Abstract
Formaldehyde cross-linking of protein complexes combined with immunoprecipitation and mass spectrometry analysis is a promising technique for analysing protein-protein interactions, including those of transient nature. Here we used integrin β1 as a model to describe the application of formaldehyde cross-linking in detail, particularly focusing on the optimal parameters for cross-linking, the detection of formaldehyde cross-linked complexes, the utility of antibodies, and the identification of binding partners. Integrin β1 was found in a high molecular weight complex after formaldehyde cross-linking. Eight different anti-integrin β1 antibodies were used for pull-down experiments and no loss in precipitation efficiency after cross-linking was observed. However, two of the antibodies could not precipitate the complex, probably due to hidden epitopes. Formaldehyde cross-linked complexes, precipitated from Jurkat cells or human platelets and analyzed by mass spectrometry, were found to be composed of integrin β1, α4 and α6 or β1, α6, α2, and α5, respectively.
1. Introduction
Protein-protein interactions are the basis for most cellular processes, including signalling, protein synthesis, and metabolism. Detailed knowledge of these protein networks is required in order to better understand diseases and develop adequate treatments. At present, protein-protein interactions are commonly investigated by yeast-two-hybrid approaches [1] and by in vitro binding studies [2]. However, these approaches are prone to false positive identifications because they do not take into account the temporal and local separations that occur in a living system. One tool to study protein-protein interactions in a physiological context is affinity enrichment of the protein of interest followed by detection of its binding partners using either immunodetection methods or mass spectrometry [3]. However, this classical immunoprecipitation method has two drawbacks. Weak interactions could be missed, if stringent wash conditions are applied. In contrast, nonstringent conditions may enable the identification of more proteins, but many of these could be false positives only binding the bait protein during sample preparation.
One approach to solve this problem is applying covalent cross-linking to intact cells and thereby stabilizing protein-protein interactions, including very weak and transient ones [3]. After this fixation step, highly stringent conditions can be used during cell lysis and affinity enrichment, minimizing the risk of identifying false positives. Several cross-linkers varying in spacer arm lengths, reaction groups, and other properties are commercially available. One of the shortest available cross-linkers is formaldehyde (2.3–2.7 Å), which has been used for a long time in histology and pathology to “freeze” the native state of tissues and cells [4]. The experimental conditions used in these applications lead to a very tight network of cross-links, which prevents the precipitation of one protein of interest as required for protein-protein interaction studies. However, lower formaldehyde concentrations (0.4–2% instead of 4%) and especially shorter reaction times (minutes instead of hours) allow the utilization of formaldehyde as a cross-linker to analyze protein-protein interactions as shown by us and others [5–8].
The application of formaldehyde as a cross-linker has several advantages. Only closely associated proteins can be cross-linked due to the small size of formaldehyde. Furthermore, its high permeability towards cell membranes enables cross-linking in the intact cell, without addition of organic solvents such as dimethyl sulfoxide as necessary for other cross-linkers. Formaldehyde is also thought to allow very fast cross-linking and the stabilization of transient interactions [4]. Finally, formaldehyde is available in almost every laboratory at costs that amount to only a fraction of other cross-linkers. However, formaldehyde cross-linking is not yet an established standard method and many questions regarding the optimal experimental conditions and the usability of antibodies for pull-down of proteins after formaldehyde treatment remain. For example, epitopes recognized by antibodies raised against endogenous proteins could be destroyed by formaldehyde modification, which would prevent their application [9]. Similarly, the physiological environment of a protein of interest, and the type and extent of its interactions may also affect the experimental outcome. Therefore, we decided to investigate different aspects of formaldehyde cross-linking in more detail using the transmembrane protein integrin β1 as a model.
Integrins are membrane spanning heterodimeric complexes that play important roles in cell adhesion and migration processes by interacting with components of the extracellular matrix [10]. Each integrin heterodimer is composed of one α and one β subunit, which are noncovalently associated. 18 α subunits and 8 β subunits are found in humans, which form 24 different heterodimers. The biggest subgroup with 12 members is formed by β1 containing heterodimers [11]. Before being able to bind a ligand, integrins have to be activated through an intracellular process termed inside-out signalling. For example, during platelet activation, thrombin triggers talin activation via a pathway involving protein kinase C, the small GTPase Rap1 and the Rap1 effector Rap1-interacting molecule (RIAM) [12]. Activated talin then binds to the intracellular tail of the integrin and causes conformational change of the two integrin chains. This allows binding of extracellular ligands, which drives the cytoplasmic tail of the integrin to bind additional adaptor proteins, establishes a connection to the cytoskeleton and leads to the delivery of the external signal (outside-in signalling).
Several intracellular interaction partners have been described for integrins, despite the shortness of the intracellular tail of integrins, which varies between 40 and 60 amino acids. 25 adaptor proteins have been reported for integrin β1, including talin, tensin, filamin and kindlin [13]. However, these interactions cannot take place simultaneously, but depend on the activation status of the cell and the integrin. The detailed binding procedures as well as the signalling processes triggered by these are not fully understood. For example, talin and kindlin both interact with integrin β and a crosstalk between them is assumed. However, it remains unclear, whether both proteins connect with the integrin at the same time or binding occurs sequentially [14]. Studying the interaction partners of integrins using the formaldehyde cross-linking approach, which should be able to identify transient and indirect interaction partners of proteins, may shed more light on these processes and would therefore be very valuable.
In the present study, we report the optimization of a protocol applying formaldehyde cross-linking combined with immunoprecipitation and mass spectrometry (Figure 1(a)) to analyze the interaction network of integrin β1.
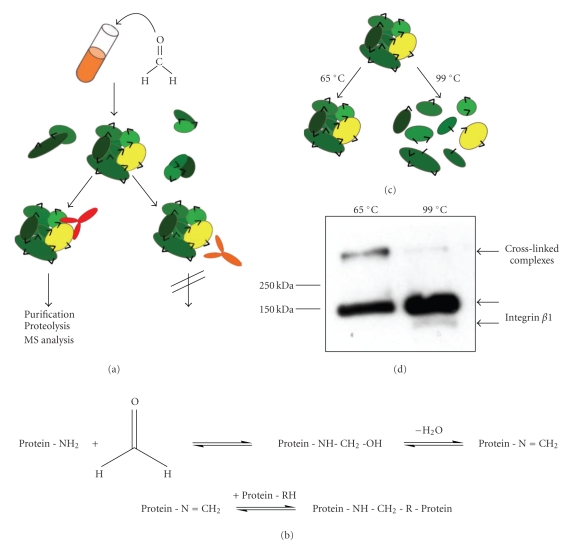
Figure 1.
Formaldehyde cross-linking. (a) Workflow of formaldehyde cross-linking. Cells are treated with formaldehyde, lysed and protein complexes (oval shapes) are precipitated by antibodies (y-shaped). Cross-links are indicated by black triangles. Only antibodies, whose epitopes are not destroyed during formaldehyde modification, can precipitate the complex. (b) Reaction scheme of formaldehyde modification, cross-linking and cross-linking reversal. (c) and (d) Formaldehyde derived cross-links are preserved, if samples are only incubated at 65°C, whereas most of the cross-links are reversed at 99°C. (c) Schematic model. Proteins are depicted as oval shapes, formaldehyde cross-links as black triangles. (d) Anti-integrin β1 immunoblot analysis of cross-linked Jurkat cells (JB1A, anti-mouse HRP).
2. Materials and Methods
2.1. Cells and Reagents
Jurkat cells were grown in Dulbecco's modified medium (GIBCO, high glucose) containing 10% fetal bovine serum (GIBCO), L-glutamine and penicillin/ streptomycin. Human platelets were isolated from healthy human volunteers as described earlier [15]. This was approved by the University of British Columbia Research Ethics Board and informed consent was granted by the donors. Briefly, whole blood was drawn from the antecubital vein into 0.15% (v/v) acid-citrate-dextrose anticoagulant. Platelets were isolated by centrifugation and washed in physiological buffer. All anti-integrin β1 antibodies were monoclonal mouse anti-human antibodies provided by John Wilkins (Manitoba). Goat anti-mouse Alexa Fluor 680 was obtained from Molecular Probes and goat anti-mouse HRP was received from BioRad.
2.2. Modeling of Integrin β1 Structure
The integrin β1 structure was modeled on the structure of human integrin β3 [16] using SWISS MODEL [17, 18] and visualization was performed using the Swiss-Pdb Viewer Deep View (http://spdbv.vital-it.ch).
2.3. Formaldehyde Cross-Linking
Formaldehyde solution was obtained by dissolving 0.4% to 4% paraformaldehyde (Fisher Scientific) in PBS for 2 h at ~80°C. The solution was filtered (0.22 μm), stored in the dark at RT and discarded after 4 weeks. For cross-linking, Jurkat cells were pelleted in a 50 ml reaction tube, resuspended in PBS and counted. Cells were centrifuged again and resuspended to 1 × 107 cells/ml in formaldehyde solution. Cells were incubated with mild agitation for 7 min at RT and then pelleted at 1800 g and RT for 3 min, resulting in 10 minutes exposure to formaldehyde. The supernatant was removed and the reaction was quenched with 0.5 ml ice-cold 1.25 M glycine/ PBS. Cells were transferred to a smaller tube, spun, washed once in 1.25 M glycine/PBS and lysed in 1 ml RIPA buffer (50 mM Tris HCl, pH 8.0, 150 mM sodium chloride, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, protease inhibitors (Complete mini, EDTA-free, Roche Diagnostics)) per 1 × 108 cells for 60 minute on ice. After 30 minutes, cell lysates were treated with 50 strokes using a Dounce homogenizer. Lysates were spun for 30 minutes at 20000 g and 4°C to remove insoluble debris. The supernatant was either used directly or stored at −80°C. Control cells were treated exactly the same way, except that they were resuspended in PBS instead of formaldehyde solution. When platelets were used for cross-linking, ~1.5 × 109 cells were resuspended in 10 ml formaldehyde solution and lysed in 1 ml RIPA buffer.
2.4. Immunoprecipitation, Western Blot Analysis and Silver Staining of SDS PAGE
Protein concentration was determined using a BCA assay (Pierce). The indicated amounts of antibodies and lysates were incubated for 1 h, after that 10 to 25 μl protein G agarose beads (Immobilized Protein G, Pierce) were added and immunoprecipitation was performed overnight. All steps were performed with mild agitation at 4°C. For mass spectrometric analysis, lysates were precleared by incubation with the same amount of beads for 2 h before the antibody was added. The supernatants of the immunoprecipitations were kept for analysis and the beads were washed either twice with PBS for western blot analysis or three times with RIPA buffer for mass spectrometric analysis. 4x reducing SDS Loading Dye (500 mM Tris HCl, pH 6.8, 8% SDS, 40% glycerine, 20% β-mercaptoethanol, 5 mg/ml bromophenol blue) was added to the beads as well as to the lysates and the supernatants and samples were incubated at either 65°C or 99°C for 5 min or 10–20 min, respectively, before they were separated by SDS PAGE using a 8% Laemmli gel [19]. For western blot analysis, proteins were transferred via a semi-dry procedure on polyvinylidene difluoride membranes (Pall Corporation), blocked for 1 h at RT with 5% milk powder in PBST (PBS, 0.1% Tween 20) and incubated with JB1A (0.1 μg/ml) overnight at 4°C. Membranes were either incubated with anti-mouse HRP or anti-mouse Alexa Fluor 680 (both 1 : 10000) for 1 h at RT. Membranes treated with anti-mouse Alexa Fluor 680 were scanned with a fluorescence scanner (Odyssey, LICOR) using excitation/emission wavelengths of 700 and 800 nm, whereas signals on anti-mouse HRP incubated membranes were detected with chemiluminescence solution (ECL, GE Healthcare). Quantitation of immunoblot analysis was performed using the software ImageJ [20]. Silver staining was performed using a modified protocol of Gharahdaghi et al. [21]. Briefly, gels were fixed with 10% acetic acid/10% methanol, washed with water and sensitized with 1 μg/ml dithiothreitol for 15 min. Gels were incubated in 0.1% (w/v) silver nitrate for 15 min and developed with 0.02% (w/v) paraformaldehyde in 3% (w/v) potassium carbonate until the desired staining had occurred. The reaction was stopped by addition of acetic acid.
2.5. In Gel Digestion and LC-MS/MS Analysis
Bands of interest in silver-stained gels were excised and destained as described [21]. Briefly, gel bands were incubated in a freshly prepared destaining solution (15 mM potassium ferricyanide/50 mM sodium thiosulfate) until the colour disappeared, washed several times with water and ammonium bicarbonate (pH 8.0) and chopped in smaller pieces. In-gel digestion was performed using standard procedures [22]. Briefly, samples were reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin (Promega) in ammonium bicarbonate overnight. Peptides were extracted twice from the gel pieces, dried down in a vacuum centrifuge, reconstituted in 5% formic acid and STAGE-tip purified [23]. Separation and identification of peptides was performed by nano-HPLC MS/MS on an Agilent 1100 (Agilent, Santa Clara, CA) coupled to an FT-ICR (LTQ-FT, Thermo Electron Corporation, Waltham, MA) using a 15 cm long, 75 μm I.D. fused silica column packed with 3 μm particle size reverse phase (C18) beads (Dr. Maisch GmbH, Germany) with water:acetonitrile:formic acid as the mobile phase with gradient elution. Proteins were identified by extracting the Mascot generic format (MGF) files from the MS data using DTA Super Charge (part of the MSQuant open source project (http://msquant.sourceforge.net/)) and searching them against the ENSEMBL database with the X!Tandem algorithm embedded in the Global Proteome Machine (http://www.thegpm.org/). The protein expect score (log(e)) indicates the probability of false assignment of a protein: log(e) = −1 equates to a 1 in 10, log(2) = −2 to a 1 in 100 chance of a stochastic protein assignment. Known contaminants such as keratin were removed from the protein lists.
3. Results and Discussion
3.1. Formaldehyde Cross-Linking Conditions
The usage of formaldehyde as cross-linking reagent has to be evaluated in order to determine the optimal balance between highest yield of complex formation and lowest artefact generation [4]. Optimization has to be performed for each protein of interest, as it is dependent on the physiological environment of the protein itself and can vary for example between cytosolic and membrane proteins. Three main parameters play a critical role during formaldehyde cross-linking: the reaction temperature, the incubation time and the formaldehyde concentration. The temperature dependency had been studied in our laboratory earlier and a difference between incubation at 37°C and 25°C could not be detected (unpublished results). Room temperature is advantageous, as it results in the most convenient and easiest approach possible. In addition, we chose not to increase the incubation time to more than 10 minutes, as the advantage of using formaldehyde as a cross-linker is the short reaction time it requires, which minimizes the formation of unspecific cross-links and allows the fixation of transient interactions. Moreover, model studies on peptides had shown that incubation time and formaldehyde concentration are complementary [24]. Therefore, we decided to limit our study to the usage of different concentrations of formaldehyde in terms of our model protein integrin β1 complex.
Jurkat cells were chosen for studying integrin β1 interactions, as this human T cell line expresses high amounts of this integrin and has been used extensively for its investigation before [25]. Different concentrations of formaldehyde (0.4% to 2%) were used to cross-link Jurkat cells. The lowest concentration was chosen as it had been shown earlier to result in the best protein loss/cross-linking yield balance [4], the highest was twice as high as formaldehyde concentrations shown to be successful earlier [5]. Cells were lysed under stringent conditions using RIPA buffer to destroy weak and noncovalent interactions, and protein amounts were determined. Lysates of formaldehyde treated cells contained lower amounts of protein than nontreated cells. This can be explained by the formation of insoluble complexes, for example, nuclear proteins being cross-linked to DNA, which were precipitated during lysis and removed in the insoluble pellet. This effect was visible during sample generation: lysis of nontreated cells using the stringent RIPA buffer led to the release of DNA, which formed a cloudy precipitate and could be easily removed. In contrast, a cloudy suspension was generated during lysis of formaldehyde treated cells and the pellet had a different consistency, which required a longer centrifugation period to become separated. Consistent with this observation, nuclear proteins were not detected in the lysate by immunoblot analysis. However, membrane proteins were overrepresented due the loss of nuclear proteins (data not shown). We recommend using this difference in appearance as an early indication of successful cross-linking.
3.2. Detection of Formaldehyde Cross-Linked Integrin β1 Complexes
Optimization of the formaldehyde cross-linking protocol involving integrin β1 required a read-out of the cross-linking efficiency, for example, the detection of a complex containing the integrin. Formaldehyde cross-links are reversible during the standard sample preparation for SDS PAGE analysis (Figure 1(b)), which includes boiling in reducing Laemmli buffer [19], thus cross-linked complexes would not be detected under these conditions [8]. However, by reducing the incubation temperature to 65°C, cross-linked complexes are not fully destroyed and remain detectable (Figure 1(c)) [5]. Membrane protein studies by gel electrophoresis are often performed at even lower temperatures (37–40°C). Initial immunoprecipitation experiments we had performed indicated that at 37°C the antibodies used for pull-downs would not dissociate into their heavy and light chains (data not shown). Instead, during gel electrophoresis the intact antibodies would migrate at molecular weights that would overlap with the cross-linked complexes and therefore interfere with their detection. Consequently, we decided not to use temperatures lower than 65°C in our experiments.
Jurkat cells treated with 2% formaldehyde were used to confirm the detection of cross-linked complexes. Lysates were incubated at 65°C and 99°C, respectively and analyzed by western blot using the antibody JB1A, which had been used for detection of integrin β1 in earlier studies [26]. We could recognize a higher molecular weight complex containing integrin β1 in samples treated for 5 min at 65°C, whereas this band was nearly undetectable after boiling for 10 minutes at 99°C (Figure 1(d)). Therefore, we concluded that we were visualizing a cross-linked complex containing integrin β1. However, integrin β1 was also found in the monomeric form at appr. 150 kDa after incubation at 65°C. This could be due to incomplete cross-linking, as the conditions applied during formaldehyde cross-linking do not lead to a high extent of protein cross-linking, leaving a large fraction of integrin β1 noncross-linked. Alternatively, incubation at 65°C may lead to partial reversal of formaldehyde cross-links and release of integrin β1 even at a lower temperature.
3.3. Balance of Cross-Linking Efficiency and Protein Loss
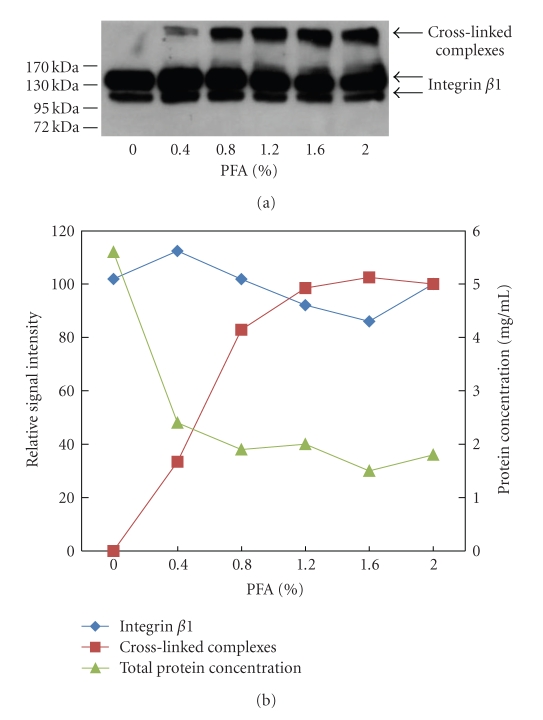
Equal amount of lysates of the samples generated using different concentrations of formaldehyde were analyzed by immunoblotting (Figure 2(a)). No complex was detected in the control, in which cells were treated under cross-linking conditions but without formaldehyde. With 0.4% formaldehyde, a small amount of complex was detected, which increased with higher formaldehyde concentrations. At 0.8% formaldehyde, much more complex was visible. The amount increased slightly at higher concentrations of formaldehyde, but then settled, with no apparent difference between 1.2 and 2%. The signal intensities of monomeric integrin β1 and the complex were quantified, the values at 2% formaldehyde were set to 100% and relative intensities were calculated. These were plotted together with the total protein concentrations of the lysates to determine the optimal cross-linking parameters (Figure 2(b)). The amount of monomeric integrin β1 did not vary significantly between the different formaldehyde concentrations, even though the amount of the complex was increasing (Figure 2(b)). This apparent contradiction can be explained by the aforementioned observation that membrane proteins are enriched during formaldehyde treatment relative to other cellular components. Thus, by loading equal total protein amounts in each lane, increasingly higher amounts of total integrin β1 were applied. Unfortunately, this variation cannot be compensated using loading controls, as the exact amount of each cross-linked protein cannot be predicted. The decrease in protein concentration due to formaldehyde modification was most pronounced between 0% and 0.4% formaldehyde treatment, increased at 0.8% but stabilized at higher formaldehyde concentrations (Figure 2(b)). Comparing the loss of protein to the gain of integrin β1 complex (Figure 2(b)) implied that using a formaldehyde concentration between 1 and 2% should lead to the best cross-linking efficiency/protein loss balance, without major differences in this range. However, increasing the extent of cross-linking may also result in the formation of larger complexes by involving proteins that do not directly bind to integrin β1, including the cytoskeleton. This would lead to the formation of more extensive, heterogeneous cross-linked complexes that would provide a larger surface to which nonspecific proteins can bind during sample processing. As a result, an increasing amount of abundant cytoskeletal proteins and common contaminants would be identified that would not be considered specific interactors. Therefore, to minimize such apparent artefact generation, we decided to perform the following investigations using more stringent conditions by applying 0.4% formaldehyde. This increases the likelihood of missed identifications of specific interactors of low abundance and of interactions of low stoichiometry. Even though higher formaldehyde concentrations would counter this effect, they would also result in more artefacts, hence differential analysis using multiple formaldehyde concentrations would still not be able to distinguish between specific interactors and artefacts. Instead, individual follow-up experiments for each identified protein would be required to determine its specificity. As the focus of this study was not to obtain an extensive list of putative interactors, but rather to demonstrate the general validity of the approach, we chose lower formaldehyde concentrations and high stringency. Users interested in maximizing the number of captured proteins should consider using higher formaldehyde concentrations instead.
Figure 2.
Optimization of formaldehyde concentration. Jurkat cells were treated with different concentrations of formaldehyde (PFA), lysed and the cell lysates were analyzed. (a) Anti-integrin β1 immunoblot of lysates (JB1A, anti-mouse HRP). (b) Western Blot results were quantified using ImageJ. Values obtained at 2% PFA were set to 100% and relative intensities were calculated. Protein concentration of the lysates had been determined using a BCA assay. Total protein concentrations and relative signal intensities of the western blot analysis were plotted in one graph to show the correlation between the parameters.
3.4. Analysis of Anti-Integrin β1 Antibodies
Studying interaction partners using formaldehyde cross-linking and affinity purification requires the antibody to still recognize its target after reaction with formaldehyde. In our earlier studies utilizing myc-tagged proteins, the antibody 9E10 was shown to be suitable for formaldehyde treatment, as it still precipitated protein complexes [5]. However, this has to be proven for every antibody/antigen pair before it can be used for interaction studies involving cross-linking. The use of a tagged, exogenous version of integrin β1 would result in additional risks coming with this approach, such as identification of false positives binding to the tag, altered protein localization, or general changes in the protein environment due to elevated expression levels. Antibodies recognizing the endogenous protein are required to exclude these influences. Not all monoclonal antibodies are expected to be suitable in this technique, as some epitopes might be destroyed by formaldehyde modification of amino acids (Figure 1(a)).
We wondered whether it is possible to predict the applicability of an antibody in the formaldehyde cross-linking approach. Therefore, we analyzed eight different and mostly well studied monoclonal anti-integrin β1 antibodies (Table 1 and Figure 3). First we wanted to know whether these antibodies were able to immunoprecipitate integrin β1 from cell lysates generated using the lysis conditions we apply during cross-linking. All antibodies were tested for affinity purification and were found to precipitate two integrin β1 bands between 100 and 150 kDa from Jurkat cell lysates (Figure 3(a)). These two bands likely represent two differently N-glycosylated integrin β1 chains that have been detected on the cell surface of several cell types [27]. In contrast to our results, Meng et al could not detect the low-mass integrin β1 in Jurkat cells, but they only investigated the cell membrane whereas we precipitated integrin β1 from whole cell lysates. This suggests that the low-mass form may be retained in the endoplasmatic reticulum.
Table 1.
Monoclonal anti-integrin β1 antibodies used in this study.
| Antibody | Activity | Epitope location | Reference |
|---|---|---|---|
| JB1B | Stimulatory | Continuous (AA 671-705) | Wilkins et al. [26] |
| JB1A | Inhibitory | Continuous (AA 82-87) | Ni and Wilkins [28] |
| B3B11 | Stimulatory | Continuous (AA 660-668) | Wilkins et al. [26] |
| N29 | Stimulatory | Continuous (AA 14-55) | Ni and Wilkins [29] |
| B44 | Stimulatory | Probably continuous/Diff. Region than N29 and JB1B/B3B11 | Wilkins et al. [26] |
| 6S6 | — | — | |
| 3S3 | Inhibitory | Probably discontinuous | Gao et al. [30] |
| 21C8 | Stimulatory | Discontinuous near JB1B/B3B11 | Wilkins et al. [26] |
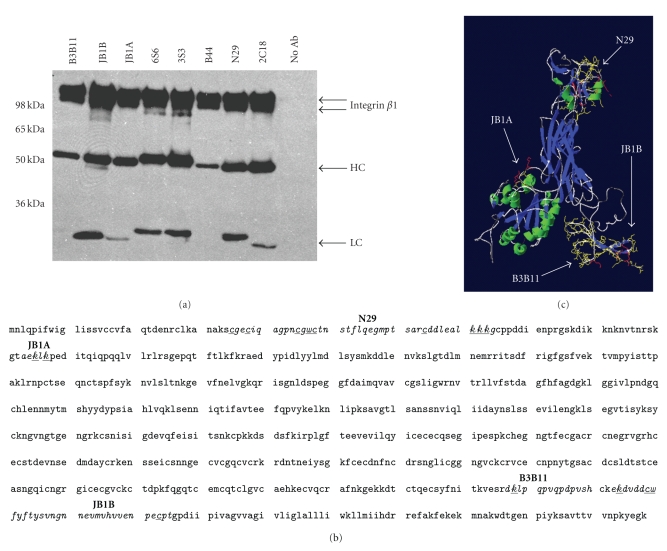
Figure 3.
Analysis of anti-integrin β1 antibodies. (a) Test of antibodies for immunoprecipitation. 0.5 mg Jurkat cell lysate was incubated with 1 μg of the indicated antibody and affinity purification was performed. Precipitations were analyzed by JB1A immunoblot (anti-mouse HRP). Two integrin β1 chains were detected in all immunoprecipitations. The heavy (HC) and light chains (LC) of the antibodies are indicated. (b) Amino acid sequence of the extracellular domain of human integrin β1. Known continuous epitopes are indicated. Underlined amino acids are known to become formaldehyde modified first. (c) Model of the extracellular part of human integrin β1, based on the crystal structure of Integrin β3 (Swiss Model). Indicated are the same epitopes as in (b).
A closer look at the known properties of the anti-integrin β1 antibodies revealed that five of them had been shown to stimulate integrins when added to living cells, while two showed inhibitory character (Table 1). However, as we were planning to use these antibodies for precipitation of integrins from cross-linked and thereby fixed cells, these properties should not affect our experiments. More importantly, the epitopes of some of these antibodies are known and located at very different positions in the extracellular part of the protein: N29 and JB1A detect continuous peptide sequences close to the protein N-terminus (Figure 3(b)). In contrast, B3B11 and JB1B detect continuous sequences at the opposite end of the extracellular part of integrin β1 (Figure 3(b)), where also the discontinuous epitope of 2C18 is found [29]. B44 is known to recognize a different part of the protein than the other antibodies but the precise epitope is unknown. Since the B44 antibody is working in immunoblotting experiments, its epitope is expected to be composed of a continuous sequence [26]. In contrast, 3S3 cannot be used for immunoblotting indicating a discontinuous epitope.
We further modelled the integrin β1 structure on integrin β3 to gain insight into the three-dimensional location of the known epitopes (Figure 3(c)). As expected the epitopes are located in very different parts of the protein. Additionally it is known that the epitopes of N29 and B44 are hidden in the inactivated form of integrin β1 as on some cell lines they can only detect the receptor after it had become activated [29]. To assess whether the antibodies would also be able to work after formaldehyde treatment, we had a closer look at the epitope sequences, which are known for four of them (Figure 3(b)). It had been shown that the amino acids, which are formaldehyde-modified first during the short incubation times used under cross-linking conditions, are lysine, cysteine and tryptophan [24]. The epitopes of N29 and JB1A are composed to a higher extent of these amino acids (20 and 30%), compared to 7% and 12% for B3B11 and JB1B, suggesting that N29 and JB1A are less likely to work after formaldehyde treatment.
3.5. Precipitation of Formaldehyde Generated Integrin β1 Containing Complex
Our next step was to determine whether the antibodies found to precipitate integrin β1 in the previous section can detect and pull down the integrin β1 complex. Jurkat cells were treated with or without 0.4% formaldehyde. Cells were lysed and the lysates were used for an immunoprecipitation using the antibody JB1B and a control pull-down which was performed without an antibody (Figure 4(a)). The higher molecular weight complex containing integrin β1 was only detected in formaldehyde treated cells and was only precipitated when the antibody was added. No complex was detected in the supernatant of the immunoprecipitation, indicating its complete pull-down. Furthermore, no loss in total integrin β1 precipitation was observed, indicating that formaldehyde treatment did not destroy the JB1B epitope. This had been expected, as the epitope does not contain many of the easily modifiable amino acids.
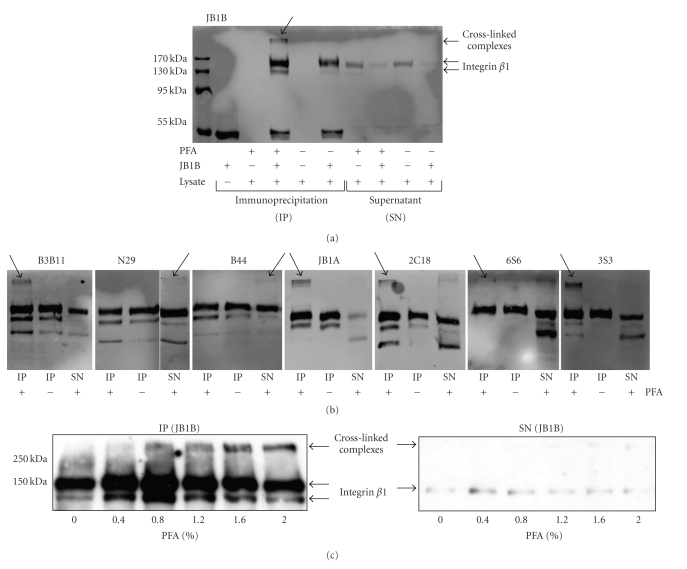
Figure 4.
Precipitation of cross-linked integrin β1 complexes. (a, b) Jurkat cells were treated with or without 0.4% formaldehyde (PFA) before lysis. Immunoprecipitation was performed using 0.5 mg of lysate and 1 μg of the indicated antibody. Immunoprecipitations (IP) and supernatants (SN) were analyzed for presence of integrin β1 complexes using JB1A for immunoblotting (anti-mouse Alexa Fluor 680). (a) JB1B precipitated the integrin β1 complex (indicated by arrow) from formaldehyde treated cells. (b) Test of the remaining seven antibodies for precipitation of integrin β1 from formaldehyde treated cells using the same parameters as in (a). (c) Jurkat cells were treated with different concentrations of formaldehyde (PFA) and lysed. 100 μg lysate were used for pull-downs with JB1B (1 μg) and immunoprecipitations (IP) and supernatants (SN) were analyzed by anti-integrin β1 immunoblotting (JB1A, anti-mouse HRP).
As a next step, all of the other antibodies were used to precipitate integrin β1 from formaldehyde treated and non-treated Jurkat cells (Figure 4(b)). All of them were able to precipitate the target protein even after formaldehyde treatment, indicating that none of their epitopes was destroyed in such a way that it would prevent precipitation. Even for N29 and JB1A, whose epitopes contain a high percentage of easily modifiable amino acids, no fundamental loss in integrin β1 precipitation was observed. However, not all antibodies were equally able to precipitate integrin β1 containing complexes: B3B11, JB1A and 3S3 precipitated the complex completely, since no integrin β1 was detected in the high molecular range of the supernatant. Only partial precipitation of a high molecular weight complex was observed using the antibodies 2C18 and 6S6, where part of the complex was detected in the supernatant of the precipitation. No precipitated complex was detected using the antibodies N29 and B44. As these antibodies were able to precipitate integrin β1 of formaldehyde treated cells, it is unlikely that their epitopes were destroyed during the process. However, as mentioned earlier N29 and B44 have both been shown to detect activated integrin β1 on cells, whereas their epitopes are disguised on non-activated cells [29]. Stringent lysis of cells destroys this conformational limitation and allows detection by the antibodies. In contrast, cross-linking preserves the conformation and therefore a non-activated complex would be undetectable and not precipitated by these two antibodies. On the other hand, they could be able to precipitate an activated complex which in turn would not be recognized by some of the other monoclonal antibodies. This limitation of antibody usage does not only apply to formaldehyde cross-linking, but restricts every approach where protein complexes are precipitated by antibodies, including the traditional coimmunoprecipitation method which uses gentle lysis conditions to preserve protein interactions.
Finally we asked whether a higher degree of modification induced by higher concentrations of formaldehyde would impede the precipitation of integrin β1 using the antibody JB1B. Therefore we tested the ability of JB1B to pull down complexes from cell lysates, which were obtained after treatment of Jurkat cells with varying formaldehyde concentrations. JB1B was able to precipitate integrin β1 and the complex regardless of the formaldehyde concentrations applied, which was shown by analysis of the pull-downs as well as the supernatants of the immunoprecipitations (Figure 4(c)): no protein was detected in the supernatants, whereas integrin β1 was detected in the range of the monomeric protein and the high molecular weight complex in the immunoprecipitations. We concluded that JB1B is the most universal of the anti-integrin β1 antibodies we had tested, and therefore the most useful for formaldehyde cross-linking.
3.6. Mass Spectrometric Analysis of Complexes Containing Integrin β1
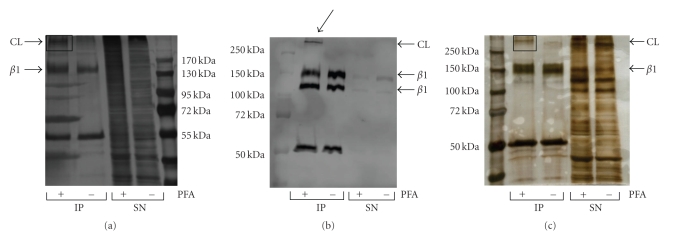
Western blot analysis had shown that we precipitate a high molecular weight complex containing integrin β1 after formaldehyde treatment. As the other components of this complex were unknown to us, we used mass spectrometry to identify them. The integrin β1 complex was precipitated by JB1B, separated by SDS PAGE and visualized by silver staining (Figure 5(a)). The indicated band was excised, in gel digested and analyzed by LC-MS/MS. Using Jurkat cells we performed this experiment in several independent repeats, and detected integrin β1 in every sample (Table S1). Furthermore we consistently identified integrin α4. The receptor formed by these integrins is VLA-4 (very late antigen 4), a known receptor found on T cells [31] and a high abundant integrin on Jurkat cells [25].
Figure 5.
Analysis of integrin β1 complexes (CL) from Jurkat cells and human platelets. (a) Jurkat cells were treated with or without 0.4% formaldehyde (PFA) before lysis. Immunoprecipitation was performed using 2 mg lysate and 5 μg of JB1B and analyzed by SDS PAGE. The indicated area was excised and analyzed by mass spectrometry. (b) Anti-integrin β1 immunoblot (JB1A, anti-mouse Alexa Fluor 680) of JB1B pull-downs performed on human platelets treated with and without 0.4% formaldehyde (PFA). 100 μg platelet lysate and 0.85 mg JB1B were used. Complexes containing integrin β1 are indicated by the arrow. (c) SDS PAGE analysis of JB1B immunoprecipitates from noncross-linked and cross-linked human platelets. Immunoprecipitation was performed using 0.7 mg lysate and 4.25 μg JB1B. The indicated band was excised and examined with mass spectrometry.
Two of these experiments also allowed the identification of additional proteins (Table 2). The listed proteins were not detected in control experiments using either non-formaldehyde treated cells or immunoprecipitations performed without the antibody when analysing gel bands at masses corresponding to the integrin β1 containing complex. Of these additionally identified proteins, integrin α6 is known to form a receptor termed VLA-6 together with integrin β1 on T cells, which has not been detected in Jurkat cells before [25]. Two additional integrin α subunits were detected only in dataset 2: integrin α5 (log(e) = −36.4) and α1 (log(e) = −1.7). Integrin α5 had been demonstrated to be expressed together with integrin β1 on Jurkat cells [25], whereas integrin α1 is known to form a collagen-recognizing receptor with integrin β1 on endothelial cells [32]. Even though only few peptides of each protein were sequenced, the identification of the integrin α subunits is reliable as they do not share high homologues sequences and all sequenced peptides were unique for each integrin α.
Table 2.
Proteins identified in the integrin β1 complex precipitated from Jurkat cells. Formaldehyde cross-linked Jurkat cell lysate was used to enrich integrin β1 with JB1B. The integrin β1 complex was identified by SDS PAGE analysis and studied by LC-MS/MS and the GPM. Only proteins with a log(e) <−1 and which were identified in both datasets and not in control samples are listed.
| Protein | Accession | Mr [kDa] | log(e) (Dataset 1) | log(e) (Dataset 2) | |
|---|---|---|---|---|---|
| Integrin β1 | ITGB1 | ENSP00000303351 | 88.4 | −39.8 | −118.3 |
| Actin, cytoplasmic 2 | ACTG1 | ENSP00000331514 | 41.8 | −23.4 | −73.1 |
| Desmoglein-1 | DSG1 | ENSP00000257192 | 113.7 | −16.4 | −42.9 |
| Filaggrin-2 | FLG2 | ENSP00000373370 | 247.9 | −15.6 | −40.1 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDHL6 | ENSP00000229239 | 36 | −7.4 | −18.2 |
| Arginase-1 | ARG1 | ENSP00000349446 | 35.6 | −7.3 | −15.1 |
| Junction plakoglobin | JUP | ENSP00000377507 | 81.7 | −6 | −7 |
| Integrin α4 | ITGA4 | ENSP00000380227 | 114.8 | −5.1 | −6.3 |
| Heat shock protein HSP 90α | HSP90AA2 | ENSP00000216281 | 84.6 | −2.9 | −4.5 |
| C1orf68 | C1orf68 | ENSP00000354769 | 26.2 | −2.1 | −3.2 |
| Integrin α6 | ITGA6 | ENSP00000264106 | 126.5 | −1.6 | −2.4 |
The other identified proteins are not known to interact directly with integrin β1 [13]. As a member of the cytoskeleton, actin is intertwined with integrin function and associated with integrins through adaptor proteins such as talin [10]. However, an adaptor was not consistently identified in these immunoprecipitations. Furthermore actin is a highly abundant protein and its identification has to be judged carefully. The same is true for GAPDH and heat shock protein HSP 90α, which are both abundant proteins and frequently found in immunoprecipitations. Thus we cannot exclude that these proteins are non-specific background, even though they were not detected in the control samples. As formaldehyde cross-linking could also occur with abundant proteins that colocalize but have lower binding affinity to integrin β1, this may explain the presence of abundant proteins. Alternatively, proteins that do not colocalize but show medium to high affinity may form a non-cross-linked complex in the lysate that is sufficiently stable not to be destroyed by the incubation temperature of 65°C that is applied prior to gel separation. This scenario may also be true for most of the other proteins (Desmoglein-1, Filaggrin-2, Junction plakoglobin and C1orf68), which are highly expressed in skin cells. These could be contaminants similar to keratins (which were removed before data interpretation) occurring during sample preparation. The last identified protein arginase-1 is found in the liver and plays an important role in arginine metabolism [33], a connection to integrin β1 is not known. On the other hand, formaldehyde cross-linking would also stabilize interactions between two proteins that do not directly interact but have a common binding partner. Therefore, such unexpected findings may still provide valuable information.
No known adaptor proteins like talin or tensin were consistently detected in the complex. This indicates that the precipitated complex is mostly in an inactive state, where the two integrin chains form a stable and very compact complex and no adaptor proteins are bound to the intracellular tail. This matches our earlier observation that N29 and B44, which were shown to be unable to detect inactive integrin β1, did not precipitate this complex. However, talin-1 was identified with high confidence (log(e) = −94.9) in one experiment (dataset 2) indicating the ability to precipitate integrin β1/adaptor complexes using our approach, provided the integrin β1 on Jurkat cells is activated. All our results imply that most of the integrin β1 found on the Jurkat cells is not activated, which is in contrast to an earlier study where Jurkat cells have been shown to express constitutively active integrin β1 [25]. However, it has to be considered that cell lines change their behaviour over time, thus it might be that the Jurkat cells used by them and by us differ in this aspect.
3.7. Detection and Analysis of Integrin β1 Complexes in Human Platelets
In addition to our studies using Jurkat cells, we wanted to apply our approach also to a different and physiologically more relevant system, and therefore used human platelets. Platelets express high amounts of integrins, which play an important role in platelet aggregation. Even though integrin β1 is not the most abundant integrin found on platelets, it forms three different receptors (α2β1, α5β1, α6β1) and is involved in platelet adhesion [34].
Human platelets were treated with and without formaldehyde and the antibody JB1B was used to precipitate integrin β1 from the lysates. In addition to the monomeric protein, a high molecular weight complex was detected in immunoblotting experiments, which was only visible after formaldehyde treatment (Figure 5(b)). We identified the contents of the complex precipitated by JB1B from human platelets by mass spectrometry (Figure 5(c), Table S2). Proteins identified in two independent experiments are listed in Table 3. Integrin β1 was detected in both pull-downs as expected. Furthermore, the integrins α6 and α2, which are known to form heterodimers with integrin β1 on platelets, were identified [34]. Thrombospondin-1, a known ligand of the integrin receptor α6β1, was also detected in the integrin β1 complex. As thrombospondin-1 is also the most abundant protein found in the α-granules of platelets [35], its identification may be merely due to its high abundance in the platelet lysate. Similarly, integrin αIIb does not form a heterodimer with integrin β1, but together with integrin β3 forms glycoprotein IIb/IIIa, the most abundant platelet receptor. Integrin β3 was only detected in dataset 2 (log(e) = −40.2), however, the presence of both of these proteins could be solely due to their high presence in the platelet lysate as well. On the other hand we cannot exclude that integrin β1 interacts with one or both of these integrins forming multi-heterodimers.
Table 3.
Proteins identified in the integrin β1 complex precipitated from human platelets. Integrin β1 was precipitated by JB1B from formaldehyde cross-linked platelets. The integrin β1 complexes were identified by SDS PAGE analysis and investigated by LC-MS/MS and the GPM. Only proteins with a log(e) <−2 and which were identified in both datasets are listed. *Two different isoforms were detected in the experiments.
| Protein | Accession | Mr [kDa] | log(e) (Dataset 1) | log(e) (Dataset 2) | |
|---|---|---|---|---|---|
| Thrombospondin-1 | THBS1 | ENSP00000260356 | 129.3 | −46.2 | −140.1 |
| Integrin α6* | ITGA6 | ENSP00000264106/ | 126.5/124 | −38.2 | −299 |
| ENSP00000364369 | |||||
| Integrin β1 | ITGB1 | ENSP00000303351 | 88.4 | −29.8 | −201.1 |
| Immunoglobulin heavy | IGHV4-31 | ENSP00000374990 | 43.9 | −9.6 | −10 |
| chain C | |||||
| Hornerin | HRNR | ENSP00000357791 | 282.2 | −9.4 | −20.2 |
| Integrin α2 | ITGA2 | ENSP00000296585 | 129.2 | −3.4 | −144.5 |
| Integrin αIIb | ITGA2B | ENSP00000262407 | 113.3 | −2.8 | −22.8 |
| Filaggrin-2 | FLG2 | ENSP00000373370 | 247.9 | −4 | −4.5 |
The identification of immunoglobulin heavy chain C is assumingly due to the immunoprecipitation, whereas hornerin and filaggrin-2 are both proteins expressed in the skin and could be contaminants caused during sample preparation. Three proteins which are known to interact with integrin β1 were only detected in dataset 2: integrin α5 (log(e) = −1.8), talin-1 (log(e) = −142.2) and kindlin-3 (log(e) = −18.5). Integrin α5 is known to form the receptor VLA-5 with integrin β1. With the detection of this α chain, we identified all known integrin β1 containing heterodimers on platelets. The identification of the adaptors talin-1 and kindlin-3 indicates again that we are able to precipitate integrin β1/adaptor protein complexes. However, these experiments were performed on platelets which were not intentionally stimulated, thus we do not expect high amounts of activated integrin β1 on these cells. Stimulation can nonetheless occur to a low degree during platelet preparation, which explains the detection of talin-1 and kindlin-3 in one of the samples.
4. Conclusion
Using the transmembrane integrin β1 complex as a model, our study addressed several critical factors in the formaldehyde cross-linking procedure and provides a practical guide to the optimization of such experiments.
By modifying SDS PAGE sample preparation conditions, we could detect an additional formaldehyde generated, high-molecular weight band containing integrin β1 by immunoblot analysis. LC-MS/MS analysis showed that this complex was mainly composed of integrins. Several integrin α subunits known to form heterodimers with integrin β1 could be detected in two different systems, Jurkat cells and human platelets (Table 4). The appearance of a broad band on the type of gel we had chosen did not provide any insights into heterogeneity and molecular weight of the corresponding integrin complex(es). Indeed, the band could have arisen from a single, multi-integrin complex containing all of the identified proteins, albeit the presence of several distinct complexes with similar molecular weight resulting in overlapping bands is more likely. Targeted experiments will be necessary to unequivocally answer this question.
Table 4.
Integrin β1 heterodimers detected in Jurkat cells and human platelets. *Only detected in one dataset.
| Detected in | Integrin heterodimers | Ligands |
|---|---|---|
| Jurkat cells | β1α4 (VLA-4) | Fibronectin, VCAM-1 |
| β1α6 (VLA-6) | Laminins | |
| β1α5 (VLA-5)* | Fibronectin | |
| β1α1* | Collagens | |
| Human platelets | β1α6 (VLA-6) | Laminins |
| β1α2 | Collagens | |
| β1α5 (VLA-5)* | Fibronectin | |
One major focus of this study was to systematically test the applicability of monoclonal antibodies in precipitating formaldehyde treated endogenous, nontagged proteins. Two out of eight antibodies were expected to be less likely to work in precipitating integrin β1 from formaldehyde treated cells, since their epitopes contained amino acids that are known to be susceptible to formaldehyde modification [24]. However, we could not detect any difference in precipitation, indicating that destruction of epitopes is not an overwhelming problem in the formaldehyde cross-linking procedure at least for the panel of antibodies we had tested.
Two antibodies, which had been shown before to recognize only activated integrin β1 [29], did not pull-down the complex probably due to hidden epitopes in the complex. This finding, together with the observation that the analyzed complexes were mainly composed of integrin α and β chains, indicates that the majority of integrin β1 containing heterodimers found on Jurkat cells and human platelets are in an inactive state. However, in two experiments out of four we could identify talin-1, which is an adaptor protein that binds activated integrin β1. This suggests that formaldehyde cross-linking is a promising tool to study integrin activation and regulation. In future experiments Jurkat cell lines or platelets will be stimulated deliberately and the composition of the integrin β1 complexes will be analyzed. A quantitative approach based on SILAC or iTRAQ labelling will then be applied to distinguish non-activated and activated integrin β1 and to allow timecourse studies.
By testing different formaldehyde concentrations for cross-linking, we realized that the balance between yield of cross-linked complex and the loss of protein was shifted in the case of integrin β1 compared to earlier studies: whereas 0.4% formaldehyde had been established as optimal concentration for studying myc-tagged soluble protein earlier [4], 1% formaldehyde seemed to be more suitable for integrin β1. While we used 0.4% formaldehyde for the current studies in order to minimize artefact generation, higher formaldehyde concentration may be used for future investigations. Indeed, varying the formaldehyde concentration is an excellent means not only to balance yield and protein loss, but also to distinguish direct and indirect interaction partners of a protein.
The multistep protocol for the optimization of formaldehyde cross-linking described here can be applied to any protein of interest. While the formaldehyde concentration is the most important variable that has to be optimized for each protein, detection of a formaldehyde cross-linked complex via western blot analysis simplifies this process. In addition, the application of antibodies for immunoprecipitations of modified proteins has to be monitored carefully, and the possibility that they do not recognize their antigen anymore has to be considered at all times. If these points are taken into account, however, formaldehyde cross-linking paired with immunoprecipitation will be a very powerful and reliable method to study protein-protein interactions, especially weak and transient ones, that can be established as a routine method in any laboratory.
Supplementary Material
Table S 1 Mass spectrometric data (GPM) for JB1B immunoprecipitations performed on Jurkat cells.
Table S 2 Mass spectrometric data (GPM) for JB1B immunoprecipitations performed on human platelets.
Acknowledgments
The authors would like to thank John Wilkins for providing the monoclonal anti integrin β1 antibodies, Jason Rogalski and Nikolay Stoynov for technical support and Geraldine Walsh for preparation of human platelets.
Abbreviations
- HRP:
Horse radish peroxidase
- IP:
Immunoprecipitation
- iTRAQ:
Isobaric tag for relative and absolute quantitation
- LC-MS/MS:
Liquid chromatography- tandem mass spectrometry
- Mr:
Molecular weight
- PBS:
Phosphate buffered saline
- PFA:
Paraformaldehyde
- RIPA:
Radio immunoprecipitation assay
- RT:
Room temperature
- SDS PAGE:
Sodium dodecylsulfate polyacrylamide gel electrophoresis
- SILAC:
Stable isotope labelling with amino acids in cell culture
- SN:
Supernatant
- VLA:
Very late antigen.
References
- 1.Young KH. Yeast two-hybrid: so many interactions, (in) so little time. Biology of Reproduction. 1998;58(2):302–311. doi: 10.1095/biolreprod58.2.302. [DOI] [PubMed] [Google Scholar]
- 2.Willander M, Al-Hilli S. Analysis of biomolecules using surface plasmons. Methods in Molecular Biology. 2009;544:201–229. doi: 10.1007/978-1-59745-483-4_14. [DOI] [PubMed] [Google Scholar]
- 3.Markham K, Bai Y, Schmitt-Ulms G. Co-immunoprecipitations revisited: an update on experimental concepts and their implementation for sensitive interactome investigations of endogenous proteins. Analytical and Bioanalytical Chemistry. 2007;389(2):461–473. doi: 10.1007/s00216-007-1385-x. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. Journal of Mass Spectrometry. 2008;43(6):699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 5.Vasilescu J, Guo X, Kast J. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics. 2004;4(12):3845–3854. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 6.Schmitt-Ulms G, Hansen K, Liu J, et al. Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nature Biotechnology. 2004;22(6):724–731. doi: 10.1038/nbt969. [DOI] [PubMed] [Google Scholar]
- 7.Bai Y, Markham K, Chen F, et al. The in vivo brain interactome of the amyloid precursor protein. Molecular and Cellular Proteomics. 2008;7(1):15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. Journal of Biological Chemistry. 2002;277(48):46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
- 9.Lindskog M, Rockberg J, Uhlen M, Sterky F. Selection of protein epitopes for antibody production. BioTechniques. 2005;38(5):723–727. doi: 10.2144/05385ST02. [DOI] [PubMed] [Google Scholar]
- 10.Harburger DS, Calderwood DA. Integrin signalling at a glance. Journal of Cell Science. 2009;122(2):159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brakebusch C, Fassler R. β 1 integrin function in vivo: adhesion, migration and more. Cancer and Metastasis Reviews. 2005;24(3):403–411. doi: 10.1007/s10555-005-5132-5. [DOI] [PubMed] [Google Scholar]
- 12.Lee H-S, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to Ras GTPase membrane-targeting sequences. Journal of Biological Chemistry. 2009;284(8):5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legate KR, Fassler R. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. Journal of Cell Science. 2009;122(2):187–198. doi: 10.1242/jcs.041624. [DOI] [PubMed] [Google Scholar]
- 14.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324(5929):895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman MD, Walsh GM, Rogalski JC, Kast J. Identification of nitroxyl-induced modifications in human platelet proteins using a novel mass spectrometric detection method. Molecular and Cellular Proteomics. 2009;8(5):887–903. doi: 10.1074/mcp.M800230-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong J-P, Stehle T, Diefenbach B, et al. Crystal structure of the extracettutar segment of integrin αVβ3. Science. 2001;294(5541):339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 18.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL repository and associated resources. Nucleic Acids Research. 2009;37:387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with imageJ. Biophotonics International. 2004;11(7):36–41. [Google Scholar]
- 21.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20(3):601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Jeno P, Mini T, Moes S, Hintermann E, Horst M. Internal sequences from proteins digested in polyacrylamide gels. Analytical Biochemistry. 1995;224(1):75–82. doi: 10.1006/abio.1995.1010. [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using stage tips. Nature Protocols. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 24.Toews J, Rogalski JC, Clark TJ, Kast J. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Analytica Chimica Acta. 2008;618(2):168–183. doi: 10.1016/j.aca.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 25.Seminario M-C, Sterbinsky SA, Bochner BS. β1 integrin-dependent binding of Jurkat cells to fibronectin is regulated by a serine-threonine phosphatase. Journal of Leukocyte Biology. 1998;64(6):753–758. doi: 10.1002/jlb.64.6.753. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins JA, Li A, Ni H, Stupack DG, Shen C. Control of β1 integrin function: localization of stimulatory epitopes. Journal of Biological Chemistry. 1996;271(6):3046–3051. [PubMed] [Google Scholar]
- 27.Meng X, Cheng K, Krohkin O, et al. Evidence for the presence of a low-mass β1 integrin on the cell surface. Journal of Cell Science. 2005;118(17):4009–4016. doi: 10.1242/jcs.02520. [DOI] [PubMed] [Google Scholar]
- 28.Ni H, Wilkins JA. Localisation of a novel adhesion blocking epitope on the human β1 integrin chain. Cell Adhesion and Communication. 1998;5(4):257–271. doi: 10.3109/15419069809040296. [DOI] [PubMed] [Google Scholar]
- 29.Ni H, Li A, Simonsen N, Wilkins JA. Integrin activation by dithiothreitol or Mn2+ induces a ligand- occupied conformation and exposure of a novel NH2-terminal regulatory site on the β1 integrin chain. Journal of Biological Chemistry. 1998;273(14):7981–7987. doi: 10.1074/jbc.273.14.7981. [DOI] [PubMed] [Google Scholar]
- 30.Gao JX, Wilkins J, Issekutz AC. Migration of human polymorphonuclear leukocytes through a synovial fibroblast barrier is mediated by both β 2 (CD11/CD18) integrins and the β 1 (CD29) integrins VLA-5 and VLA-6. Cellular Immunology. 1995;163(2):178–186. doi: 10.1006/cimm.1995.1114. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu Y, Van Seventer GA, Horgan KJ, Shaw S. Regulated expression and binding of three VLA (β1) integrin receptors on T cells. Nature. 1990;345(6272):250–253. doi: 10.1038/345250a0. [DOI] [PubMed] [Google Scholar]
- 32.Abair TD, Bulus N, Borza C, Sundaramoorthy M, Zent R, Pozzi A. Functional analysis of the cytoplasmic domain of the integrin {alpha}1 subunit in endothelial cells. Blood. 2008;112(8):3242–3254. doi: 10.1182/blood-2007-12-126433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christianson DW. Arginase: structure, mechanism, and physiological role in male and female sexual arousal. Accounts of Chemical Research. 2005;38(3):191–201. doi: 10.1021/ar040183k. [DOI] [PubMed] [Google Scholar]
- 34.Rivera J, Lozano ML, Navarro-Nunez L, Vicente V. Platelet receptors and signaling in the dynamics of thrombus formation. Haematologica. 2009;94(5):700–711. doi: 10.3324/haematol.2008.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung J, Gao A-G, Frazier WA. Thrombspondin acts via integrin-associated protein to activate the platelet integrin αIIbβ3. Journal of Biological Chemistry. 1997;272(23):14740–14746. doi: 10.1074/jbc.272.23.14740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S 1 Mass spectrometric data (GPM) for JB1B immunoprecipitations performed on Jurkat cells.
Table S 2 Mass spectrometric data (GPM) for JB1B immunoprecipitations performed on human platelets.