Abstract
The arenaviruses include a number of important pathogens including Lassa virus and Junin virus. Presently, the only treatment is supportive care and the antiviral Ribavirin. In the event of an epidemic, patient triage may be required to more effectively manage resources; the development of prognostic biomarker signatures, correlating with disease severity, would allow rational triage. Using a pair of arenaviruses, which cause mild or severe disease, we analyzed extracts from infected cells using SELDI mass spectrometry to characterize potential biomarker profiles. EDGE analysis was used to analyze longitudinal expression differences. Extracts from infected guinea pigs revealed protein peaks which could discriminate between mild or severe infection, and between times post-infection. Tandem mass-spectrometry identified several peaks, including the transcriptional regulator prothymosin-α. Further investigation revealed differences in secretion of this peptide. These data show proof of concept that proteomic profiling of host markers could be used as prognostic markers of infectious disease.
1. Introduction
Viral hemorrhagic fevers (VHFs) represent an important class of emerging diseases and are classified as category A priority pathogens by the National Institute for Allergy and Infectious Diseases. The VHFs Lassa fever and the Argentine, Bolivian, Venezuelan, and Brazilian hemorrhagic fevers are caused by members of the Arenaviridae [1]. Despite their status as emerging infectious diseases, little is known about the molecular basis of the pathology of these diseases, and this has hindered the design of antiviral therapeutics. Arenavirus pathogenesis is believed to involve the dysregulation of cytokines; for example, infection with the virulent Lassa virus (LASV) prevents macrophage activation and TNF-α production, whereas infection with the avirulent Mopeia virus induces proinflammatory cytokine production [2], a similar effect has been shown with attenuated and virulent isolates of Pichindé virus (PICV) [3]; proteomic and kinomic level analyses have also shown differential cell signaling events induced by attenuated and virulent arenavirus infection [4–6].
Currently, the only drug used in the treatment of these infections is the broad-spectrum antiviral, Ribavirin, which requires administration within the first week of infection to be most efficacious [7]. As only a small proportion of patients infected with LASV develop a hemorrhagic fever, indicators of prognosis will be critical in the efficient management of healthcare resources during epidemics. Currently, the best prognostic marker for severe disease is viral load, with high levels of virus correlating with increased likelihood of severe disease [8]. However, quicker methods of prognosis determination would be of significant use in a clinical setting.
We have previously used surface-enhanced laser desorption ionization time-of-flight (SELDI-TOF) mass spectrometry to characterize proteins bound to a thioaptamer (oligonucleotides with thiophosphate backbone substitutions) on a ProteinChip array [9]. SELDI is a high-throughput profiling platform that combines retentive chromatography and mass spectrometry in one platform REFS. The process involves selective extraction and retention of proteins on chromatographic chip surfaces arranged in an array of spots and their subsequent analysis by mass spectrometry.
In this study, we have used SELDI-TOF to compare cell extracts for representative protein “signatures” following infection. PICV is a genetically characterized model for Lassa fever and recent construction infectious clones may allow the role of individual sequence changes in determining pathogenesis to be elucidated [10, 11]. Using two variants of PICV, P2, which causes a mild, self-limiting infection, and P18, which causes a lethal hemorrhagic disease [12], we can identify signatures unique to mild and severe infection, and which reflect common responses.
In this report, we describe the host-cell protein “signatures” following mock, P2, and P18 PICV infection over time. We show that, consistent with previous data from several groups, the virulent variant more closely resembles mock-infection, suggesting a suppression of cellular responses to infection [2, 5, 13]. In addition, we identify one of these protein peaks by tandem mass spectrometry as prothymosin-α, which may indicate a potential therapeutic target for the treatment of hemorrhagic arenavirus infection.
2. Materials and Methods
2.1. Cell Culture and Viruses
Murine monocyte-like P388D1 cells were maintained in RPMI supplemented with 5% fetal bovine serum and 5 mM glutamine. Cells were cultured in the absence of antibiotics to ensure no underlying contamination. Cells were infected in triplicate with PEG purified P2 or P18 PICV at a multiplicity of infection of 5 or mock infected with virus purification medium. Cells were harvested at 8 timepoints and cytoplasmic and nuclear extracts prepared. For animal experiments, male Hartley guinea pigs were infected intraperitoneally with 1000 plaque forming units of the P2 or P18 variant of PICV or mock-infected with phosphate buffered saline (n = 7 per group/timepoint) and sacrificed at 1 and 6 days post infection. Animal experiments were performed following institutional animal care and use approved guidelines and protocols.
2.2. Cytoplasmic and Nuclear Fractionation of Cells
Cells were fractionated into cytoplasmic and nuclear fractions as previously described [14] with the addition of a nuclear purification step using Optiprep (Axis-Shield, Oslo, Norway) gradients. Briefly, lysates were underlayered with 10 mL 30% Optiprep and 5 mL 35% Optiprep and centrifuged at 4°C for 30 minutes at 4300 × g. The interface was removed and placed in a fresh tube which was filled with sucrose buffer I (described in Dyer & Herzog, 1995) plus 1.5 mM CaCl2. Following centrifugation at 4°C for 15 minutes at 1900 × g, the pellet was resuspended in sucrose buffer I and the centrifugation repeated. Nuclear lysis was completed following the referenced protocol.
2.3. Fractionation of Cytoplasmic Portion of P388D1 Cells
5.4 mL buffer containing 10 mM Tris and 9 M urea (pH 9) was added to 2.7 mL combined cytoplasmic portion of mock-, P2-, and P18-infected macrophages, and the mixture was incubated at 4°C for 20 minutes. It then was dialyzed against 50 mM Tris (pH 8), after which the dialyzed solution was added to 30 μL packed Q HyperD F beads (Pall corporation, PA), and incubated for 30 minutes at 4°C. The beads were collected by centrifugation (3000 × g, 1 minutes) and the supernatant was saved as flow-through. The proteins bound to the beads were eluted by lowering the pH of the buffer in five fractions (90 μL of each eluent in 50 mM of each pH buffer): Q1 (pH 6.5), Q2 (pH 5.5), Q3 (pH 4), Q4 (pH 3) and Q5 (pH 3 + 1 M NaCl). 10 μL of each fraction was added to 90 μL 50 mM Tris (pH 8) buffer and was captured on Q10 anion exchange ProteinChip array surfaces (Bio-Rad Laboratories, Hercules, CA) with a bioprocessor Biomek 3000 (Beckman Coulter, Fullerton, CA).
2.4. SELDI-TOF Mass Spectrometry and Spectral Analysis
The mass spectra of the proteins captured on Q10 chips were recorded on the PCS 4000 ProteinChip array reader (Bio-Rad laboratories, Hercules, CA). Experiments were run in triplicate at the 8 time points mentioned above, leading to a total of 72 samples each for the cytoplasmic and nuclear fractions. SELDI spectra were recorded on the PCS4000 ProteinChip System at a laser intensity setting of 2600. Spectra were analyzed with the Ciphergen Express (CE) software. Analysis was limited to the molecular weight (MW) range 2 kDa–20 kDa. Peak intensities were scaled using CE.
2.5. Statistical Analysis
Log-transformed peak intensity values were subjected to a 2-way ANOVA test with Matlab v. R2007a (The Mathworks, Natick, MA). P-values were calculated for three null hypotheses: (a) that all samples from the Mock, P2, and P18 infections came from the same population, (b) that all samples from the eight different time points came from the same population, and (c) that the effects due to the two factors, infection and time, are additive.
Spotfire DecisionSite 9.0 (TIBCO Software Inc., Palo Alto, CA) was used for hierarchical clustering (HC) to group the biomarkers detected by the timecourse analysis described above. SELDI peak intensities were first Z-score normalized and HC was then run using the unweighted average method, with Euclidean distance as the similarity measure and average value as the ordering function. Following HC, Principal Components Analysis (PCA) was performed transforming the data into a small number of dimensions to visualize the clustering of the biomarkers.
We also analyzed data using “Extraction of Differential Genomic Expression” (EDGE) as this technique is designed for the analysis of timecourse data [15, 16]. The method builds on the false-discovery rate (FDR) developed by Benjamini and Hochberg to control the expected proportion of falsely rejected null hypotheses [17]. For the multiple significance testing performed in the analysis of-omics data, the Q-values provide a more meaningful and practical measure than the P-value [18]. Appropriate cutoffs are imposed on the Q-value to control the desired proportion of falsely rejected hypotheses.
2.6. Protein Identification and Verification
The cytoplasmic samples were treated with 9 M urea to denature the proteins, fractionated by Q HyperD F beads, and eluted into flow-through (FT), and five pH factions at pH 6.5, 5.5, 4.0, 3.0, and pH 3 + 1 M NaCl. The fractions were captured on Q10 chips. The chip was loaded on the high-resolution QSTAR XL spectrophotometer and data collection was performed. The fragment masses were used as probes to search the mouse protein database for protein identification.
2.7. Biomarker Identification Using SELDI Quadrupole Time of Flight Tandem Mass Spectrometry
Tandem mass spectrometric peptide sequencing was accomplished using QSTAR XL (Applied Biosystems Inc., Foster City, CA) quadrupole time of flight instrument equipped with Ciphergen ProteinChip Interface PCI 1000. The instrument was calibrated externally using the known MS/MS spectrum of ACTH peptide (1–24) at 2465.2 m/z, where four fragment ions and the parent ion were used as calibration points. All mass spectra were acquired in positive ion mode with collision energy that follows the rule of approximate 50 eV/kDa parent peptide mass. Raw data were analyzed using the instrument's software. Fragmentation information from tandem MS spectrum was first submitted to the sequence query program in Matrix Science (http://www.matrixscience.com/) for possible identities with a precision tolerance of 0.4 Da for MS/MS fragments and 70 ppm for the parent peptide.
2.8. Verification of the Identified Biomarkers
60 uL of packed protein G agarose beads (Santa Cruz Biotechnology, CA) were incubated with 30 uL mouse monoclonal antibody against prothymosin alpha (Alexis Biochemicals, CA) for 1 hour at 4C, followed by three times wash of 1XPBS, 50 uL combined cytoplasmic portion of mock-, P2-, and P18-infected macrophages was then added to the beads, and the solution was incubated at 4C for 2 hours, followed by three times wash of 1XPBS + 0.1% Triton X-100. After a brief wash with water, proteins bound to the antibody against prothymosin alpha were eluted with 60 uL of elution buffer containing 0.3% TFA and 50% acetonitrile. The eluant was analyzed by PCS 4000 ProteinChip array reader (Bio-Rad laboratories, Hercules, CA).
2.9. Enzyme-Linked Immunosorbant Assay
Supernatants from infected triplicate cultures were added to 96-well plates in triplicate wells (Nunc) and incubated overnight at 4°C. Wells were then blocked with 5% w/v bovine serum albumin for 1 hour. Primary antibody (antiprothymosin-α1, Santa Cruz, Santa Cruz, CA, 1/2000) was added to wells in blocking solution and incubated for 2 hours. Wells were then washed four times in phosphate buffered saline + 0.05% tween-20 for 5 minutes prior to incubation with secondary antibody (anti-rabbit Ig HRP conjugate, 1/2000) in blocking solution for 1 hour. Wells were washed as before and levels of [pro] thymosin are indirectly assayed by the addition of the colorimetric substrate TMB (Sigma, St Louis, MO).
3. Results
3.1. SELDI Mass Spectrometry of Guinea Pig Peritoneal Cells
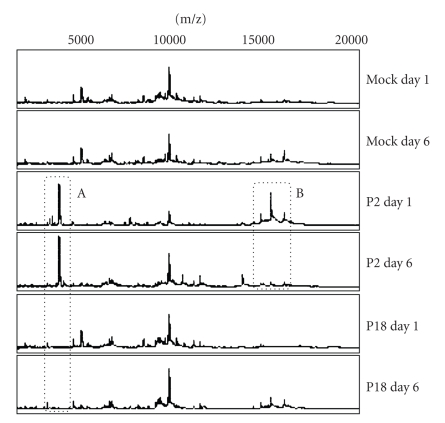
Peritoneal cell extracts from infected guinea pigs were analyzed by surface enhanced laser desorption ionization (SELDI) mass spectrometry (Figure 1). Peaks were identified that could discriminate between type of infection (e.g., Figure 1, box A) and stage of infection (e.g., Figure 1, box B). Consistent with our previous findings, infection with the virulent P18 variant of the virus resulted in a spectrum that more closely resembled mock infection than infection with the attenuated P2 virus.
Figure 1.
SELDI spectra reveal virus and time-based peak differences. Guinea pigs were mock-infected or infected with the P2 or P18 variant of Pichinde virus. At 1 day and 6 days post infection, peritoneal cells were harvested and cytoplasmic and nuclear extracts prepared and analyzed using surface enhanced laser desorption ionization time-of-flight mass spectrometry. Experiments were performed in triplicate. Protein peaks were identified which showed patterns specific to type and stage of infection. The figure shows a representative set of spectra from cytoplasmic extracts.
We next infected triplicate cultures of murine P388D1 macrophage-like cells for a timecourse of infection from 15 minutes to 16 hours. We used a cell line for this study in order to increase the sample quantity for downstream assays and database searching for identification by mass spectrometry. We have previously used this cell line to investigate PICV-host interactions and are familiar with how cellular responses in this cell line model those seen in primary guinea pig macrophages. Triplicate nuclear and cytoplasmic samples were prepared following mock-infection or infection with P2 or P18 PICV at 15 m, 30 m 1, 2, 4, 8, 12, and 16 hours post infection. Samples were analyzed by SELDI mass spectrometry. Statistical analysis by ANOVA and EDGE identified 27 and 35 significantly different peaks, respectively.
Hierarchical cluster analysis revealed similarities in peak patterns between P2 and P18 infection at late times (12 and 16 hours) post infection (data not shown), but that P18 was most similar to mock infection at early times post infection, consistent with our previous results. We next selected several significant peaks for identification by mass spectrometry. We identified several peaks as two SH3-domain binding proteins, stathmin, prothymosin-α, thioredoxin, and various ribosomal proteins. Prothymosin-α was identified as a significantly differentially expressed protein peak with and EDGE Q value of 0.0164. We have previously identified prothymosin-α as a node in a signaling network constructed following kinomics analysis of PICV-infected cells [5] and so was selected for additional study.
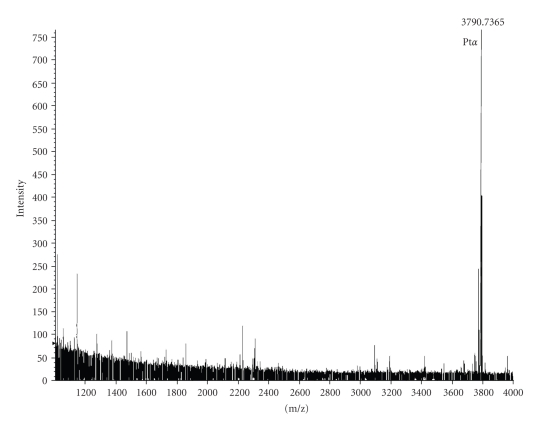
3.2. Verification of the 3787 Peak as Prothymosin-α from On-Chip Digestion and MS/MS
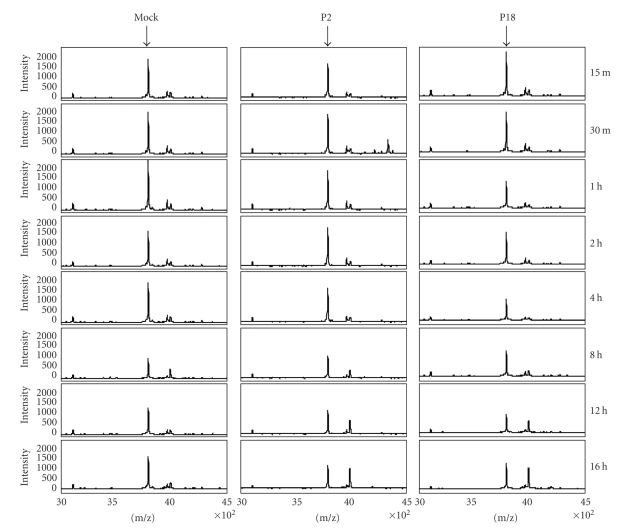
The pattern of prothymosin-α peak expression in a representative sample of cytoplasmic extracts is shown in Figure 2. Lower peak intensity following infection with P18 PICV can be seen at 1, 2, and 4 hours post-infection. However, prothymosin-α can shuttle between the cytoplasm and the nucleus where it functions as a histone H1 binding protein and is associated with cell growth [19–21]. Prothymosin-α and the peptide thymosin-α1, produced by proteolytic processing of prothymosin-α and an immune-modulator, can also be secreted [22–24]; for these reasons, the changes in peak intensities may not reveal the functional changes and effects in prothymosin-α activity and regulation.
Figure 2.
Differential intensity of prothymosin-α peak. SELDI mass spectrometry analysis of triplicate cytoplasmic extracts from mock-, P2-, or P18 PICV-infected P388D1 cells over a time course of infection shows differential intensity of a protein peak (indicated by the arrows) at m/z approximately 3790 (Q = 0.0164). This peak was identified as prothymosin-α by tandem mass spectrometry. The figure shows one representative set of spectra from three independent experiments.
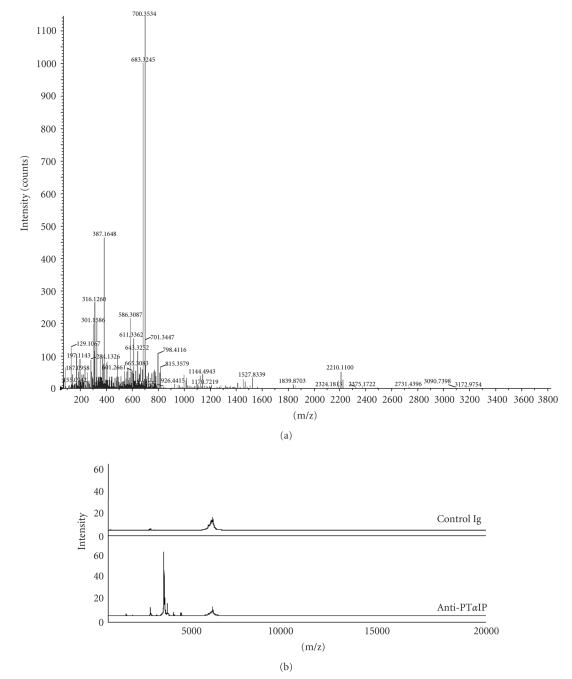
In order to verify the marker with molecular mass at 3787 Da, the cytoplasmic portion of the combined mock-, P2-, and P18-infected macrophages was fractionated on Q HyperD F beads with six fractions including the flow-through; each fraction was further captured on Q10 Protein Chip array surfaces. The marker protein was highly enriched in Q3 fraction on Q10 array surfaces (data not shown). The molecular mass of the marker is only 3787 Da; since the QSTAR XL instrument can analyze parent ion up to 5400 m/z, the peak could be identified as prothymosin-α without any further purification (Figure 3). The MS/MS spectrum is shown in Figure 4(a). We confirmed the identification of prothymosin-α using immunoprecipitation. Protein G agarose beads coupled to a monoclonal antibody against prothymosin alpha were used to immunoprecipitate prothymosin-α from extracts; the same protein peak at 3787 Da was identified (Figure 4(b)).
Figure 3.
Identification and validation of peak 3790 as prothymosin-α. Mass spectrum of proteins from fraction Q3 that bound to Q10 ProteinChip array and was analyzed on QSTAR XL instrument (mass range from 1000 to 4000 m/z).
Figure 4.
(a) The peptide with m/z approximately 3790 was identified as the fragment of prothymosin alpha by the following MS/MS microsequencing. (b) Mass spectrum of proteins pulled out from cytoplasmic fraction of murine macrophage samples by antiprothymosin alpha which was coupled to protein G agarose beads (mass range from 1000 to 20,000 m/z). Antiprothymosin alpha-coupled protein G agarose beads were incubated with cytoplasmic fraction and eluted proteins were analyzed on NP 20 ProteinChip array.
3.3. Verification of Protein Identification and (Pro)Thymosin-α Secretion
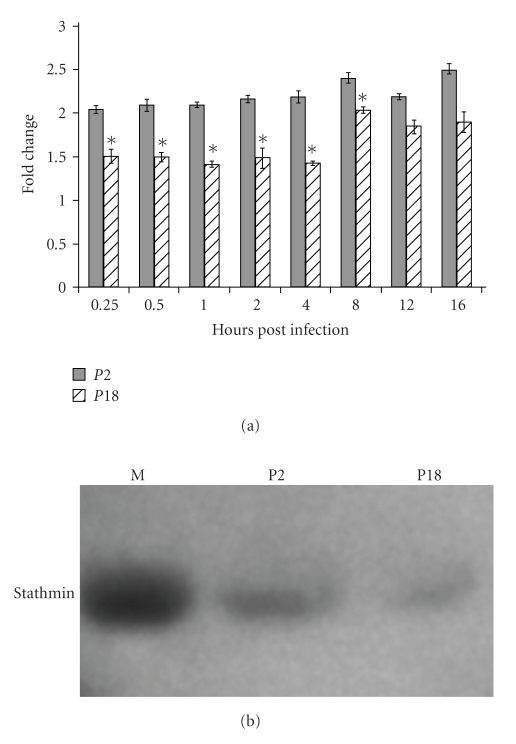
Prothymosin-α contains the secreted peptide thymosin-α1 [25–27]. We harvested supernatants from triplicate infected cultures over a 16-hour timecourse of infection and assayed the levels of (pro)thymosin-α in the supernatants (Figure 4(a)). As the thymosin peptide is contained within prothymosin-α, we are unable to distinguish between these peptides. Infection with the attenuated P2 variant of PICV induced consistently higher levels of (pro)thymosin-α in supernatants than mock infection or infection with the virulent P18 variant, ranging a 2–2.5-fold increase over baseline (mock-infected) levels (Figure 5(a)). Levels of secreted (pro)thymosin-α were significantly higher (P < .05) from 15 minutes to 8 hours postinfection. Consistent with many of the significant peaks in the SELDI analysis and many of our previous observations, the response induced by P18 infection more closely resembled mock-infection than infection with P2 PICV [3–5], although P18 did begin to induce higher levels of secreted (pro)thymosin-α than mock-infection at late times postinfection.
Figure 5.
(a) Induction of (pro)thymosin-α expression by PICV infection. P388D1 cells were infected with either P2 or P18 PICV in triplicate and tissue culture supernatants collected at various times post infection for determination of (pro)thymosin-α levels by ELISA. Fold increases are compared to levels from triplicate samples at time = 0 and show the results from three experiments. Error bars show the standard error of the mean, and asterisks indicate significant (P < .05) differences in levels induced between P2 and P18 infection as determined by Student's t-test. (b) Changes in stathmin expression following infection by P2 and P18 PICV. Triplicate cultures of P388D1 cells were infected with PICV and whole cell lysates assayed or stathmin levels by immunoblotting.
We infected triplicate cultures of P388D1 cells with P2 or P18 PICV or mock infected with PEG purification medium and prepared whole cell extracts at 24 hours post infection. Stathmin expression was assayed by immunoblot. Stathmin expression was significantly reduced in both P2 and P18 infected cells compared to mock, with P18 infection showing reduced stathmin expression compared to P2 across triplicate experiments (Figure 5(b)). Stathmin expression is controlled by a number of transcription factors, including a signaling module based around NF-κB, AP-1, c-Myc, and p53, which we have previously shown to be differentially regulated in PICV infection [4, 5], and which may suggest the molecular basis for differential stathmin expression.
4. Discussion
Many virus infections present with nonspecific flu-like symptoms complicating diagnosis; additionally, very few markers exist for determining the prognosis of infection. In particular cases, such as in the response to an epidemic or an intentional release, the ability to quickly triage patients on the basis of likely outcome could play a significant role in the management of what might be limited healthcare resources. The development of biomarkers that correlate with disease prognosis would be a valuable clinical tool. Additionally, certain biomarkers may reveal an underlying mechanism of pathogenesis. These can feed back into basic research to identify potential therapies.
Biological phenomena, such as the progression of viral infection studied here, are temporal in nature, and the resulting observations need to be analyzed with statistical techniques that can reveal temporal differential expression. EDGE analysis of our data uncovered 35 differentially expressing proteins as compared to 27 by 2-way ANOVA. Thus our results here indicate for proteomic data, as Storey et al. (2005) first demonstrated with genomic data, that dynamic methods such as EDGE are more effective at discovering candidate biomarkers that may be hidden to static methods. It is worth noting, however, that EDGE is a univariate method and it examines the data protein by protein (peak by peak, in the case of the present method) to evaluate differential expression. These studies demonstrate that SELDI mass spectrometry can be used to determine differences between attenuated and virulent virus infection, and that the development of characteristic diagnostic and prognostic “biomarker signatures” may be of use in managing these infections. In addition, we have shown that this approach can be used as a discovery tool to determine potential mechanisms of pathogenesis and identify targets for therapeutic intervention at the level of modulation of the host immune response.
In this model system, we have used two variants of a PICV which result in a mild infection, from which the animals recover, or a severe hemorrhagic disease. Further studies will use only the virulent virus variant and investigate the differential responses between animals that succumb to infection and those which recover. This study represents an important proof of principle that mass spectrometry methods can be used to differentiate between infections that result in severe disease and those that are self-limiting, suggesting that diagnostic and prognostic assays can be developed which function by detecting host responses to infection, rather than virus material directly. This may allow the development of assays which can identify the agent of infection a priori and which may be less prone to reduction of assay sensitivity by viral mutation.
We also identified differential secretion of a host immunomodulatory peptide between infection with attenuated and virulent virus variants although we could not discriminate between prothymosin-α and thymosin-α1 in this study. As infection with the attenuated virus leads to a mild-infection and viral clearance, it is likely that the correct immune signaling events are activated, leading to the development of protective immunity, events which may be suppressed in virulent infection. The use of immunomodulation and targeting of the host-responses to, and critical cellular-factors for, infection may be a viable therapeutic option in the treatment of infectious disease and may allow existing drugs to be repositioned [28–32]. Decreased thymosin-α1 secretion in virulent infection may contribute to the lack of development of the protective immune response, potentially through failure of proper induction of NF-κB, MAP kinase, and MyD88 pathways [33, 34]. Thymosin-α1 is already being tested as a therapeutic against HIV/AIDS, sepsis, hepatitis B and C viruses, and several cancers [24, 35–40]. Its characterized pharmacodynamics and kinetics may facilitate its development as a therapeutic for emerging infectious diseases.
Prothymosin-α can also function as a transcriptional regulator. Interestingly, prothymosin-α has been shown to interact with cAMP response element-binding (CREB) binding protein and can stimulate NF-κB and AP-1 driven transcription [41]. We have previously identified these transcription factors as being centrally involved in mediating host-responses to PICV infection [3–5]. NF-κB and AP-1 have also been implicated in mediating host-responses to other Lassa fever models and hemorrhagic fever infections such as Ebola and Rift Valley fever [42–45]. These pathways have also suggested further targets for immunomodulatory therapies [31]. Modulation of CREB binding protein function is the mechanism used by the A238L protein of the hemorrhagic DNA virus African swine fever virus, to modulate pro-inflammatory gene transcription. The protein translocates to the nucleus and alters CREB binding protein and p300 function, preventing recruitment to enhanceosomes and inhibiting expression of genes including TNF-α and COX-2 [46–49].
We have used SELDI mass-spectrometry to compare protein “signatures” following infection of guinea pigs with attenuated and virulent virus variants. This study demonstrates proof of principle that proteomic methods can distinguish between attenuated and virulent forms of infection in vivo. We have shown that, consistent with previous studies, infection with the virulent virus results in responses more similar to mock-infection, suggesting an inhibition of host responses. We identified a peptide-prothymosin-α-which contains the soluble immune mediator thymosin-α1 as being differentially expressed between infections and confirmed differential secretion. These studies shed further light on the mechanisms of hemorrhagic arenavirus pathogenesis, identify a potential strategy for therapy, and illustrate how proteomic approaches for biomarker discovery can generate additional hypotheses for further, targeted investigation.
Disclosure Statement
H. Wang and L. O. Lomas disclose employment by Vermillion Inc. during the period of performance of this study.
Acknowledgments
This research was supported by the Defense Advanced Research Projects Agency (P42296LS0000) and the National Institutes of Health (UO1 A1054827). The authors are grateful to Barry Elsom for technical assistance.
References
- 1.Cummins D. Arenaviral haemorrhagic fevers. Blood Reviews. 1991;5(3):129–137. doi: 10.1016/0268-960x(91)90029-c. [DOI] [PubMed] [Google Scholar]
- 2.Lukashevich IS, Maryankova R, Vladyko AS, et al. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells: different effects on IL-8 and TNF-α gene expression. Journal of Medical Virology. 1999;59(4):552–560. [PMC free article] [PubMed] [Google Scholar]
- 3.Fennewald SM, Aronson JF, Zhang L, Herzog NK. Alterations in NF-κ B and RBP-J κ by arenavirus infection of macrophages in vitro and in vivo. Journal of Virology. 2002;76(3):1154–1162. doi: 10.1128/JVI.76.3.1154-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowick GC, Fennewald SM, Elsom BL, et al. Differential signaling networks induced by mild and lethal hemorrhagic fever virus infections. Journal of Virology. 2006;80(20):10248–10252. doi: 10.1128/JVI.01384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowick GC, Fennewald SM, Scott EP, et al. Identification of differentially activated cell-signaling networks associated with pichinde virus pathogenesis by using systems kinomics. Journal of Virology. 2007;81(4):1923–1933. doi: 10.1128/JVI.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowick GC, Spratt HM, Hogg AE, et al. Analysis of the differential host cell nuclear proteome induced by attenuated and virulent hemorrhagic arenavirus infection. Journal of Virology. 2009;83(2):687–700. doi: 10.1128/JVI.01281-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCormick JB, King IJ, Webb PA, et al. Lassa fever: effective therapy with ribavirin. The New England Journal of Medicine. 1986;314(1):20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- 8.Johnson KM, McCormick JB, Webb PA, Smith ES, Elliott LH, King IJ. Clinical virology of Lassa fever in hospitalized patients. Journal of Infectious Diseases. 1987;155(3):456–464. doi: 10.1093/infdis/155.3.456. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Yang X, Bowick GC, et al. Identification of proteins bound to a thioaptamer probe on a proteomics array. Biochemical and Biophysical Research Communications. 2006;347(3):586–593. doi: 10.1016/j.bbrc.2006.06.132. [DOI] [PubMed] [Google Scholar]
- 10.Lan S, Schelde LM, Wang J, Kumar N, Ly H, Liang Y. Development of infectious clones for virulent and avirulent pichinde viruses: a model virus to study arenavirus-induced hemorrhagic fevers. Journal of Virology. 2009;83(13):6357–6362. doi: 10.1128/JVI.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan S, McLay L, Aronson J, Ly H, Liang Y. Genome comparison of virulent and avirulent strains of the Pichinde arenavirus. Archives of Virology. 2008;153(7):1241–1250. doi: 10.1007/s00705-008-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson JF, Herzog NK, Jerrells TR. Pathological and virological features of arenavirus disease in guinea-pigs—comparison of 2 pichinde virus-strains. American Journal of Pathology. 1994;145(1):228–235. [PMC free article] [PubMed] [Google Scholar]
- 13.Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot M-C, Deubel V. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. Journal of Immunology. 2004;172(5):2861–2869. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 14.Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. BioTechniques. 1995;19(2):192–195. [PubMed] [Google Scholar]
- 15.Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8(2):414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- 16.Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(36):12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 18.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papamarcaki T, Tsolas O. Prothymosin α binds to histone H1 in vitro. FEBS Letters. 1994;345(1):71–75. doi: 10.1016/0014-5793(94)00439-0. [DOI] [PubMed] [Google Scholar]
- 20.Karetsou Z, Sandaltzopoulos R, Frangou-Lazaridis M, et al. Prothymosin α modulates the interaction of histone H1 with chromatin. Nucleic Acids Research. 1998;26(13):3111–3118. doi: 10.1093/nar/26.13.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eschenfeldt WH, Berger SL. The human prothymosin α gene is polymorphic and induced upon growth stimulation: evidence using a cloned cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(24):9404–9407. doi: 10.1073/pnas.83.24.9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco FJ, Diaz C, Barcia M, et al. Synthesis and apparent secretion of prothymosin α by different subpopulations of calf and rat thymocytes. Immunology. 1989;67(2):263–268. [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein AL, Cohen GH, Thurman GB, Hooper JA, Rossio JL. Regulation of immune balance by thymosin: potential role in the development of suppressor T-cells. Advances in Experimental Medicine and Biology. 1976;66:221–228. doi: 10.1007/978-1-4613-4355-4_34. [DOI] [PubMed] [Google Scholar]
- 24.Romani L, Bistoni F, Montagnoli C, et al. Thymosin α1: an endogenous regulator of inflammation, immunity, and tolerance. Annals of the New York Academy of Sciences. 2007;1112:326–338. doi: 10.1196/annals.1415.002. [DOI] [PubMed] [Google Scholar]
- 25.Haritos AA, Tsolas O, Horecker BL. Distribution of prothymosin α in rat tissues. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(5):1391–1393. doi: 10.1073/pnas.81.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haritos AA, Goodall GJ, Horecker BL. Prothymosin α: isolation and properties of the major immunoreactive form of thymosin α1 in rat thymus. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(4):1008–1011. doi: 10.1073/pnas.81.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freire M, Rey-Mendez M, Gomez-Marquez J, Arias P. Evidence for the synthesis of thymosin α1 by calf thymocytes and for the production of this peptide by natural processing. Archives of Biochemistry and Biophysics. 1985;239(2):480–485. doi: 10.1016/0003-9861(85)90715-5. [DOI] [PubMed] [Google Scholar]
- 28.Lu M, Menne S, Yang D, Yu Y, Roggendorf M. Immunomodulation as an option for the treatment of chronic hepatitis B virus infection: preclinical studies in the woodchuck model. Expert Opinion on Investigational Drugs. 2007;16(6):787–801. doi: 10.1517/13543784.16.6.787. [DOI] [PubMed] [Google Scholar]
- 29.Vela EM, Bowick GC, Herzog NK, Aronson JF. Exploring kinase inhibitors as therapies for human arenavirus infections. Future Virology. 2008;3(3):243–251. doi: 10.2217/17460794.3.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vela EM, Bowick GC, Herzog NK, Aronson JF. Genistein treatment of cells inhibits arenavirus infection. Antiviral Research. 2008;77(2):153–156. doi: 10.1016/j.antiviral.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fennewald SM, Scott EP, Zhang L, et al. Thioaptamer decoy targeting of AP-1 proteins influences cytokine expression and the outcome of arenavirus infections. Journal of General Virology. 2007;88(3):981–990. doi: 10.1099/vir.0.82499-0. [DOI] [PubMed] [Google Scholar]
- 32.Bowick GC, Fennewald SM, Zhang L, et al. Attenuated and lethal variants of pichinde virus induce differential patterns of NF-κB activation suggesting a potential target for novel therapeutics. Viral Immunology. 2009;22(6):457–462. doi: 10.1089/vim.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozza S, Gaziano R, Bonifazi P, et al. Thymosin α1 activates the TLR9/MyD88/IRF7-dependent murine cytomegalovirus sensing for induction of anti-viral responses in vivo. International Immunology. 2007;19(11):1261–1270. doi: 10.1093/intimm/dxm097. [DOI] [PubMed] [Google Scholar]
- 34.Peng X, Zhang P, Wang X, et al. Signaling pathways leading to the activation of IKK and MAPK by thymosin α1. Annals of the New York Academy of Sciences. 2007;1112:339–350. doi: 10.1196/annals.1415.025. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, He M-Y, Li Y-M. Treatment of patients with severe sepsis using Ulinastatin and Thymosin α1: a prospective, randomized, controlled pilot study. Chinese Medical Journal. 2009;122(8):883–888. [PubMed] [Google Scholar]
- 36.Zhang Y, Chen H, Li Y-M, et al. Thymosin α1- and ulinastatin-based immunomodulatory strategy for sepsis arising from intra-abdominal infection due to carbapenem-resistant bacteria. Journal of Infectious Diseases. 2008;198(5):723–730. doi: 10.1086/590500. [DOI] [PubMed] [Google Scholar]
- 37.Camerini R, Ciancio A, De Rosa A, Rizzetto M. Studies of therapy with thymosin α1 in combination with pegylated interferon α2a and ribavirin in nonresponder patients with chronic hepatitis C. Annals of the New York Academy of Sciences. 2007;1112:368–374. doi: 10.1196/annals.1415.047. [DOI] [PubMed] [Google Scholar]
- 38.Moody TW. Thymosin α1 as a chemopreventive agent in lung and breast cancer. Annals of the New York Academy of Sciences. 2007;1112:297–304. doi: 10.1196/annals.1415.040. [DOI] [PubMed] [Google Scholar]
- 39.Shuqun C, Mengchao W, Han C, et al. Antiviral therapy using lamivudine and thymosin α1 for hepatocellular carcinoma coexisting with chronic hepatitis B infection. Hepato-Gastroenterology. 2006;53(68):249–252. [PubMed] [Google Scholar]
- 40.Chadwick D, Pido-Lopez J, Pires A, et al. A pilot study of the safety and efficacy of thymosin α1 in augmenting immune reconstitution in HIV-infected patients with low CD4 counts taking highly active antiretroviral therapy. Clinical and Experimental Immunology. 2003;134(3):477–481. doi: 10.1111/j.1365-2249.2003.02331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karetsou Z, Kretsovali A, Murphy C, Tsolas O, Papamarcaki T. Prothymosin α interacts with the CREB-binding protein and potentiates transcription. EMBO Reports. 2002;3(4):361–366. doi: 10.1093/embo-reports/kvf071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Djavani M, Crasta OR, Zhang Y, et al. Gene expression in primate liver during viral hemorrhagic fever. Virology Journal. 2009;6, article 20 doi: 10.1186/1743-422X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djavani MM, Crasta OR, Zapata JC, et al. Early blood profiles of virus infection in a monkey model for Lassa fever. Journal of Virology. 2007;81(15):7960–7973. doi: 10.1128/JVI.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez O, Valmas C, Basler CF. Ebola virus-like particle-induced activation of NF-κB and Erk signaling in human dendritic cells requires the glycoprotein mucin domain. Virology. 2007;364(2):342–354. doi: 10.1016/j.virol.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Billecocq A, Spiegel M, Vialat P, et al. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. Journal of Virology. 2004;78(18):9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Granja AG, Nogal ML, Hurtado C, et al. The viral protein A238L inhibits TNF-α expression through a CBP/p300 transcriptional coactivators pathway. Journal of Immunology. 2006;176(1):451–462. doi: 10.4049/jimmunol.176.1.451. [DOI] [PubMed] [Google Scholar]
- 47.Granja AG, Sanchez EG, Sabina P, Fresno M, Revilla Y. African swine fever virus blocks the host cell antiviral inflammatory response through a direct inhibition of PKC-θ-mediated p300 transactivation. Journal of Virology. 2009;83(2):969–980. doi: 10.1128/JVI.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Granja AG, Perkins ND, Revilla Y. A238L inhibits NF-ATc2, NF-κB, and c-Jun activation through a novel mechanism involving protein kinase C-θ-mediated up-regulation of the amino-terminal transactivation domain of p300. Journal of Immunology. 2008;180(4):2429–2442. doi: 10.4049/jimmunol.180.4.2429. [DOI] [PubMed] [Google Scholar]
- 49.Silk RN, Bowick GC, Abrams CC, Dixon LK. African swine fever virus A238L inhibitor of NF-κB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. Journal of General Virology. 2007;88, part 2:411–419. doi: 10.1099/vir.0.82358-0. [DOI] [PubMed] [Google Scholar]