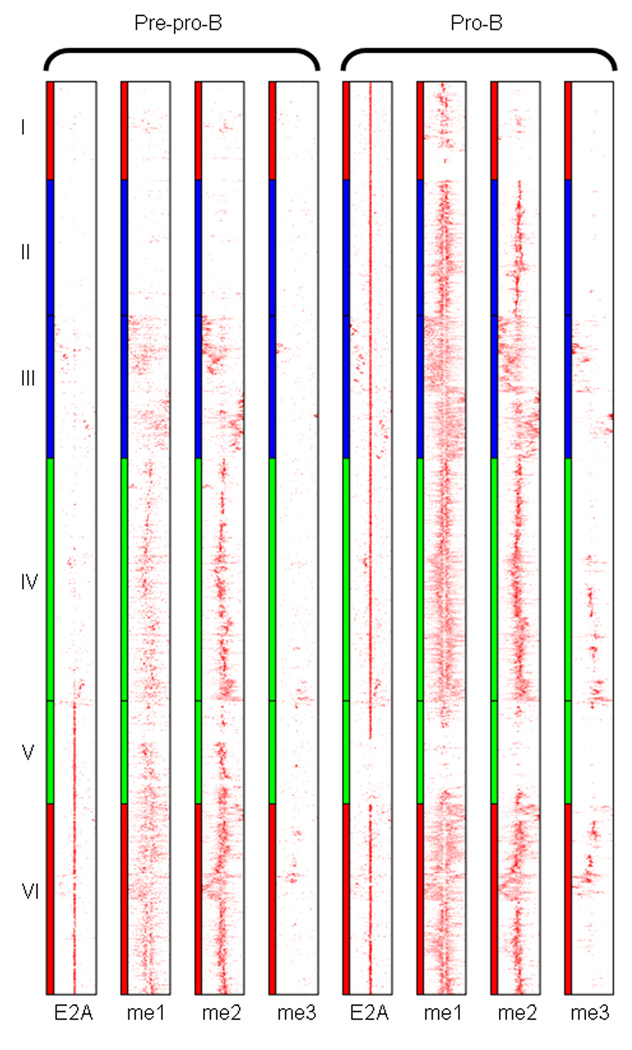
Since we observed multiple patterns of E2A occupancy and H3K4 methylation in pre-pro-B and pro-B cells, hierarchical clustering was performed to categorize patterns of E2A occupancy and H3K4 methylation across the two developmental stages (Fig. 3). To identify such clusters a Ward hierarchical clustering method was performed23. This analysis revealed distinct groups that can be distinguished from each other based on E2A occupancy and its associated patterns of H3K4me. A subset of DNA segments (clusters I-IV) showed E2A occupancy in pro-B cells but not in pre-pro-B cells. Cluster V showed E2A DNA binding at the pre-pro-B cell stage, whereas another subset exhibited E2A occupancy at both the pre-pro-B and pro-B cell stage (cluster VI).
Figure 3.
E2A occupancy and patterns of H3K4 methylation in cultured EBF1-deficient pre-pro-B and RAG1-deficient pro-B Cells. Heat map of E2A occupancy and distributions of H3K4me1, H3K4me2 and H3K4me3 patterns for promoter-distal (>3 kb from nearby transcription initiation sites) region (7296 sites). Left, distributions for pre-pro-B cells; right, distributions for pro-B cells. Each row represents a 6 kb window centered across the E2A binding site. The heat map reflects the enrichment of E2A binding and H3K4 methylation and grouped as determined by Ward hierarchical clustering method. Clustered E2A sites that comprise a group, occupied in either pre-pro-B or pro-B cells or both, are more closely related to each other than those assigned to different clusters. Clusters I, II, III, IV, V, and VI represent 785, 1083, 1140, 1940, 824, and 1524 binding sites, respectively. Eight independent ChIP-Seq experiments were performed in order to determine global E2A occupancy as well as patterns of H3K4 methylation.
The difference in the patterns of H3K4me between pre-pro-B and pro-B cells was quite distinct, but overall correlated well with E2A occupancy across the entire spectrum of binding sites (Fig. 3). The large majority of E2A occupancy correlated well with H3K4me1 and H3K4me2 in both pre-pro-B and pro-B cells. In pre-pro-B cells, H3K4me1 symmetrically flanked the majority of E2A binding sites (clusters V and VI). However, a subset of E2A binding sites in pre-pro-B cells was not associated with H3K4me1. In pro-B cells we observed multiple patterns of H3K4me that correlated with E2A occupancy. Cluster I either lacked H3K4me1 or showed a diffuse and subtle pattern of H3K4me. Cluster II and IV exhibited a distribution of H3K4 that was focused and symmetrically positioned. In contrast, cluster III was associated with a diffuse and asymmetric distribution of H3K4me1. Cluster VI showed a mixed pattern of H3K4me1, primarily focused and symmetric but also regions that showed an asymmetric distribution.
To explore the possibility that binding patterns are associated with unique cis-regulatory codes, we examined for the presence of enriched DNA sequences that companion E2A occupancy using both MEME (motif finding) as well as HOMER (statistical values). The E2A consensus motif was largely invariant across each of the clusters (Fig. 4). However, within each cluster distinct cis-regulatory sequences were associated with E2A occupancy.
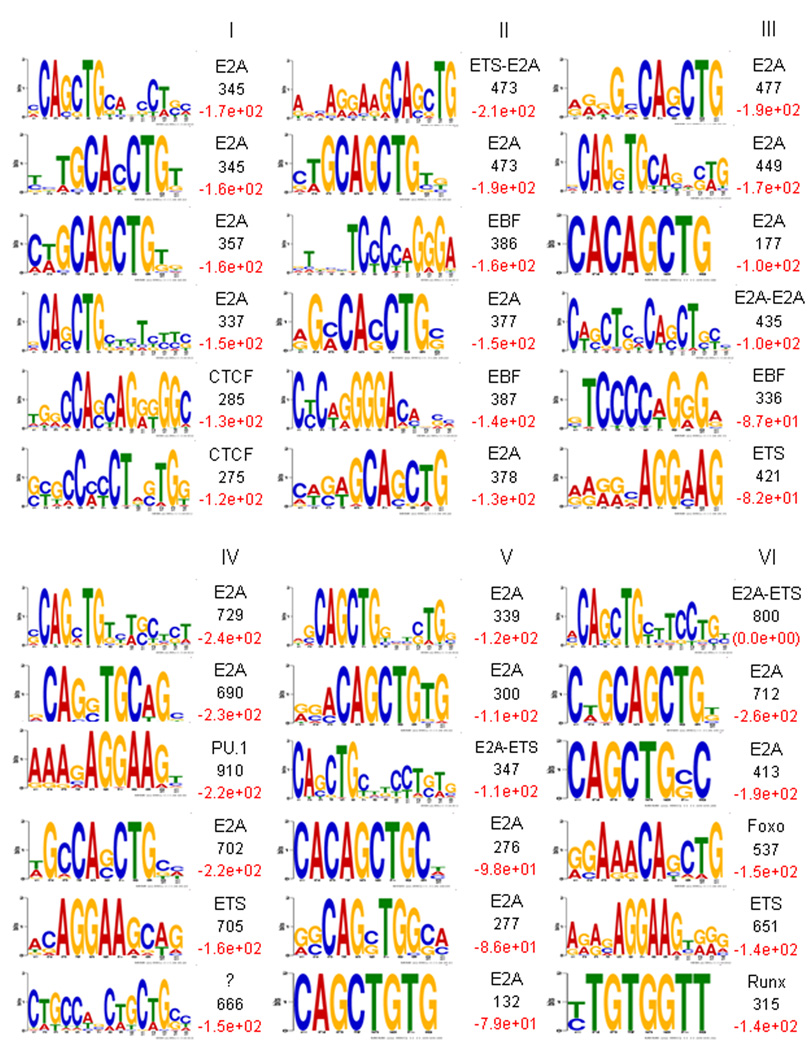
Figure 4.
Distinct cis-regulatory DNA sequences associate with E2A occupancy in cultured RAG1-deficient pro-B cells. Cis-regulatory sequences associated with E2A occupancy and patterns of H3K4 monomethylation were identified by comparison of enriched DNA binding sites (clustered according to Fig. 3) to random selected genomic DNA sequences. Cis-regulatory sequences were gated on DNA sequences located plus or minus 100 bp from the E2A binding sites. Sequences distinct from each cluster were pooled and analyzed by MEME algorithm. The top 6 scoring motifs (ranked by log P-value as determined by HOMER are indicated in red, right) are listed for each cluster. The number of non-redundant motif occurrence is indicated in black (right). Transcription factors associated with cis-regulatory sequences (CTCF, ETS, EBF, Pu.1, Foxo, Runx) are indicated.
Notably, cluster I showed a substantial enrichment for cis-regulatory sequences closely associated with CTCF DNA binding. The potential association of E2A with the architectural protein CTCF is intriguing and it raises the possibility that E2A and CTCF act in concert to modulate long-range chromatin structure. Consistent with them acting as modulating long-range chromatin structure, cis-regulatory elements that contain both E2A and CTCF binding sites predominantly lacked the enhancer mark, H3K4me1 (Fig. 3, 4). Clusters II and III were primarily associated with a composite ETS-E2A consensus DNA sequence (AGGAAG-CAGCTG) as well as EBF1 (TCCCNNGGGA). Cluster IV showed substantial enrichment for a PU.1 consensus DNA sequence (AGGAAG). Cluster V, primarily associated with E2A occupancy in pre-pro-B but not in pro-B cells, was linked to ETS-E2A consensus DNA sequences. Foxo1 (G/AGAAACAGCTG) as well as Runx (TGTGGTT) consensus DNA sequences appeared primarily linked with cluster VI, a group of DNA segments that showed E2A occupancy in both pre-pro-B and pro-B cells (Fig. 3, 4). Taken together, the clustering analysis revealed that in pro-B cells, E2A occupancy is associated with a wide spectrum of cis-regulatory elements.
In pro-B cells E2A, EBF1, FOXO1 and CTCF are wired together by distinct cis-regulatory codes
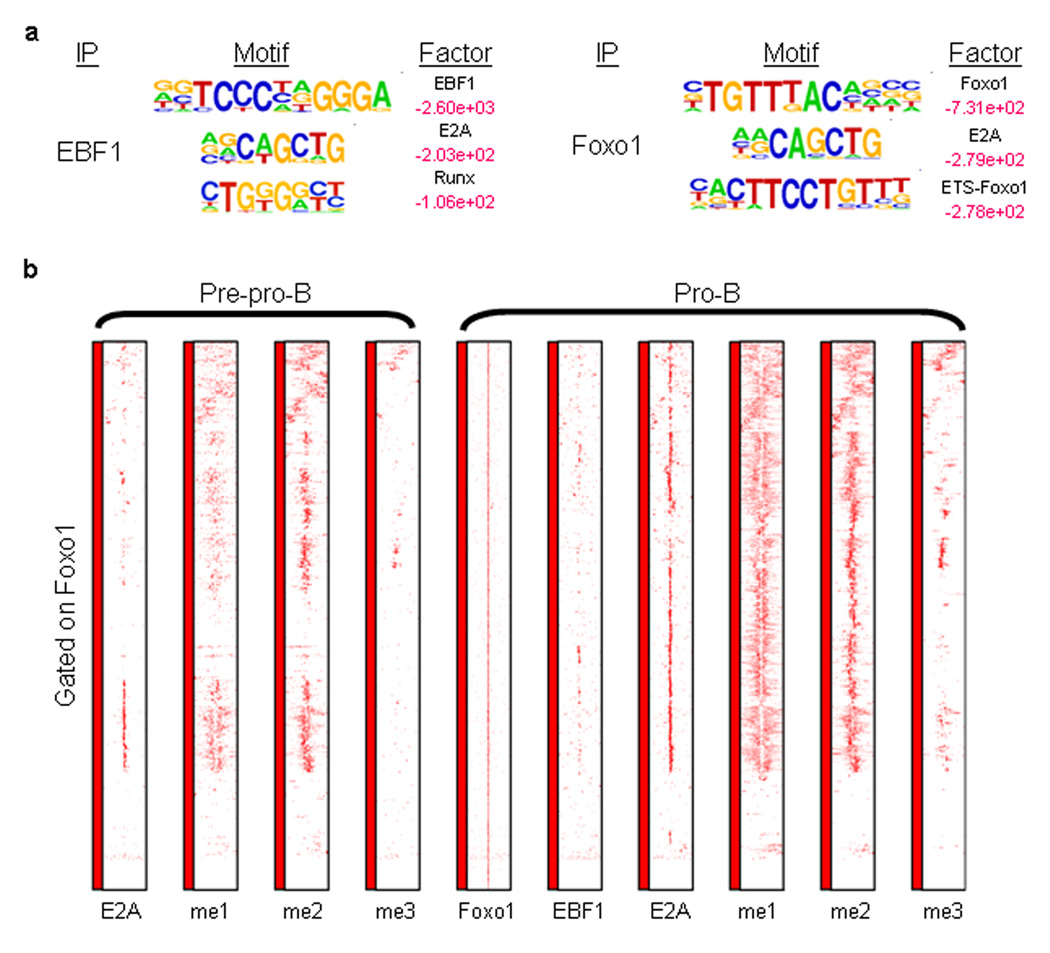
The E2A-associated motif identification analyses presented above suggest coordinate DNA binding activity of E2A and EBF, E2A and Foxo1 as well as E2A and CTCF to a subset of regulatory elements. To confirm the concerted DNA binding activities of E2A and its putative partners, cell lysates derived from RAG1-deficient pro-B cells were immunoprecipitated with antibodies directed against EBF1, Foxo1 and CTCF and analyzed by ChIP-Seq. EBF1, Foxo1and CTCF occupancy was primarily associated with intergenic and intronic regions (data not shown).
The top-scoring motif associated with EBF1 occupancy was the EBF1 consensus-binding site (Fig. 5a, left; Supplementary Table 1). Consistent with the data described above, the E2A binding site was frequently associated with EBF1 occupancy (Fig. 5a and Supplemental Fig. 2). The clustering analysis predicts that E2A and Foxo1 also bind in a coordinate manner to a subset of cis-regulatory elements. To test this prediction directly, RAG1-deficient pro-B cells lysates were immunoprecipitated with an antibody directed against Foxo1 and analyzed by ChIP-Seq. In addition to the expected enrichment for the Foxo1-consensus site, composite ETS-Foxo1 and E2A-Foxo1 binding sites were also enriched (Fig. 5a, right). Normalized tag densities for distally located regulatory elements were plotted for E2A, H3K4me1, H3K4me2 and H3K4me3 for a 6 kb window centered across Foxo1 DNA binding sites (Fig. 5c). A sizable fraction of Foxo1 sites were associated with E2A DNA binding (Fig. 5c)
Figure 5.
Distinct patterns of H3K4 monomethylation are associated with coordinate E2A and EBF1 and FOXO1 occupancy. (a) Cis-regulatory sequences associated with EBF1 and FOXO1 occupancy. RAG1-deficient pro-B cell lysates were immunoprecipitated with antibodies directed against EBF1 and FOXO1 and analyzed by ChIP-Seq. Motifs that companion E2A occupancy were identified by comparing enriched peaks to random selected genomic DNA sequences. Log-p-values are indicated in red. Smaller values represent relative high enrichment for a given motif as compared to randomly selected regions. (b) Coordinate E2A and FOXO1 DNA binding in pro-B cells. Heat map of FOXO1 occupancy and H3K4me1, H3K4me2 and H3K4me3 patterns centered across 1118 promoter distal FOXO1 binding sites. Each row represents a 6 kbp genomic region centered across FOXO1 occupancy. The heat map reflects the enrichment of FOXO1 and E2A DNA binding as well as H3K4 methylation by color.
As described above, the computational analysis showed that a small fraction of E2A binding sites was enriched for CTCF consensus-binding sequences (Cluster 1) (Fig. 4). These data raise the possibility that a tiny but significant fraction of E2A DNA binding is associated with CTCF occupancy. To directly test whether E2A and CTCF bind coordinately to a subset of cis-regulatory seqences, CTCF was immunoprecipitated form pro-B cell lysates and analyzed by ChIP-Seq. As predicted from the computational analysis, a subset of CTCF occupancy showed coordinate DNA binding with E2A (Supplementary Fig. 3a,b and Supplemental Fig. 4). Taken together, in pro-B cells E2A binds coordinately with EBF1, Foxo1 and CTCF to a wide spectrum of cis-regulatory elements.
Global transcription factor occupancy and B cell development
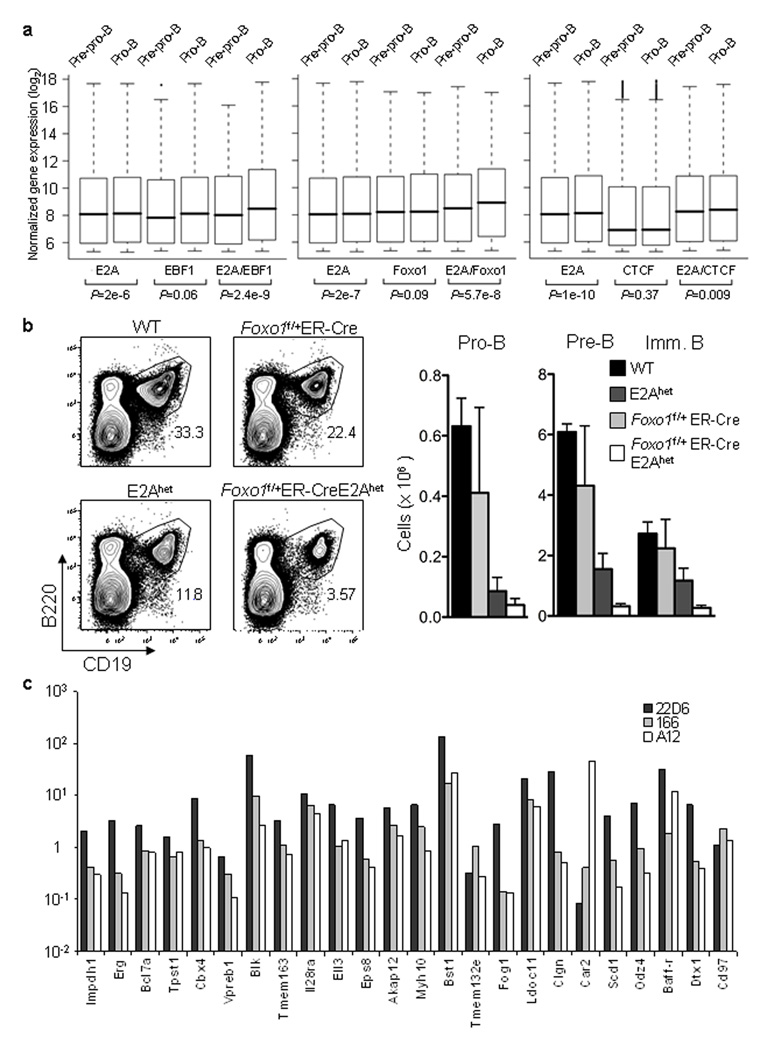
To determine the extent of coordinate E2A and EBF1 binding, genomic distances were measured between the two closest binding sites. Close to 50% of EBF1 binding sites were flanked by E2A binding sites within 150 bp (Supplementary Fig. 5a). To determine whether coordinate E2A and EBF1 DNA binding directly relates to differences in gene expression, pre-pro-B and pro-B cell transcript abundance were correlated with E2A and EBF occupancy. E2A and EBF1 binding sites were assigned to a specific locus based on the genomic distance separating them from the nearest transcription start site. To this end, mRNA was derived from pre-pro-B cells and pro-B cells and analyzed for absolute gene expression by microarray analysis. The distributions of transcript abundance in pre-pro-B and pro-B cell stage were displayed as a whisker-box plot (Fig. 6a). A substantial fraction of genes that were enriched for coordinate E2A and EBF1 DNA binding was positively associated with an increased abundance of pro-B cell specific transcripts as compared to genes that contain binding sites for either E2A or EBF1 binding alone (Fig. 6a). We note that the P-values are highly significant because of the large number of genes that were analyzed. Among the loci that showed coordinate E2A and EBF1 DNA binding were a large set of loci known to play critical roles in early B cell development, including Vpreb1, Vpreb3, Cd19, Bst1, Foxo1, Pou2f1, Cd79a and Pax5 (Supplementary Table 2). Seventeen hundred and fifty three genes analyzed were highly expressed in pro-B versus pre-pro-B cells ( > 1.5 fold change, P-value < 0.01). Thirteen percent of these showed coordinate E2A and EBF1 DNA binding, within a 150 bp DNA segment (Supplementary Table 2). Less than two percent of the genes contained one EBF1 binding site but lacked E2A occupancy (Supplementary Table 3). Forty-three percent of loci was companioned by at least one E2A binding site but did not show coordinate EBF1 occupancy. Taken together, these data indicate that E2A and EBF1 bind coordinately to activate a B lineage program of gene expression. These data are consistent with genetic data indicating that E2A and EBF1 act synergistically to modulate early B cell development11.
Figure 6.
Coordinate E2A, EBF and FOXO1 DNA binding is associated with a B-lineage specific program of gene expression. (a) Box plots depicting the distribution of RNA expression levels of different groups of genes in pre-pro-B and pro-B cells are shown. The genes closest to the binding sites (measured from TSS) were categorized into 3 groups: E2A and EBF1 binding sites localized within 150 bp of each other, genes in the vicinity of EBF1 binding sites with no E2A peak within 150 bp and genes in the vicinity of E2A binding sites with no EBF1 peak within 150 bp. The same analysis was performed for E2A and FOXO1, and E2A and CTCF binding sites. The p-values of paired t-tests are indicated below each pair. (b) B cell development in E2Ahet;FOXO1 f/+;ER-Cre mice is severely perturbed. Bone marrow derived from wild-type, E2Ahet, FOXO1 f/+;ER-Cre as well as E2Ahet;FOXO1f/+;ER-Cre mice were analyzed for the expression of B220 and CD19. Four mice of each group were analyzed by flow cytometry. Two independent experiments were performed with similar results. (c) Enhancer activity of 24 selected H3K4 monomethylated DNA segments that show coordinate E2A and EBF1 DNA binding in pro-B cells were measured in a pro-B cell line (22D6) and two T cell lines (A12 and 166).
The degree of co-occupancy was also observed between Foxo1 and E2A (Supplementary Fig. 5b) but to a much lesser degree between Foxo1 and EBF1 (Supplementary Fig. 5c). The fraction of genes that were enriched for coordinate E2A and Foxo1 binding activity was positively associated with an increase in the abundance of pro-B cell specific transcripts as compared to loci that contained either E2A or Foxo1 DNA binding (Fig. 6a). Among the loci that showed co-occupancy of E2A and Foxo1 were Blk, TdT (Dntt), and Erg (Supplementary Tables 4, 5). Notably, a well known E2A and Foxo1 target gene, Bcl2l11 (Bim) showed coordinate E2A and Foxo1 DNA binding (Supplementary Table 4). Interestingly, CTCF binding sites were less frequently associated with loci showing pro-B lineage specific transcription as compared to the E2A binding sites (Fig. 6a).
To further explore the coordinate activities of E2A and Foxo1 in B cell development, we adopted a genetic strategy. Double as well as single heterozygous E2A; Foxo1;ER-Cre mice were analyzed for abnormalities in B cell development after treatment with 4-hydroxytamoxifen (4-OHT). The pro-B cell compartment in compound heterozygote E2A+/- Foxo1f/+ ER-Cre mice was modestly affected as compared to E2A+/− mice two weeks post tamoxifen treatment (Fig. 6b). On the other hand, the pre- and immature-B cell compartments were substantially reduced (both percentage and absolute numbers) in E2A+/− Foxo1f/+ ER-Cre mice when compared to single heterozygotic E2A+/− or Foxo1 f/+ ER-Cre mice, reflecting a critical role for the coordinate activities of E2A and Foxo1 during the pro-B to pre-B cell transition (Fig. 6b).
The coordinate DNA binding activities of E2A and EBF1 to sites in H3K4me1 islands is consistent with them acting as enhancer binding proteins. To determine directly whether the binding patterns indeed reflect lineage-specific enhancer activity, a total of twenty-four segments that were H3K4 monomethylated and that showed coordinate E2A and EBF1 DNA binding, were assayed for enhancer activity. Putative enhancer elements were inserted upstream of a basal promoter (adenovirus major late promoter in a luciferase reporter construct and transfected into both a pro-B (22D6) and two T cell lines (166 and A12). Luciferase activities were recorded one day post-transfection. Luciferase activity of each reporter was first normalized to renilla activity. The fold changes compared to the basal promoter activity are reported in logarithmic scale (y-axis). The genes closest to the respective enhancer elements (measured from TSS) are indicated on the x-axis. As predicted, the majority of reporter constructs (20 out of 24) containing coordinate E2A and EBF1 binding sites showed substantial enhancer activity in the pro-B cell line 22D6 (Fig. 6c). In contrast, only a minor proportion (9 out of 24) showed enhancer activity in the T cell lines 166 and A12 (Fig. 6c). Three out of twenty-four regulatory elements containing both E2A and EBF1 binding sites suppressed reporter gene expression in which cell line(s) (Fig. 6c). Additionally, several of the reporters showed activation in B lineage cells whereas repression was observed in T cells (Fig. 6c). Thus, regulatory elements carrying E2A and EBF1 binding sites can function either as transcriptional enhancers or as silencers dependent on cell context.
Genome-wide H3K4 monomethylation by E47
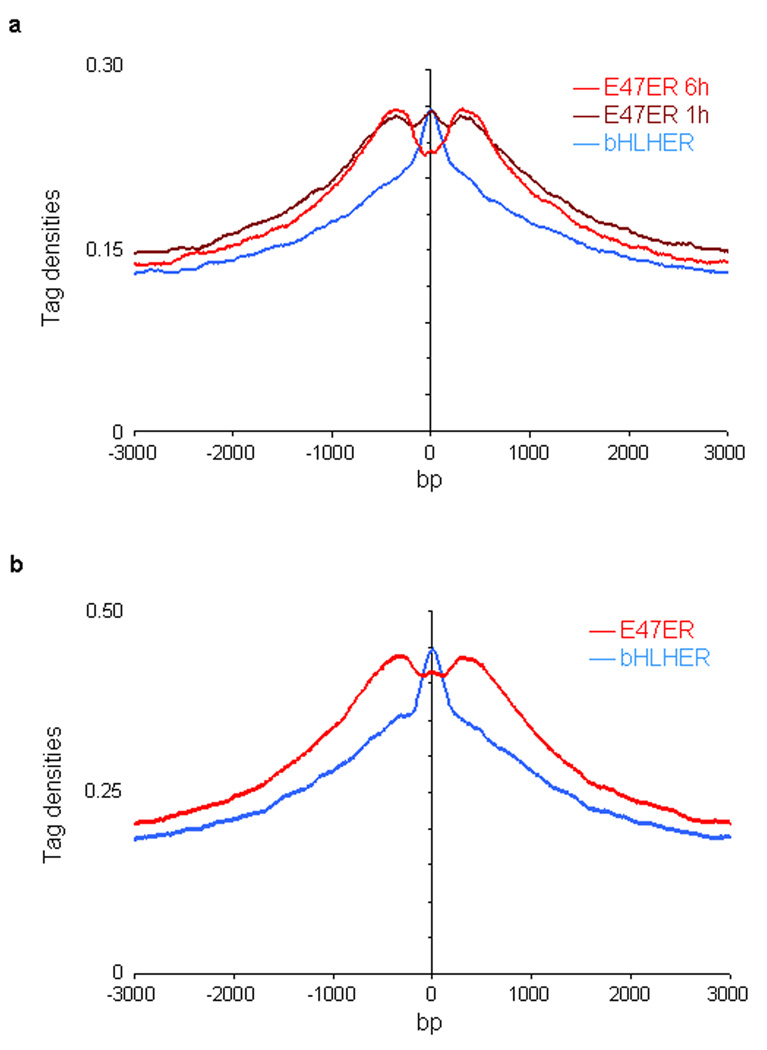
Previous knock-in studies in the Igk locus, have described a critical role for E2A in enhancer function. Here we demonstrate that the E2A proteins bind to a wide repertoire of putative enhancers. We are now faced with the question as to how the E2A proteins modulate enhancer function on a global scale. As a first approach to this question we examined whether the E2A proteins have the ability to modulate chromatin structure. To this end, E2A deficient pre-pro-B cells were transduced with a virus carrying an E47-estrogen receptor (E47ER) fusion protein as well as an hCD25 reporter. As a control, cells were transduced with a virus carrying the bHLH (basic helix-loop-helix) region of E47 but lacking the N-terminal transactivation domain (bHLHER). 21 h post infection cells were cultured for an additional 1 h or 6 h in the presence of 4-OHT. Transduced cells were enriched using magnetic beads, and E2A as well as H3K4me1 were immunoprecipitated and analyzed by massive parallel DNA sequencing. In the presence of E47 lacking the N-terminal transactivation domains, the pattern of H3K4 monomethylation peaked across the E47 binding sites, indicating that H3K4 monomethylated histone residues were closely associated with the E47 bHLH domain (Fig. 7a). Notably, upon forced expression of E47 containing the bHLH as well as the N-terminal transactivation-domains, a bimodal distribution of H3K4 monomethylation was observed (Fig. 7a). The bimodal distribution was observed within 1 h post treatment with 4-OHT, indicating that the E2A proteins acted directly to alter the pattern of H3K4me1 (Fig. 7a). A similar redistribution of H3K4me1 was observed upon forced E47 expression in an E2A-deficient thymoma cell line (Fig. 7b). To exclude the possibility that the E2A binding sites where histone is remodeled were different from the bHLH binding sites where histone is not remodeled, H3K4me1 patterns were directly compared for E47bHLH as well as full-length E47 (Supplementary Fig. 6). Please state the results here. These data show that E47 acts directly to elevate the abundance as well as the pattern of H3K4me1 across a wide repertoire of putative enhancer regions.
Figure 7.
E47 DNA binding alters the pattern of H3K4 monomethylation. (a) Forced E47 expression in E2A-deficient pre-pro-B cells promotes the establishment of a bimodal distribution of H3K4me1 centered across E2A occupancy. E2A-deficient pre-pro-B cells were transduced with virus carrying the full-length coding DNA sequence of E47 fused to the estrogen receptor domain (E47ER). As a control, cells were transduced with virus expressing the E47 bHLH domain fused to the ER domain (bHLHER). Twenty-one hours post infection, cells were incubated with tamoxifen to induce E47 activity for either one or six hours, harvested, formaldehyde cross-linked, immunoprecipitated with an anti-E2A or anti-H3K4me1 antibody, and analyzed by ChIP-Seq. X-axis shows the genomic distance from E2A bound regions. Y-axis shows the individual immunoprecipitated mapped reads (or tags) per base pair. (b) Forced E47 expression in E2A-deficient T cells promotes the establishment of a bimodal distribution of H3K4me1 centered across E2A occupancy. E2A-deficient A12 cells were transduced with virus and analyzed by ChIP-Seq as described above.
Network construction
The data described raise the raise the question as to how the genome-wide DNA binding analysis of individual transcription factors can be linked into a global network of interacting components that collectively promote the B cell fate. We divided the genome into cis-coding elements that show E2A, EBF1 and Foxo1 occupancy, using a computational approach, named Cytoscape24,25. Putative regulatory targets of E2A were identified based on two criteria. First, genes that showed a significant (≥ 1.5 fold change, P ≤ 0.05) change in transcript abundance between pro-B cells and E2A-deficient pre-pro-B cells were selected. This subset of genes was subsequently examined for the presence of sites that showed E2A occupancy. For the analysis of EBF1 targets, transcript abundance in pro-B cells was compared to EBF1-deficient pre-pro-B cells. This subset of genes was subsequently examined for the presence of sites that showed EBF1 occupancy. Loci showing Foxo1 occupancy were not directly analyzed for differences in transcript abundance between pro-B cells and Foxo1-ablated cells. We note that differences in transcript abundance between E2A- and EBF1-deficient pre-pro-B cells versus wild-type pro-B cells may not necessarily reflect direct regulation by either E2A or EBF1, but rather reflect the developmental stages where B cell maturation is arrested. We also note that co-occupancy relates here to binding within a genomic locus, rather than coordinate DNA binding activity within putative enhancer elements.
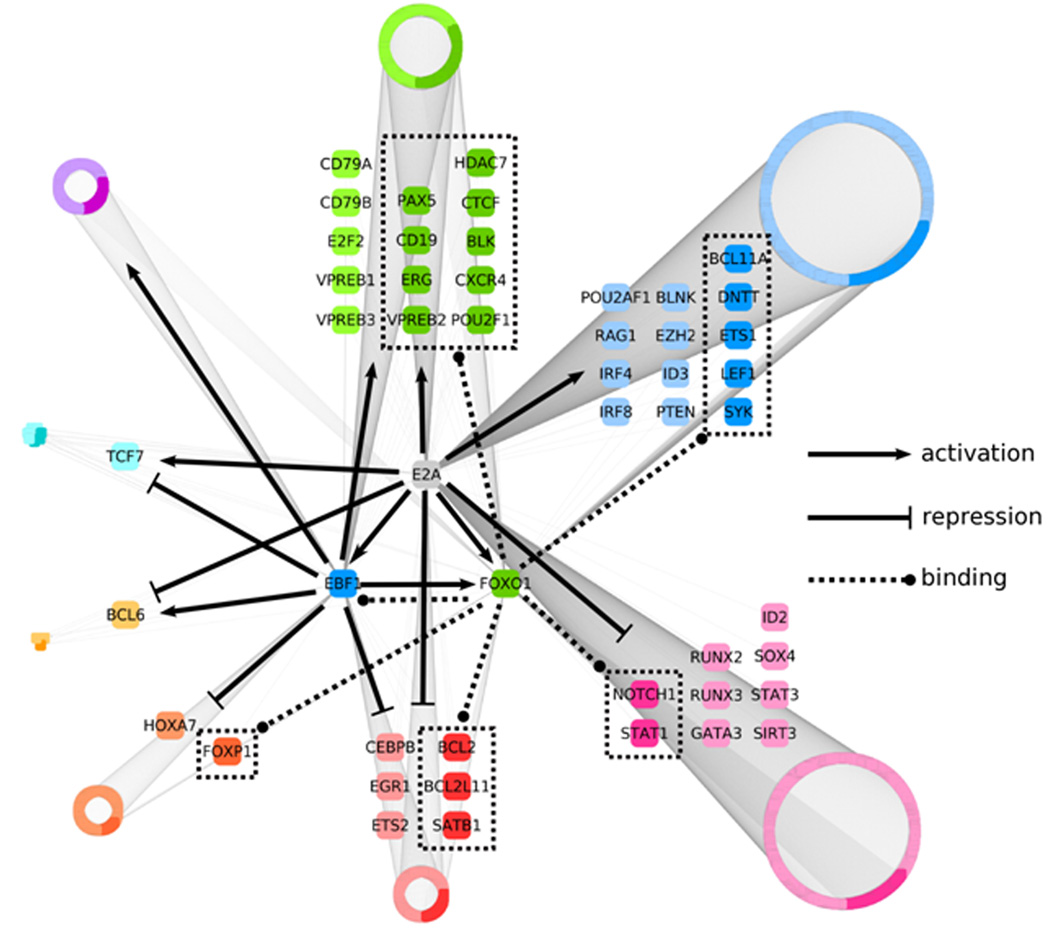
We identified 2045 putative E2A, EBF1 and Foxo1 target sites that were divided into eight groups based on the criteria described above (Fig. 8). The overlap of E2A and EBF1 targets was highly significant (P = 4.8 × 10−255), with over two-thirds of EBF targets being potential regulatory targets of E2A (Supplementary Fig. 7a). Many of genes that show E2A and EBF1 occupancy also showed enrichment for Foxo1 binding (P = 9.88 × 10−324) (Supplementary Fig. 7b). In terms of gene function, as expected we found that the network containing all potential target genes was significantly enriched for genes encoding for proteins involved in lymphocyte development (Supplementary Figure 7c).
Figure 8.
Regulatory network that links the activities of an ensemble of transcriptional regulators, signaling components and survival factors in developing B cells into a common pathway. An integrative genome-wide analysis, using Cytoscape software, of protein-DNA binding and changes in transcript levels across E2A-deficient pre-pro-B, EBF1-deficient pre-pro-B and pro-B cells was used to identify transcriptional regulatory targets of E2A and EBF1, as well as genes bound by FOXO1. Resulting 2045 regulatory targets were divided into eight groups based on positive or negative changes in transcript levels in pro-B cells as compared to E2A or EBF1-deficient cells. Arrows represent gene activation whereas barred lines indicate transcriptional repression. Dotted lines represent FOXO1 occupancy. Selected genes from each group either known to play critical roles in B cell commitment or potential novel regulators are enlarged. All other genes are grouped into rings where the size of each ring indicates the relative number of members. Darker colors present loci that show FOXO1 occupancy.
This analysis raises the question as to what extent the identified regions show indeed enhancer activity. First, several of these binding sites were validated by ChIP followed by quantitative PCR (Supplementary Fig. 8). Second, the putative regulatory elements identified by the network of several loci were inserted into a luciferase reporter and assayed for transcriptional activity (Supplementary Fig. 9a). As predicted by the presence of H3K4 monomethylation as well as E2A, EBF1 and/or Foxo1 occupancy, the regulatory elements showed high reporter activity in pro-B cells (Supplementary Fig. 9b). Moreover, the majority of the elements elevated transcription to a much higher degree as observed than in T lineage cells (Supplementary Fig. 9b). In sum, we have used a computational approach to divide the pro-B cell genome into clusters of loci that are characterized by the occupancy of E2A, EBF and Foxo1. From this analysis we have generated a global network that we propose orchestrates the B cell fate (Fig. 8).
DISCUSSION
Regulatory networks have been proposed previously that link the activities of E2A, EBF1, PU.1 and Pax5 (refs. 10,26,27). Here we have extended this analysis using genome-wide screens and generated a global network in which the genome has been divided into clusters of genes on the basis of distinct sets of cis-regulatory codes. The network shows links between E2A, EBF1 and Pax5, consistent with previous observations17. The network also includes Foxo1, which until now has not yet been directly connected to other factors, within the context of early B cell development. Based on these as well as previous observations, we propose that E2A activates EBF1 and Foxo1 expression. E2A and EBF1 and possibly Foxo1, in conjunction with IRF4, IRF8 and PU.1, then act in concert to induce Pax5 transcription and promote the B cell fate9.
The linkage involving E2A and Foxo1 is of significant interest since they connect E2A and the PI3K cascade into a common module. Previous studies have suggested that the PI3K-PTEN-AKT- Foxo1 pathway and E-proteins are linked. Depletion of the LT-HSC and CLP compartments has been observed in both E2A and Foxo1, 2 and 4 triple null mutant mice28,29,30,31. The roles of E2A and Foxo1 in early B cell development also overlap13,32. At the immature-B cell compartment both PI3K signaling and E2A play critical roles in receptor editing33,34. The marginal versus follicular zone B cell fate decision is also modulated by the PI3K and E2A pathways34,35. Class switch recombination is severely perturbed in both E2A and PTEN-deficient activated B cells36,37. In thymocyte development both E2A and PTEN enforce the pre-TCR checkpoint38,39. Finally, PTEN, Foxo and E2A act to prevent the development of T cell lymphoma39,40,41,42. The data described here now provide a mechanism that underpins the linkage between the E2A and FOXO1 proteins: E2A acts immediately upstream of FOXO1 and E2A and FOXO1 are wired together by a wide spectrum of cis-regulatory codes.
The analysis described here also revealed that E2A, FOXO1 and EBF1 bind to a large subset of genes involved in pre-BCR signaling.. Additionally, LEF1, ETS1 and Bcl11a are potential novel targets of E2A and FOXO1, linking these key regulators into a common framework. Of particular interest is LEF. LEF mediates Wnt signaling and modulates B cell proliferation43. Thus, these data raise the interesting possibility that E2A, PI3K and Wnt signaling are connected by LEF to coordinate cell growth and developmental progression. As expected, a substantial number of genes appear to be repressed during the developmental transition from the pre-pro-B to the pro-B cell stage. Consistent with previous observations, these include genes expressed in alternative lymphoid cell lineages such as CEBPβ, Notch1, RUNX2, RUNX3 and GATA3 6, 16.
The network includes a small proportion of loci, for example TCF7, that show elevated transcript abundance in pro-B cells as compared to E2A-deficient pre-pro-B cells whereas the reverse pattern is observed in EBF-deficient pre-pro-B cells. We note that it remains to be determined whether E2A and/or EBF indeed modulate the expression of loci assigned to such clusters. Rather then comparing transcript abundance of wild-type pro-B cells to E2A- or EBF-deficient pre-pro-B cells, it will be important to ablate E2A, EBF1 and Foxo1 expression in pro-B cells.
In sum, we have generated a global network that involves the transcription factors E2A, EBF1 and Foxo1. We suggest that this network of transcription factors orchestrates the B cell fate. The global network described here confirms previous connections and reveals novel links and new players. The network that is presented is merely a starting point since key regulators, including Pax5 and PU.1, were not included in the analysis. Ultimately, the approach can be applied to other stages of B cell development and other developmental pathways. Global networks that reflect distinct stages of a developmental pathway provide insight into how enhancers and cis-regulatory codes change in developing cells to induce lineage- and stage-specific programs of gene expression. For example, within the context of the B cell lineage, it will be of particular interest to determine how global networks change in response to pre-BCR signaling, to self-reactivity and invading pathogens. Ultimately, it will be possible to describe the entire B lineage developmental pathway in terms of global networks using the strategy described here.
METHODS
Mice
C57Bl/6, Foxo1 and RAG1-deficient mice were maintained in ventilated cages. Animal studies were approved by the IACUC-UCSD.
Culture conditions, viral infection, and luciferase reporter assay
Ebf1−/− hematopoietic progenitors were isolated and grown in the presence of IL-7, SCF and FLT3L as previously described6. RAG1-deficient pro-B cells were grown in the presence of Opti-MEM, 10% FCS, 2% penicillin/streptomycin and glutamine, 50 µM beta-mercaptoethanol with IL-7 and SCF. E2A deficient pre-pro-B cells were grown in conditions similar to EBF1-deficient cells. Retroviral supernatants, containing E47ER or bHLHER control were prepared and transduced into E2A-deficient cell lines as previously described6. E47-ER and bHLH-ER were activated for 1 h or 6 h with 1 µM tamoxifen and transduced cells were isolated by magnetic selection (Miltenyi) for co-expressed human CD25. For transfection studies reporter constructs were transiently transfected into 22D6 (pro-B cell line), A12 (E2A-deficient T cell line) and 166 (p53−/− T cell line) using TransIT®-LT1 reagent (Mirus). 22D6 and A12 cells were grown in the presence of RPMI, 10% FCS, 2% PSG, 50 µM beta-mercaptoethanol. 166 cells were grown in the presence of Opti-MEM, 10% FCS, 2% PSG, 50 µM beta-mercaptoethanol. The cloning of luciferase constructs was described36. Dual-luciferase reporter assays were performed as described by the manufacturer (Promega). Two to three independent experiments were performed. Primers for generating constructs, which yield between 500 to 600 bp long putative enhancers, are indicated in Supplementary Table 7. Each construct was verified by DNA sequencing.
Profiling of expression arrays
Total RNA derived from E2A-deficient, EBF-deficient pre-pro-B and RAG1-deficient pro-B cells was purified using RNeasy Kit (Qiagen). Samples (250 ng RNA) were amplified and labeled using the Quick AMP Labeling kit (Agilent) and hybridized to Mouse Oligo Microarrays (Agilent) as described by the manufacturer. Images were quantified the using Agilent's Feature Extraction software.
Chromatin immunoprecipitation and genome-wide DNA sequencing
Chromatin immunoprecipitation was performed as described previously21. Briefly, cells were fixed with 1% formaldehyde for 5 to 20 min at 21°C, lysed and sonicated. Sonicated chromatin was immunprecipitated with 10 µg of anti-E2A (Santa Cruz Biotechnology; SC-349), anti-EBF1 (Abnova; H00001879), anti-Foxo1 (Santa Cruz Biotechnology; SC-11350) or CTCF (Millipore 07–0729). Antibodies directed against anti-H3K4me1 (ab8895), H3K4me2 (ab7766), and H3K4me3 (ab8580) were obtained from Abcam. Antibodies directed against H3K27me3 (07–449) and acetylated H3 (06–599) were obtained from Millipore. Samples were washed and bound chromatin was eluted and incubated at 65 °C overnight to reverse the cross-linking. Samples were subsequently treated with RNase A, Proteinase K, and purified using PCR Purification Kit (Qiagen). Immunoprecipitated chromatin was adapter-ligated and amplified by PCR according to the manufacturer’s protocol (Illumina). Amplified ChIP DNA was sized selected (150 to 250 bp) by 8% polyacrylamide (PAGE) gel electrophoresis and sequenced using an Illumina Genome Analyzer II. The flow-cell was sequenced for 36 cycles to generate 23–25-bp reads and aligned to the mm8 assembly (NCBI Build 36) using ELAND allowing two nucleotides mismatches. Tags that mapped to unique DNA sequences were further analyzed (Supplemental Table 6). Visualization was performed by generating custom tracks for the UCSC Genome browser.
DNA sequence analysis
Data was analyzed using the HOMER software freely available at http://biowhat.ucsd.edu/homer/. The identification of ChIP-Seq bound elements was performed using HOMER. The position of each tag 3’ of its position was adjusted by 75 bp. One tag from each unique position was examined to eliminate peaks generated by clonal amplification. Binding sites were identified by searching for groups of tags located within a sliding 200 bp window. Adjacent clusters were required to be at least 500 bp separated from each other. The threshold for the number of tags that generate a bound element was selected for a false discovery rate of 0.001. Additionally, required peaks were to have at least 4-fold more tags (normalized versus total number) as compared to input control samples. We also required 4-fold more tags relative to the local background region, within a 10 kb region to avoid identifying DNA segments containing genomic duplications or non-localized occupancy. Peaks were associated with gene products by identifying the nearest RefSeq TSS. To identify H3K4 methylated regions, the peak finding procedure was modified. The peak region was required to show 4-fold more tags (as compared to total tag count) than controls. Additionally, DNA bound regions were required to contain 4-fold more tags within a 1 kb region relative to the flanking 10 kb DNA segments. To enrich for H3K4me1 occupancy located within putative enhancer regions, coordinate H3K4me1 and H3K4me3 regions (within a 3 kb region) were eliminated. In addition, H3K4me1 regions located within 3 kb of an annotated TSS were removed from further analysis.
FACS analysis
ER-Cre mediated deletion of floxed alleles was induced in mice 9–10 weeks old by intraperitoneal injections of 1 mg tamoxifen (Sigma) emulsified in 200 µl of sunflower seed oil (Sigma) for five consecutive days. Ten days after the start of injections, bone marrow were harvested and Fc-blocked (CD16/CD32, 2.5G2) cells were stained with antibodies against CD11B (M1/70) FITC, GR1 (RB6-8C5) FITC, TER119 (Ter119) FITC, NK1.1 (PK136) FITC, CD3 (145-2c11) FITC, B220 (RA3-6B2) Alexa700, CD19 (ID3) PECY5.5, KIT (2B8) APC, IgD (11/26) Pacific Blue IgM (11/41) biotin (visualized with streptavidin QD655) and propidium iodine. Analytical flow cytometry was conducted using a LSRII (BD Bioscience). Data was analyzed using FlowJo™ software (Tree Star Inc.).
Network construction
Input gene expression and ChIP-Seq data was pre-processed using in-house Python scripts. The network was prepared and analyzed using Cytoscape platform24. Gene ontology enrichment analysis was performed using Bingo plugin25. The significance of the overlap between target sets was calculated based on the Fisher's exact test. Statistical analysis was done using R statistical environment (http://www.R-project.org).
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Hardiman, C. Ludka, L. Edsall and Z.Ye for help with Solexa DNA Sequencing. We thank R. DePinho (Harvard Medical School) for the generous gift of Foxo1-ablated mice. We are grateful to J. Sprague and R. Sasik for microarray analysis. Y.C.L. was supported by the Ruth L. Kirschstein Research Service Award (1F32CA130276), C.B. by NIH P01DK074868 and S.H. by F32HL083752. We thank the members of the Murre laboratory for comments on the manuscript. The studies were supported by grants from the NSF (NSF IIS-0803937) to T.I., BIOGEM DK063491 to the UCSD Core Facility and from the NIH to J.H., C. K.G. (CA52599) and C.M. (CA054198-20).
Footnotes
AUTHORS CONTRIBUTIONS
Y.C.L. designed and performed experiments, analyzed data and wrote the manuscript. S.J. and C.B. wrote programs and analyzed data. S.H. performed CTCF ChIP-Seq and H3K4 monomethylation of RAG-deficient pro-B cells. J.H. generated EBF-deficient pre-pro-B cells. M.S. provided anti-EBF antibody. E.W. and R.M. performed the analysis of E2A–Foxo1 deficient mice. C.E. performed ChIP-Seq experiments during the initial phase of the study. J.D. and T.I. applied computational approaches to generate a global network. C.K.G. analyzed data and edited the manuscript. C.M. designed experiments, analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Cosgrove MS. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev. Proteomics. 2007;4:465–478. doi: 10.1586/14789450.4.4.465. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;26:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 5.Kee BL, Murre C. Induction of EBF and multiple B lineage genes by the helix-loop-helix protein E12. J Exp. Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- 7.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Devel. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promogter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7, Pax5. Mol. Cell. Biol. 2007;2:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors E2A and EBF. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 12.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel R, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat. Immunol. 2003;4:525–531. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 13.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008;9:396–404. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–572. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 16.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5Nat. Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 17.Murre C. Developmental trajectories in early hematopoiesis. Genes Dev. 2009;15:2366–2370. doi: 10.1101/gad.1861709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui K, Zang C, Roh T, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visel A, et al. ChIP-Seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;12:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintzman N, et al. Histone modifications at human enhancers reflect global cell-type specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agata Y, et al. Regulation of T cell receptor beta gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Barski A, et al. High resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Ward JH. Hierarchical grouping to optimize an objective function. J. American Statistical Association. 1963;58:236–242. [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang TJ, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maere S, Heymans K, Kuiper M. Bingo: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 26.Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate T lineage specific progression and commitment. J. Exp. Med. 2006;15:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, et al. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J. Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphid and myeloerythorid progenitors. Proc. Natl. Acad. USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bain G, et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997b;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 33.Llorian M, Stamataki Z, Hill S, Turner M, Martensson IL. The PI3K p110delta is required for down-regulation of RAG expression in immature B cells. J. Immunol. 2007;178:1981–1985. doi: 10.4049/jimmunol.178.4.1981. [DOI] [PubMed] [Google Scholar]
- 34.Quong MW, et al. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein E2A. J. Exp. Med. 2004;199:1101–1112. doi: 10.1084/jem.20031180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anzelon AN, Wu H, Rickert RC. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat. Immunol. 2003;4:287–294. doi: 10.1038/ni892. [DOI] [PubMed] [Google Scholar]
- 36.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki A, et al. Critical roles of Pten in B cell homeostasis and immunoglobulin class switch recombination. J. Exp. Med. 2003;197:657–667. doi: 10.1084/jem.20021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagenbeek TJ, et al. The loss of PTEN allows TCR alphabeta lineage thymocytes to bypass IL-7 and Pre-TCR-mediated signaling. J. Exp. Med. 2004;200:883–894. doi: 10.1084/jem.20040495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel I, Murre C. Disruption of pre-TCR expression accelerates lymphomagenesis in E2A-deficient mice. Proc. Natl. Acad. Sci. USA. 2002;99:11322–11327. doi: 10.1073/pnas.162373999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;28:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bain G, et al. E2A deficiency leads to abnormalities in alpha/beta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 1997a;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reya T, O;Riordan M, Okamura R, Devaney E, Willert K, Nusse R, Grosschedl R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.