Abstract
Arenaviridae comprises 23 recognized virus species with a bipartite ssRNA genome and an ambisense coding strategy. The virions are enveloped and include nonequimolar amounts of each genomic RNA species, designated L and S, coding for four ORFs (N, GPC, L, and Z). The arenavirus Junín (JUNV) is the etiological agent of Argentine Hemorrhagic Fever, an acute disease with high mortality rate. It has been proposed that Z is the functional counterpart of the matrix proteins found in other negative-stranded enveloped RNA viruses. Here we report the optimized expression of a synthetic gene of Z protein, using three expression systems (two bacterial and a baculoviral one). One of these recombinant proteins was used to generate antibodies. A bioinformatic analysis was made where Z was subdivided into three domains. The data presented contributes methodologies for Z recombinant production and provides the basis for the development of new experiments to test its function.
1. Introduction
Junín virus (JUNV) is a South American arenavirus, the etiological agent of a severe endemo-epidemic disease called Argentine Hemorrhagic Fever (AHF).
Arenaviridae is a family composed of a growing number of enveloped viruses with a bipartite single stranded RNA genome. Members of the Arenaviridae family were subdivided into two groups based on the geographical site of isolation, serological cross-reactivity and genetic data. The prototype of the family, lymphocytic choriomeningitis virus (LCMV) is a member of the Old World arenavirus group, which also includes Ippy (IPPV), Lassa (LASV), Mobala (MOBV), and Mopeia (MOPV) viruses. JUNV is a member of the New World arenavirus group, that also includes Allpahuayo (ALLV), Amapari (AMAV), Bear Canyon (BCNV), Flexal (FLXV), Guanarito (GTOV), Latino (LATV), Machupo (MACV), Oliveros (OLVV), Paraná (PARV), Pichindé (PICV), Pirital (PIRV), Sabiá (SABV), Tacaribe (TCRV), Tamiami (TAMV), and Whitewater Arroyo (WWAVs) viruses [1]. In addition, there are several recently described species that have not yet been classified by the ICTV (http://www.ictvonline.org/virusTaxonomy.asp?version=2008).
The two RNA segments of the Junín virus genome are designated L and S and have approximate sizes of 7.2 and 3.5 kb, respectively. Each RNA segment directs the synthesis of two proteins; their open reading frames are arranged in opposite orientations (ambisense coding strategy) [2] and are separated by a noncoding intergenic region that folds into a stable stem-loop structure [3]. The S RNA codes for the major structural proteins of the virion: the precursor of the envelope glycoproteins (GPC) and the viral nucleocapsid protein (N). Posttranslational cleavage of GPC renders a signal peptide and the two viral glycoproteins (GP1 and GP2). The L RNA segment codes for the viral RNA-dependent RNA polymerase (L) and the small protein (Z). Z is a 94 residue long polypeptide, and its central portion is predicted to fold into a RING finger domain. Moreover, Z has been implicated in several aspects of arenavirus biology [4–6].
Based on nucleotide sequence data, a better comprehension of the taxonomy and evolution of the Arenaviridae has been achieved. It has been suggested that New World arenaviruses should be classified into four different lineages, named A, B, C, and Rec [7]. Lineage A would contain ALLV, FLXV, PARV, PICV and PIRV; lineage B contains AMAV, GTOV, JUNV, MACV, SABV, and TCRV, lineage C contains LATV and OLVV, and lineage Rec, with the WWAV, TAMV, and BCNV. Members of this last clade are thought to have a recombinant S RNA. Currently (ICTV website) the last three viruses are classified with Clade A.
The role of Z in the virus life cycle is not completely elucidated, and homologues of Z are not found in other ambisense or negative-stranded RNA viruses. Z is a structural component of the virion [8], and by means of in vivo and in vitro experiments, the interaction of Z with several cellular factors has been reported, including the promyelocytic leukemia protein and the eukaryotic translation initiation factor 4E [9, 10]. Because of this latter interaction, it was proposed that Z inhibits Cap-mediated translation [11, 12]. Other researchers suggested that Z could be a transcriptional regulator of the viral cycle [13] or even an inhibitor of viral replication [14]. Furthermore, Pérez and coworkers [15] proposed, for LCMV and LASV, that Z is the functional counterpart of the matrix proteins found in other negative-stranded enveloped RNA viruses. Late domains (LDs), found in matrix proteins from negative-stranded RNA viruses and in Gag protein from retroviruses, have an essential role in the viral budding process [16]. Three types of motifs have been defined within viral LDs: P[TS]AP, PPxY, and YxxL [17], where ‘‘x” is any amino acid. Later, Martín Serrano and coworkers [18] redefined the last as: YPxL/LxxLF. LDs are highly conserved and have been shown to mediate interaction with host cell proteins, in particular with members of the vacuolar protein-sorting pathway [19, 20]. For instance, the PTAP motif from Ebola virus VP40 matrix protein and from HIV Gag protein interacts with Tsg101, a member of the vacuolar protein-sorting pathway.
In this work, we show the strategies employed for the expression, purification, and specific antibody generation against Z protein from Candid#1 strain of Junín virus [21]. Here we report the optimized expression from a synthetic gene of Z protein tagged with different peptides, using three expression systems (two bacterial and a baculoviral one), in order to obtain recombinant Z protein suitable for functional characterization studies.
2. Materials and Methods
2.1. Virus and Cell Culture
The passage history of JUNV Candid#1 strain was described previously [21]. The parental Junín virus XJ strain was isolated in Junín city (Buenos Aires, Argentina) from a human AHF patient [22]. The BHK21 cells were cultured in growth medium (Dulbecco's minimal essential medium—DMEM—supplemented with 10% fetal bovine serum and 2 mM L-glutamine) to 50% confluence and infected at a multiplicity of infection (moi) of 1 plaque forming unit (pfu) per cell. After virus adsorption for 1 h at room temperature, infected cells were washed with phosphate-buffered saline (PBS) and maintained at 37°C in MEM containing 2% fetal bovine serum. Virions were recovered and purified from the supernatant media; viral and total infected cell RNAs were isolated according to procedures described previously [3]. Virus titers were determined by plaque assay on Vero E6 (ATCC, CCL 1586, C1008) cell monolayers as described elsewhere [3]. The cell monolayers were harvested 72 h post infection by scraping and recovered by centrifugation.
2.2. RNA Isolation, Amplification, and Cloning
RNA was isolated from pelleted virions or infected cells, using the QIAmp Viral RNA Mini Kit or RNeasy Mini Kit (Qiagen, Valencia, CA). The Z open reading frame (ORF) from JUNV Candid#1 strain, was amplified by RT-PCR (SuperScript III Reverse Transcriptase for RT, and Platinum Taq DNA Polymerase High Fidelity for PCR, Invitrogen), from Junín virus RNA using the forward primer JZv 5′-ATGGGCAACTGCAACGGGGCATC-3′ (Z ORF translation initiation sequence is underlined), and the reverse primers JZvc 5′- TGGTGGTGGTGCTGTTGGCTCCAC -3′, and JZvc-Stop: 5′-CTATGGTGGTGGTGCTGTTGGCTCCAC -3′ (sequence complementary to the Z stop codon is underlined). The amplicon was cloned into pZErO and the recombinant plasmid (ZpZ) was amplified in Escherichia coli strain TOP10 (Invitrogen). The recombinant plasmids were confirmed by nucleotide sequencing.
For cloning Z ORF into pET102/D-TOPO (Invitrogen), a variant from JZv primer was used, named JZvClamp: 5′-CCAC ATGGGCAACTGCAACGGGGCATC-3′ (the four nucleotides, added to facilitate cloning, upstream of the ATG initiation codon, are indicated in italics). The amplicon obtained with JZvC and JZvClamp was cloned following the manufacturer's instructions. The recombinant clone was named pET-Z and used for expression experiments in bacterial cell culture (E. coli BL21). The produced protein was fused to different tags: Thioredoxine by the N-terminus, and His Tag and V5 epitopes by the C-terminus. This fusion protein was called Tio-Z-V5-His.
For the subsequent cloning procedures the plasmid ZpZ was digested with XhoI and HindIII. This digestion rendered a product of 366 bp, comprising Z ORF and a portion of the alpha fragment of lacZ (from pZErO MCS), hereafter designated as z′. This fragment was cloned into different expression vectors:
pGZ (pGEX-B, Novagen). The z′ fragment was ligated into pQE-30 (Qiagen, Valencia, CA) previously digested with HindIII and SalI. The obtained plasmid, pQZ was digested with SmaI and HindIII and the 374 pb fragment was ligated into pGEX-B, previously digested with the same enzymes. The recombinant plasmid (pGZ) was amplified in E. coli DH5α (Invitrogen) and confirmed by nucleotide sequencing. pGZ was used for expression experiments in bacterial cell culture. The recombinant protein, called GST-Z, contains Glutathione-S-transferase (GST) fused to the N-terminus of Z.
pBAC-Z (pFastBac-HTa, Invitrogen). The z′ fragment was ligated into the vector pFastBac-HTa, previously digested with HindIII and SalI. Z ORF was fused to a His-Tag to the amino end of Z. This His-Tagged version of Z was named Z-His. The recombinant plasmid (pBAC-Z) was amplified in E. coli TOP10 and the insert was confirmed by nucleotide sequencing. The Bac-to-Bac system (Invitrogen) was used to generate recombinant baculoviral genomes (AcMNPV) harboring Z-His gene (Bacmid_Ac-Z). Briefly, Z ORF in pBAC-Z was transposed to an AcMNPV genome using E. coli strain DH10 BAC, which carries the target baculoviral genome and the machinery required for transposition. The recombinant AcMNPV genomes (Bacmid_Ac-Z) were purified by minipreparation from bacterial culture and then used to generate recombinant baculoviruses (v_Ac-Z) by transfecting different insect cell lines (Cellfectin, Invitrogen).
2.3. Protein Expression in Insect Cell Lines
The recombinant baculovirus (v_Ac-Z) was amplified by infection of insect cells. Viruses were recovered from the supernatant media of infected cell monolayers and purified by ultracentrifugation. Virus titers were determined by plaque assay. Optimization procedures of the expression levels consisted in testing different multiplicities of infection and insect cell lines. Two cell lines were used, SF9 from Spodoptera frugiperda [23] and High Five (BTI-TN-5B1-4) from Trichoplusia ni [24]. These cells were cultured on 25 cm2 polystyrene flasks with Grace medium (Invitrogen), supplemented with 10% fetal bovine serum and 0.1% gentamicin, to 70% confluence and infected with the recombinant baculovirus (>107 pfu/ml) at different multiplicities of infection (moi). After virus adsorption for 1 h at room temperature, infected cells were washed with phosphate-buffered saline (PBS) and maintained at 25°C in Grace containing 10% fetal bovine serum. The protein expression was analyzed by SDS-PAGE and Western blotting using Z protein-specific antibodies and His-Tag antibodies.
2.4. Protein Expression in Bacterial Cells
A variety of expression strains were tested: Rosetta pLys and Origami B (DE3) from Novagen; BL21 (DE3), BL21 (pLysS), BL21-CodonPlus (DE3) RIPL and XL1Blue from Stratagene; BL21 SI from Invitrogen; OverExpress C41 (DE3) and C43 (DE3) from Lucigen. Cells were grown on Luria broth (LB) growth medium [25] and supplemented with the proper antibiotic when required (34 μg/ml kanamycin, 100 μg/ml ampicillin, 50 μg/ml chloramphenicol, 12.5 μg/ml tetracycline).
Tio-Z-V5-His (pT-Z). For Tio-Z-V5-His fusion protein, the best expression level was observed with BL21 (DE3) strain. Transformed cells were cultured in 500 ml of LB medium containing 100 μg/ml ampicillin, at 37°C. When the culture absorbance at 600 nm reached a value of 1.5, cells were induced with 600 μM isopropyl-β-D-thiogalactopyranoside (IPTG) for 4 h at 37°C. Bacteria were then harvested by centrifugation at 4000 × g for 15 min, and resuspended in 25 ml of 50 mM NaH2PO4 pH 8.2; 300 mM NaCl; 20 mM Imidazole; 1% Triton X-100 and complete Protease Inhibitor Cocktail Tablets 1X (Roche). Cells were disrupted by French Press and, after centrifugation at 20,000 × g for 30 min, the soluble fraction was collected and analyzed by SDS-PAGE (4X Loading sample buffer containing 200 mM Tris-HCl, 8% SDS, 40% glycerol, and 0.4% Bromophenol Blue and 400 mM DTT, pH 6.8). SDS-PAGE analyses were performed according to Laemmli [26], using 12–15% polyacrylamide gels stained with R250 Coomassie blue. The running buffer was Tris-Glycine [27].
GST-Z (pGEX-Z). Very similar expression levels were obtained from the different host cells studied for the GST-Z fusion protein, as detected by Tris-Glycine 15% SDS-PAGE. The selected strain and culture conditions were the same as employed by Volpon and coworkers [28], with buffers containing Zn+2. Bacteria were harvested by centrifugation at 4,000 × g for 15 min, and the pellet was resuspended in 25 ml of PBS, 200 mM NaCl, pH 8; 0.1% Triton X-100, 100 μM DTT, and complete Protease Inhibitor Cocktail Tablets 1X (Roche). Cells were disrupted by French Press and, after centrifugation at 20,000 × g for 30 min, the soluble fraction was collected and analyzed by SDS-PAGE. Analyses were performed according to Laemmli [26], using 12–15% polyacrylamide gels stained with R250 Coomassie blue. The running buffer was Tris-Glycine.
The protein quantification was made using Bradford method and by densitometry.
2.5. Purification of Z from Bacteria
Tio-Z-V5-His —
The soluble fraction (25 ml) obtained after the induction of pET-Z was clarified by centrifugation at 12,000 × g for 30 min and loaded on a His-Trap 5 ml column (Amersham), using a running buffer 50 mM NaH2PO4 pH 8.2, 300 mM NaCl. After column wash, a step gradient of 500 mM Imidazole was applied at 1 ml/min flow for 20 min using an AKTA FPLC (Amersham). Each fraction collected was analyzed by SDS-PAGE and those containing Tio-Z-V5-His were pooled, and concentrated down to 1 ml on a spin concentrator (Vivaspin 15, 10,000 MWCO, Sartorius). This was then run on a gel filtration column Superdex 200 10/300 GL (Amersham) using running buffer 50 mM NaH2PO4 pH 8.2, 300 mM NaCl at 0.5 ml/min flow. Again, every fraction was analyzed by SDS-PAGE and those containing Tio-Z-V5-His were pooled and dialyzed overnight against Tris-HCl 20 mM pH 8.0 at 4°C.
Cleavage and Purification of Tio-Z-V5-His —
Purified Tio-Z-V5-His was dialyzed against enterokinase cleavage buffer (50 mM Tris-HCl pH 8; 1 mM CaCl2; 0.1% Tween-20) and then concentrated with a spin concentrator (Vivaspin 6 10,000 MWCO, Sartorius) down to 1 ml at a final concentration of 0.3 mg/ml. A fraction of 500 μl of this sample was subjected to proteolysis with enterokinase on a final volume of 600 μl overnight at 4°C. After this, the sample was loaded on a His-Trap 5 ml column (Amersham). The running buffer was 50 mM NaH2PO4 pH 8.2; 300 mM NaCl. Finally, a 500 mM Imidazole step gradient was applied for 20 min. Every fraction was analyzed by SDS-PAGE.
GST-Z —
The soluble fraction (25 ml), obtained after the induction of pGZ and cell disruption by Cell Press, was clarified by centrifugation at 12,000 × g for 30 min, loaded into 1 ml of Glutathione Sepharose 4B (GE Healthcare), and incubated at 4°C overnight. After five washes with 50 ml of PBS, the sample was eluted with 2 ml of 25 mM reduced glutathione (Sigma) in 50 mM Tris-HCl pH 9.5. The eluted fraction was dialyzed against phosphate buffer 20 mM Na2HPO4 pH 7.2 at 4°C overnight, concentrated down to 1 ml (Vivaspin 2, 10,000 MWCO, Sartorious), and loaded into a Superdex 200 column (Amersham) using the same buffer conditions at 0.5 ml/min flow.
Cleavage and Purification of GST-Z —
GST-Z was dialyzed against Factor Xa cleavage buffer (50 mM Tris-HCl pH 7.5; 150 mM NaCl; 1 mM CaCl2) and 600 μl of this sample were subjected to proteolysis with Factor Xa protease at room temperature overnight. The reaction product was incubated with glutathione sepharose for 1 h at room temperature to retain the cleaved GST fusion peptide, while Z protein was recovered in the supernatant. Finally the bound GST protein was eluted with 25 mM reduced glutathione in 50 mM Tris-HCl pH 9.5. The reaction products were analyzed by SDS-PAGE and Western blotting with a polyclonal antiserum specific for the Z protein.
2.6. Gel Filtration
Columns packed with 24 ml bead volumes of Superdex 200 GL and Superdex 75 GL were used to obtain the target protein in its monomeric form. Columns were loaded with 1 ml of sample and run at 0.5 ml/min flow using an AKTA FPLC (Amersham). For Tio-Z-V5-His recombinant protein, the running buffer was 50 mM NaH2PO4, 300 mM NaCl pH 8. For the GST-Z recombinant protein it was 20 mM Na2HPO4 pH 7.2.
Before each protein separation, a molecular weight marker (Bio-Rad) was run for calibration purposes using the corresponding running buffer.
2.7. Limited Proteolysis
A serial dilution 1/10 to 1/10,000 from a stock of 0.1 mg/ml of chymotrypsin (Roche) and trypsin (Roche) were incubated with a 0.5 μg/μl of GST-Z recombinant protein, for 2 h at room temperature. The reaction was stopped by adding SDS-PAGE sample buffer or by incubating the reaction at −20°C.
2.8. Sequencing of the N-Terminus
The samples subjected to limited proteolysis with chymotrypsin were loaded on a 15% SDS-PAGE and blotted to a PVC membrane (Amersham). The N-terminal sequence of relevant peptides was then obtained using an Applied Biosystems Sequencer ABI494.
2.9. Production of Z Polyclonal Antibody
Polyclonal antibodies were produced in rabbits according to standard procedures. Briefly, the purified Tio-Z-V5-His fusion protein was used to inoculate two female New Zealand white rabbits with an average weight of 2.5 kg. Before each inoculation, the animals were bled from the marginal ear vein for evaluation of antibody titers. For the first injection, 50 μg of purified recombinant protein/kg of body weight were emulsified in an equal volume of complete Freund's adjuvant (Sigma Chemicals Co) and administered subcutaneously. After 25 days, the animals were bled as described above and inoculated with 50 μg of purified recombinant protein/kg of body weight emulsified in an equal volume of incomplete Freund's adjuvant (Sigma Chemicals Co). This procedure was repeated twice with a time interval of 15 days and a last bleeding was carried out by cardiac puncture [29]. The antibody titer was estimated by inhouse EIA as described in Argüelles et al. [30]. Tio-Z-V5-His fusion protein was used as antigen in different assays, with the exception of the cutoff determination, where GST-Z was used as antigen against the heterologous sera. Purification of G type immunoglobulin (IgG) fractions from antiserum was carried out by affinity chromatography on Protein G-Sepharose fast flow (Pharmacia) according to the manufacturer's recommendations. IgG yield, determined by absorbance at 280 nm, was 3.2 mg/ml serum.
For inhouse EIA, microtiter plates (Nunc, Roskilde, Denmark) were coated overnight at 4°C with Tio-Z-V5-His fusion protein diluted to 10 μg/ml in 0.1 M bicarbonate buffer (pH 9.6) as antigen. After this, and after each of the following steps, the plates were washed three times with washing solution, that is, phosphate-buffered saline (PBS), 0.5 M NaCl, and 0.2% (v/v) Triton X-10.
Fifty microliters of each serum sample diluted 1/300 in sample buffer (i.e., 1% wt/vol bovine serum albumin in washing solution) were added to the antigen-coated wells, and the plates were incubated at 37°C for 1 h. Peroxidase-conjugated antirabbit IgG (Santa Cruz Biotechnology) diluted 1/16,000 in sample buffer was added at 50 μl/well and incubated further for 1 h at 37°C. Bound antibodies and conjugates were then developed with orthophenylenediamine (Sigma Chemical Co), 30% H2O2,and citrate buffer pH 5.0 at a ratio of 1 mg/μl/ml according to standard procedures.
The optical density (OD) was measured at a wavelength of 490 nm (OD490) (Max Line TM enzyme-linked immunosorbent assay reader; Molecular Devices, Sunnyvale, CA). Each serum was simultaneously tested with corresponding control antigens, that is, preimmune serum. The results were expressed as the value of the OD of the control antigen subtracted from the OD of the fusion protein antigen for each serum sample. The enzyme-linked immunosorbent assay cutoff value was estimated as the mean OD obtained with 20 certified negative specimens plus 3 standard deviations. These negative specimens were heterologous sera, diluted 1/300, and the antigen used was the GST-Z fusion protein. The peroxidase-conjugated anti-rabbit IgG was diluted 1/16,000 [30].
2.10. Immunoblotting
The extracts of bacterial cells and insect cell lines infected with recombinant baculovirus were separated by SDS-PAGE (12–15% polyacrylamide gel) and blotted onto nitrocellulose membranes (Hybond P+, Amersham Pharmacia) in Tris-glycine buffer containing 20% (v/v) methanol. To avoid nonspecific binding of the antibodies, the membranes were blocked by incubation with 5% wt/vol skimmed powder milk in PBS for a minimum of 2 h at 37°C. For the primary antibody incubation the blocked membrane was probed with a 1/1,000 dilution of the polyclonal antiserum specific for the Z protein on PBS 2% casein 0.1% Tween-20 at 37°C for 1 h, followed by an incubation with horseradish-peroxidase conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology), diluted 1/10,000 on PBS 0.1% Tween-20 at 37°C for 1 h. After each step the membrane was washed three times with PBS 0.1% Tween-20, 5 minutes each time.
2.11. Proteinase K Protection Assay
Insect cell suspensions were clarified by centrifugation. Supernatants were then ultracentrifuged in order to pellet down small particles, and these fractions were treated with Proteinase K (0.1 mg/ml, 37°C, 30 min), or a combination of Proteinase K plus 1% Triton X-100 in the same conditions. Proteolysis was stopped by adding 100 mM PMSF and boiling the samples for 10 min. Samples were then analyzed by SDS-PAGE followed by immunoblotting.
2.12. Sequences and Bioinformatic Tools
Sequences —
Amino acid sequences of the following arenavirus were obtained from the Genbank database (accession numbers are indicated between brackets): AMAV-BeAn70563 (ABY59841.1), CPXV-BeAn119303 (ABY59842.1), GTOV-CVH-961104 (AAT77691.1), GTOV-VHF-3990 (AAT77689.1), JUNV-MC2 (ABY59838.1), JUNV-Candid#1 (AAU34182.1), MACV-MARU-222688 (AAY27821.1), MACV-9530537 (AAY27823.1), TACV (NP_694847.1), LATV-MARU-10924 (AAY27824.1), OLVV-3229 (ABY59840.1), PIRV-VAV488 (ABY59836.1), PIRV-1743 (AAT77682.1), ALLV-CLHP2472 (ABY59833.1), PICV-AN3739 (YP_138535.1), BCNV-A0060209 (ABY59834.1), BCNV-AVA0070039 (AAX99343.1), WWAV-AV9310135 (AAX99351.1), TAMV-W10777 (AAX99348.1), LCMV-Clone#13 (ABC96003.1), LCMV-MX (CAA10342.1), LASV-CSF (AAO59514.1), LASV-AV (AAO59508.1), IPPY-DakAnB188 (YP_516232.1), MOBV-Acar3080 (YP_516228.1), MOPV-AN20410 (YP_170707.1), MOPV-Mozambique (ABC71136.1), Lujo (YP_002929492), and Morogoro (ACJ24975.1).
Alignments —
Sequence alignments were done using the CLUSTAL X program [31, 32].
Sequence Logos —
Determination of Sequence Logo was made with the server available at http://weblogo.berkeley.edu site [33]. Synthetically, the Sequence Logo is a graphic representation of a multiple alignment of sequences [34]. Each logo is constructed from characters stacking in each sequence alignment position. The stacking height indicates the local information content in this position, while the height of each symbol inside the stacking is the fraction of information content representing the frequency of each residue in that position. Because of the characteristics of the software, and to avoid bias towards particular species that are found over-represented in the sequence data bank, at most two strains for each viral species were selected. Moreover, to get a more accurate representation, gap positions within the sequences in the alignment were considered as another character. The usual behavior of the software does not take into account the gaps, calculating the values relative to the number of sequences with an amino acid in that position. All the motif descriptions were annotated according to the syntax rules of Prosite [35], and represent more than 60% of all Z protein sequences.
Biochemical Properties of Different Recombinant Proteins —
The different biochemical parameters were calculated from the amino acid sequence data using the ProtParam tool found at the ExPASy Proteomics Server (www.expasy.ch, Proteomics Tools, Primary Structure Analysis; [36]).
3. Results
3.1. Pattern Search Using Sequences Logos
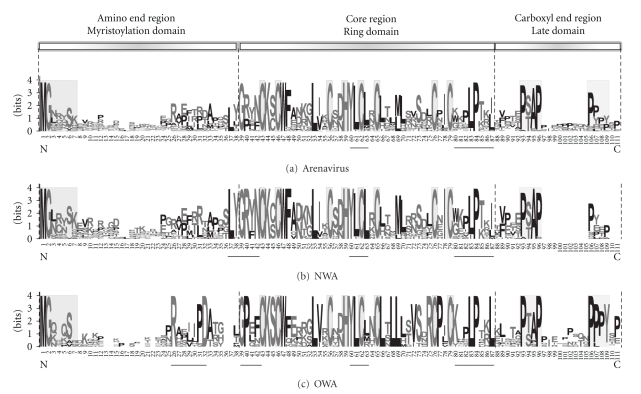
In Figure 1, three Z protein alignments (Old World, New World, and All Arenaviruses) are shown separately. In the top of the figure a diagram of the Z protein is shown, identifying three characteristic regions. We observe a myristoylation recognition motif in the amino end region, a RING finger domain, characterized by the seven cysteines (at positions 43, 46, 56, 62, 65, 76, and 79 in the alignment) and one histidine at the 59 position in the central core region, and different late domains (found in other viral matrix proteins) at the carboxyl end region.
Figure 1.
Sequence Logos of Arenavirus Z proteins. The upper diagram shows Z protein and the three regions in which it was divided. The other three are sequence logos, in which the X-axis indicates the relative position of amino acids in the multiple alignments. The length of 111 residues is due to the inclusion of gaps in the multiple alignment. The different amino acid domains described in the text are shadowed and the newly proposed domains are underlined. (a) Sequence Logo from all selected arenavirus Z proteins. (b) Sequence Logo from selected New World arenavirus Z proteins (ANM; AMAV-BeAn70563, CPXV-BeAn119303, GTOV-CVH-961104, GTOV-VHF-3990, JUNV-MC2, JUNV-Candid#1, MACV-MARU-222688, MACV-9530537, TACV, LATV-MARU-10924, OLVV-3229, PIRV-VAV488, PIRV-1743, ALLV-CLHP2472, PICV-AN3739, BCNV-A0060209, BCNV-AV-A0070039, WWAV-AV9310135, TAMV-W10777). (c). Sequence Logo from selected Old World arenavirus Z proteins (AVM; LCMV-Clone#13, LCMV-MX, LASV-CSF, LASV-AV, IPPY-DakAnB188, MOBV-Acar3080, MOPV-AN20410, MOPV-Mozambique, Lujo, and Morogoro).
In the amino end region an island configuration can be observed for each alignment. The most conserved island contains 7 amino acids comprising the first two amino acids (MG) that include the glycine modified by the myristic acid aggregate (Gly2, completely conserved) (gray shadowed in Figure 1). The other amino acids in this region are basic residues (K or R), or amino acid residues with amide (Q or N), sulfhydryl (C), or hydroxyl (S or Y) groups, probably constituting additional factors for membrane attachment [37]. The rest of the fragment has poor conservation degree, with the exception of another island for the Old World arenavirus group (Rx4PD, where x is any amino acid, underlined in Figure 1).
In the Z protein core region, a homogenous distribution of conserved sites is observed. Particularly, the 7 cysteines and the histidine residues required for Zn2+ binding and folding of the predicted RING finger domain (gray shadowed in Figure 1). These residues are totally conserved in the viral family, indicating strong selection pressure for keeping the predicted RING finger domain. Other positions within this region also show high conservation degree between both groups (New and Old World arenavirus). For example residues K44, W47, and L53 are completely conserved among all arenaviruses, suggesting they might play a critical role in protein folding, or maybe protein-protein interactions required for function, such as eIF4E interaction [12].
Furthermore, the amino end of this domain, shows a sequence motif characteristic for each group (L[YH]GR[YF]N, for the New World arenavirus, and GP[ELQ][FNS], for the Old World arenavirus). Until the moment, the role of this region is unknown, but it is interesting to highlight that the degree of conservation is unique within the Z protein, although differing among groups. It is therefore possible to hypothesize that this region could be associated with the arenavirus host range. In addition, at the carboxyl end of the RING finger domain, another conserved site within groups is detected, described as Kx(0,1)PLPTx[IL] (where x can be any amino acid). This putative site is underlined in Figure 1, where it can be seen that the conservation is stronger in the Old World arenavirus group. Furthemore, by modifying the first conserved leucine in this site (L83), it was demonstrated that this residue is involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious virus-like particles [38].
Finally, a cluster of conserved sites, in an island configuration, can also be found at the carboxyl end region. This island includes the characteristic motifs found in viral matrix proteins of retroviruses (late domains): P[TS]AP and PPPY (gray shadowed in Figure 1). These two motifs are present in the Old World arenavirus group, while in the New World arenavirus group only the first one is found. Another late domain found but situated in the core region was YLCL (YxxL [18, 19, 39, 40]) (underlined in Figure 1). The YLCL motif is also present in the Tacaribe virus Z protein and was observed not to be involved with budding of virus like-particles [41]. Interestingly, Tacaribe Z protein does not posses the P[TS]AP motif present in the majority of arenaviruses including Junin Z. Another late domain frequently identified in matrix proteins of negative-stranded RNA viruses is ΦPxV (where Φ indicates a hydrophobic residue and x any aminoacid, [42]), however the same was not found in the sequence of arenaviruses Z protein.
3.2. Biochemical Properties of the Different Recombinant Proteins
For the different fusion proteins, several parameters were calculated and are shown in Table 1. These values were taken into account for expression and purification optimization. The Z′ protein indicates the product of translation from the 366 bp fragment (z′).
Table 1.
Biochemical properties of the different expressed proteins.
| Protein | Number of amino acids | MW (Da) | pI | Extinction coefficients1 |
|---|---|---|---|---|
| Z′ | 106 | 11560.1 | 8.44 | 15970/15470 |
| Tio-Z-V5-His | 251 | 27339.1 | 6.29 | 30075/29450 |
| Tio | 120 | 12814.5 | 4.71 | 14105/13980 |
| Z-V5-His | 131 | 14542.5 | 8.43 | 15970/15470 |
| GST-Z | 331 | 37855.6 | 6.79 | 59080/58330 |
| GST | 225 | 26293.2 | 6.00 | 43110/42860 |
| His-Z | 136 | 15432.4 | 7.09 | 21930/21430 |
1Calculated extinction coefficients in M−1 cm−1 units, measured to 280 nm in water. Left numbers correspond to the calculation assuming that all Cys residues appear as half cystines, while the right numbers correspond to the calculation assuming that no Cys residues appear as half cystines.
3.3. Polyclonal Serum Titer and Cutoff Determination
Near 90 days after first inoculation with the Tio-Z-V5-His protein, the inoculated rabbits were heart bled, obtaining 30 ml of blood. After serum separation, the same was used to determine the cutoff value, yielding a number of 0.165. Afterwards, the polyclonal serum titer was determined, defined as the bigger serum dilution that renders a positive reaction (more than the cutoff value). For this determination, the assay conditions were the same as those in the assay cutoff determination, obtaining an antiserum titer of 128,000. Then, the IgGs were purified by affinity chromatography to later obtain more specific Z detection. The different fractions were analyzed by SDS-PAGE (data not shown) and quantified by Bradford method using a goat IgG as concentration reference in a calibration curve. The IgG final concentration was 3.19 mg/ml. These purified immunoglobulins were later used in Western blotting assays.
3.4. Tio-Z-V5-His
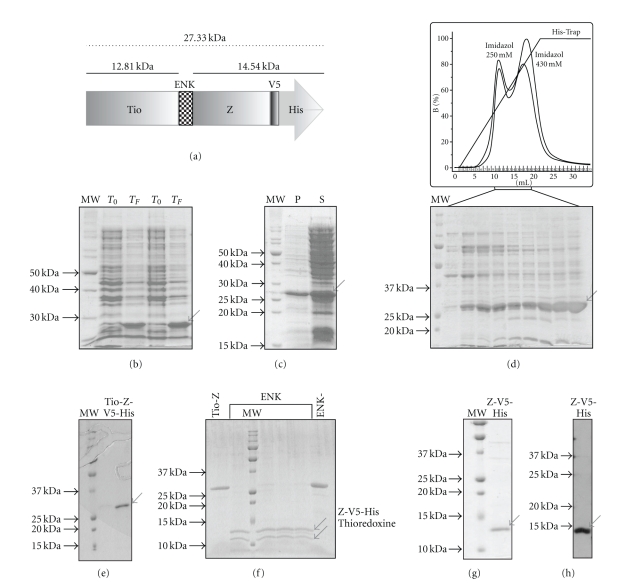
The expression profile of the recombinant Tio-Z-V5-His protein is shown in Figure 2. In Figure 2(a) a diagram of the expressed recombinant ORF is shown. In Figure 2(b) the lanes corresponding to T0 indicate the moment when the IPTG was added to the culture, while the TF lanes indicate the final culture time. After induction, a protein of expected size was expressed (Figure 2(b)). Following cell disruption, pellet and soluble fractions were separated. SDS-PAGE analysis confirmed it was possible to obtain a considerable amount of Tio-Z-V5-His in the soluble fraction (Figure 2(c)). The soluble fraction of Tio-Z-V5-His was loaded into a His-Trap column and monitored at UV280 and UV254. The peak fractions were collected and analyzed by 12% SDS-PAGE, containing a main band corresponding to the molecular weight of Tio-Z-V5-His (ca. 27 kDa) (Figure 2(d)). Final yield of pure Tio-Z-V5-His protein was between 10 and 12 mg per liter of culture. In order to further purify Tio-Z-V5-His, selected fractions were pooled, concentrated and loaded into a Superdex 200 gel filtration column (Figure 2(e)). The obtained Tio-Z-V5-His was dialyzed, concentrated and subjected to proteolysis with enterokinase. The cleaved products, thioredoxine (12.8 kDa) and Z-V5-His (14.5 kDa) were visible after 2 h at 37°C incubation (Figure 2(f)). The products of this reaction were loaded into a His-Trap column, as previously described and two major peaks were observed after elution. The first peak corresponded to thioredoxine, which eluted at 100 mM imidazole concentration, and the second peak corresponded to Z-V5-His protein, which eluted at 250 mM imidazole concentration (data not shown). The fractions of the second peak were pooled, concentrated (Vivaspin 2 5,000 MWCO, Sartorius) and analyzed by SDS-PAGE and Western blotting with anti-Z IgG (Figure 2(g) and 2(h)). In both cases a band of ca. 14 kDa corresponding to Z-V5-His was detected. The quantification data is showed in Table 2.
Figure 2.
Expression, purification and cleavage of Tio-Z-V5-His. (a) Diagram for the recombinant Tio-Z-V5-His protein expressed in E. coli. The dark gray region represents the thioredoxine (Tio) fusion protein; the striped block indicates the specific protease Enterokinase (Gly-Gly-Gly-Gly-Lys↓) target sequence; the light gray region corresponds to Z protein; the black line shows the V5 epitope localization; and the white arrow indicates the His-tag. The molecular weight of the complete recombinant protein is 27.3 kDa, the cleavage products being 12.8 kDa (thioredoxine) and 14.5 kDa (Z-V5-His). (b) 12% SDS-PAGE of samples of the whole cell culture at initial time (T0) and after 4 h at 37°C (TF). The arrow indicates Tio-Z-V5-His recombinant protein. (c) 15% SDS-PAGE of the centrifuged cell lysate of E. coli expressing Tio-Z-V5-His. S: supernatant from the whole cell lysate expressing Tio-Z-V5-His (soluble fraction). P: pellet from the whole cell lysate (insoluble fraction). The arrow indicates Tio-Z-V5-His. (d) The chromatography profile (upper part) corresponds to the elution of the bound protein with an imidazole gradient (black line). Fractions 10 to 18 (indicated with a bracket) were loaded on 12% SDS-PAGE. The arrow indicates Tio-Z-V5-His. In the lower part, is shown the 12% SDS-PAGE from Immobilized Ni2+ affinity chromatography (IMAC) His-trap (Amersham) fractions of the soluble fraction of Tio-Z-V5-His (S, from (c) part). (e) 12% SDS-PAGE of the gel filtration (Superdex 200) fractions 13 to 21 (pooled), of the Tio-Z-V5-His purification. This sample was the substrate of the cleavage with the enterokinase. (f) 15% SDS-PAGE of the cleavage of Tio-Z-V5-His with the protease enterokinase. The first lane is the purified Tio-Z-V5-His. The following lanes correspond to an incubation of 2:30 h at 37°C with different dilutions of the enterokinase (indicated with a bracket). The last lane is Tio-Z-V5-His protein without treatment. The arrows indicate the two peptides produced by the cleavage, shown in diagram A. (g) 15% SDS-PAGE of the purification of Z-V5-His with IMAC. (h) Western blot of the cleavage products of Tio-Z-V5-His with Enterokinase. The arrow indicates Z-V5-His.
Table 2.
Quantification data for expression and purification of GST-Z and Tio-Z-V5-His recombinants proteins.
| GST-Z | Tio-Z-V5-His | ||||
| Step | Relative Z-GST(%)1 | Purification factor | Step | Relative Tio-Z (%)1 | Purification factor |
| Total lysate | 21 | 0.0 | Total lysate | 19 | 0.0 |
| GST-affinity2 | 88 | 4.1 | Tio- affinity | 63 | 3.3 |
| Gel filtration2 | 91 | 4.2 | Gel filtration | 94 | 4.9 |
1Soluble protein ratio (Z fussion protein/total protein). 2 Include degradation products of Z.
3.5. GST-Z
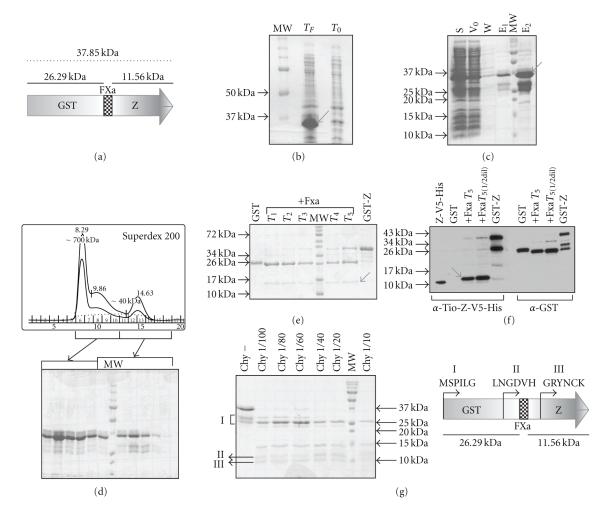
The expression profile of the recombinant GST-Z protein is shown in Figure 3. A diagram of the expressed recombinant ORF can be seen in Figure 3(a). In Figure 3(b), the overexpression of a protein with a size of about 37 kDa is shown, where the lane T0 indicates the moment when the IPTG was added to the culture, while the TFlane indicates the final culture time. The soluble fraction, after cell lysis, was mixed with Glutathione Sepharose resin for batch purification of the GST-Z protein. A protein of about 37 kDa was present in the first wash fraction; nevertheless, it was possible to obtain significant quantities of a ca. 37 kDa protein retained in the Glutathione Sepharose (Figure 3(c), lanes E1 and E2). Also, there were two extra bands of approximately 29 kDa and 25 kDa. These two peptides, retained together with the bigger protein, were probably degradation products of the ca. 37 kDa GST-Z protein. Final yield of pure GST-Z protein was between 5 and 8 mg per liter of culture. The eluted fractions were pooled, concentrated and loaded into a Superdex 200 gel filtration column. The molecular weight of the peaks was estimated from the chromatogram profile (Figure 3(d)). SDS-PAGE analysis of the collected fractions, showed that the ca. 37 kDa band and the other two, of approximately 29 kDa and 25 kDa, were present in different proportions. Most of the 37 kDa polypeptide was present at the high molecular weight peak (ca. 700 kDa), indicating that most of the overexpressed GST-Z protein was aggregated (Figure 3(d)), lanes 6 to 10). In the collected fractions of the second peak, the 37 kDa band was also detected, indicating that a portion of the overexpressed protein was soluble and monomeric (Figure 3(d), lanes 11 to 16). To obtain the Z protein without the GST fusion peptide, the purified GST-Z obtained after the glutathione sepharose purification was dialyzed against cleavage buffer and subjected to proteolysis with Factor Xa. The reaction was analyzed by SDS-PAGE (Figure 3(e)). The expected cleavage products, GST fusion peptide and Z protein were observed. To confirm the identity of the last product the sample was analyzed by Western blot with polyclonal antiserum specific for the Z protein (α-Tio-Z-V5-His) and antibodies specific for fusion protein (α-GST). Finally, as shown in Figure 3(f), it was possible to detect a ca. 11.5 kDa protein corresponding to Z. However, it was evident that the cleavage reaction was incomplete, because the full-length ca. 37 kDa polypeptide was also detected. The ca. 29 kDa peptide was also recognized by the Z polyclonal antiserum, strengthening the hypothesis that it was a degradation product of GST-Z. To confirm this hypothesis, the purified GST-Z (obtained after gel filtration in the 40 kDa peak) was subjected to a limited proteolysis with different dilutions of Chymotrypsin. The peptides thus obtained were subjected to N-Terminal sequencing (Figure 3(g)). The bands indicated with I match with the first six amino acids of the GST protein sequence, and the bands pointed out with II and III matched the C-terminal of GST and the middle region of Z protein, respectively. The quantification data is showed in Table 2.
Figure 3.
Expression, Purification, and Cleavage of GST-Z. (a) Diagram of the recombinant GST-Z protein expressed in E. coli. The dark gray region represents the glutathione-S-Transferase (GST) fusion protein; the striped block indicates the specific protease Factor Xa. (IleGluGlyArg↓) target sequence; and the light gray arrow shows the Z protein. The molecular weight of the complete recombinant protein is approx. 37 kDa, the cleavage products being ca. 25 kDa (GST) and ca. 11 kDa (Z). (b) 12% SDS-PAGE of a sample of the whole cell culture at initial time (T0) and after 20 h at 20°C (TF). (c) 15% SDS-PAGE of the purification of GST-Z with glutathione Sepharose 4B (Amersham). S: Supernatant from the whole cell lysate expressing GST-Z; V0: Lysate that ran through the column upon loading; W: Wash with PBS; E1 and E2: elution with 1 X PBS plus 25 mM reduced glutathione (Sigma). The arrow indicates GST-Z (37 kDa). (d) Gel Filtration (Superdex 200 GL, Amersham) and 12% SDS-PAGE. The first peak on the chromatogram includes fractions 6 to 10, analyzed in the SDS-PAGE, while the second peak corresponds to fractions 11 to 16. (e) 16% SDS-PAGE GST-Z cleavaged with the protease Factor Xa. The arrow indicates the cleavage product corresponding to Z′ (11.56 kDa). T1: 1 h; T2: 2 h; T3: 4 h; T4: 6 h; and T5: 16 h, at 37°C. (f) Western-blot of the cleavage products of GST-Z with Factor Xa. The arrow indicates the Z′ protein. (g) 15% SDS-PAGE of the limited proteolysis of GST-Z with Chymotrypsin (Roche). The protein products indicated with the symbols I, II, and III were sequenced at the N-terminus. The obtained sequence is indicated adjacent to the diagram of GST-Z.
3.6. His-Z
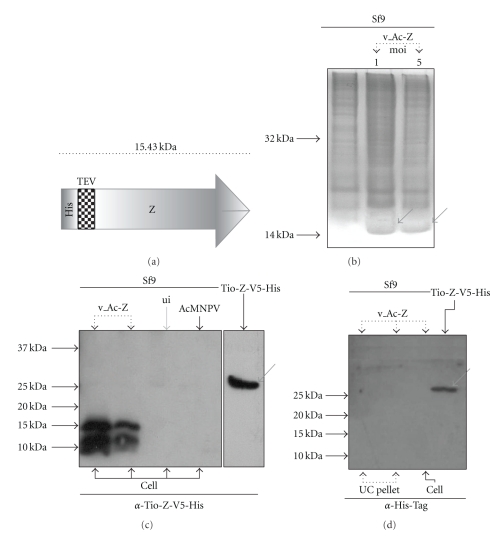
Expression results obtained for the recombinant His-Z protein are shown in Figure 4. A diagram of the tagged protein obtained using the Bac-to-Bac expression system is shown in Figure 4(a). The v_Ac-Z construct (recombinant bacmid) was transfected into High Five or Sf9 insect cells, and the resulting viral stock was amplified by infection in the same type of cells. The infection assays, optimized to obtain high virus titers, rendered a stock with a titer of 1.2 × 109 pfu/ml. In addition to infecting different cell lines, different multiplicities of infection (moi) were tested, as shown in Figure 4(b). The assayed moi's did not render significant differences in the protein expression levels. Sf9 insect cells were infected and, after 36 h, the monolayer fraction was collected and analyzed by 16% SDS-PAGE. In this case, it was not possible to observe an overexpressed product.
Figure 4.
Z Protein Expression in Sf9 Insect Cell Line. (a) Diagram of the recombinant His-Z protein expressed on Sf9 Insect cell line. The white region represents the His-tag; the squared block indicates the specific protease TEV (GluAsnLeuTyrPheGln↓Gly) target sequence; and the gray arrow shows the Z protein. The molecular weight of the complete recombinant protein is 14 kDa, the cleavage products being 11 kDa (Z) and 3 kDa (His-TEV). (b) 16% SDS-PAGE of Sf9 cells infected with v_Ac-Z. Lane 1 shows the Sf9 cells without infection. Lanes 2 and 3 correspond to the Sf9 cells infected with v_Ac-Z at different moi's (1 and 5 resp.) after an incubation of 36 h. The arrows have shown the possible Z protein expression. (c) Western blotting with α-Tio-Z-V5-His. Total protein extracts from Sf9 cells infected with v_Ac-Z (recombinant AcMNPV expressing Junín Z protein; lanes 1 and 2) and AcMNPV (wild type baculovirus; lane 4) were loaded in the indicated lanes. Lane 3 corresponds to the control of uninfected cells (ui). Tio-Z-V5-His purified sample was used as a positive control (indicated with a red arrow). (d) Western blotting with α-His Tag polyclonal serum (Santa Cruz Biotechnology). Ultracentrifugated supernatant and cellular extract of Sf9 cells infected with recombinant v_Ac-Z was loaded in the indicated lanes. At the bottom of each lane the origin of the sample is shown; Cell: Sf9 protein extract, UC pellet: ultracentrifugation pellet. Tio-Z-V5-His positive control is indicated with a red arrow.
Two immunological approaches to certify His-Z expression were used. Z-His protein was detected in the cellular fraction (lane 1 and 2, Figure 4(c)) when the purified α-Tio-Z-V5-His IgG was used in a Western blot. In this experiment, two Z corresponding bands appeared, same as observed previously by Jácamo and coworkers when Tacaribe virus recombinant Z protein was expressed [5]. Nevertheless, by using anti-His polyclonal serum for Western blotting it was not possible to visualize His-Z expression (Figure 4(d)). This suggests that possibly in insect cell lines His-Z could be subjected to a post-translational process that modifies the amino end of His-Z.
In order to ascertain the location of this protein in the supernatant fraction, the same was analyzed by Western Blot and a Proteinase K protection assay (Figure 5).
Figure 5.
Western Blotting with α-Tio-Z-V5-His. (a) The ultracentrifugated supernatant (UC pellet) and cellular extract (Cell) of Sf9 cell fractions infected with recombinant v_Ac-Z were loaded in lanes 1 and 2. The same fractions were loaded for uninfected cells (ui, lanes 3 and 4) and control v_dsRed (recombinant baculovirus expressing dsRed protein, lanes 5 and 6). (b) Proteinase K protection assay. The treatment of the UC pellet samples (Triton X-100 1% and Proteinase K 0,1 mg/ml) is indicated on the top of the figure.
4. Conclusions
The general aim of any heterologous expression system is to obtain a purified protein in its native conformation. Z is a small (approx. 11 kDa) viral protein normally expressed in the late phases of the arenavirus infection in mammalian cells. The recombinant overexpression of native Z protein requires an adequate eukaryotic expression system. However, there are some applications for which it is not necessary to obtain the protein in the native conformation, for example for antibodies production. Also, it has been reported that post-translational modifications are few in many proteins; therefore in these cases the bacterial expression systems are really advantageous because they have higher expression levels and lower costs when compared to eukaryotic expression systems [43]. Thus, the system will be selected depending on the application of the recombinant product.
Using the sequence logo (Figure 1) the conservation of amino acids for the arenaviral Z proteins was described. Z proteins could be divided into three characteristic regions named Amino, Core and Carboxyl regions. Each region comprises previously described Z protein domains. The Myristoylation domain into the amino region, the RING finger domain into the Core, and the Late Domains into the Carboxyl region. Furthermore, other conserved islands both between groups and within each group were described. Although the role of these last domains is today unknown, they could be a likely target for studies associated with the arenavirus host range.
Simple methodologies that allowed Z protein expression and facilitated its subsequent purification were employed. During the analysis of the Z protein expression in bacteria it was not possible to obtain the product without a fusion protein or tags. Probably, the reason for low Z protein yield during purification is its hydrophobicity, causing it to form relatively insoluble intracellular inclusion bodies that must be denatured. The Borden laboratory [28] discovered that including Zn+2 in the IPTG-induction media or in the lysis buffers greatly improved yields of recombinant Z protein. Although we included Zn+2 in the purification process, we have not yet tested a purification protocol that uses denaturing buffers.
Several expression strategies were tested, including cell hosts with different biological properties; it was only possible to achieve the goal when Z's amino terminus was fused to a bacterial protein. This fusion stabilized the recombinant protein and improved the expression levels.
In this work the over-expression of three different recombinant variants of Z protein: (1) Tio-Z-V5-His, (2) GST-Z, and (3) His-Z, was achieved. This last fusion protein was obtained from a baculoviral system in an insect cell line, while the other two were obtained from bacterial systems. The three variants of Z protein obtained can be cleaved from their N-fusion protein; Tio-Z-V5-His after proteolysis keeps the V5 epitope and the His tag.
Any of the expressed recombinant Z proteins could be used to obtain polyclonal or monoclonal antibodies that would allow immunological experiments to answer questions about the molecular biology of Junín virus. For example, studies of biological activity in different cell lines, cellular localization and protein interactions. Moreover, this serum could be useful for affinity chromatography designed to allow simple purification of Z protein without tags, or Z protein obtained after proteolysis of its fusion partner.
Tio-Z-V5-His recombinant protein was selected for antibody production, mainly because it showed the best expression level and solubility. This fusion protein was used for the generation of rabbit hyperimmune serum. Purified IgG fraction for subsequent Western blotting and EIA, was also obtained. On the other side, the purified Tio-Z-V5-His was used as substrate for the ENK reaction, in order to obtain Z-V5-His, employed as positive control in different assays.
Tio-Z-V5-His and His-Z fusion proteins can be purified by IMAC, since both peptides have His-Tags. However, this purification method could be counterproductive for the purification of native Z, since it is not known how Co+2 or Ni+2 ions could modify Z structure by binding and affecting its RING finger domain. It was observed that the native structure of Z protein contains two Zn+2 atoms, and other transition metals could interfere with these sites [44].
On the other hand, the GST-Z fusion protein can be purified by affinity chromatography with glutathione resins, without using immobilized metals. Its expression and purification was optimized with the aim to obtain the viral protein without tags on its native conformation, ready for crystallographic studies. The proteolysis of GST-Z protein with Factor Xa, produced only two peptides: GST and Z. The proteolysis reaction was successful because: (1) in SDS-PAGE analysis, at different times of the proteolysis reaction with Factor Xa, decreasing of GST-Z and increasing of individual GST and Z was observed; and (2) by Western blot analysis, the Z protein identity after the proteolysis reaction was asserted (Figures 3(e) and 3(f)). Consequently, high concentrations of Z protein (necessary for crystallographic studies) could be obtained.
In addition, after GST-Z purification, the presence of two proteins that copurified with GST-Z was observed (indicated with I in the Figure 3(g)). A similar phenomenon has been previously reported [45]. The Western blot, with IgG purified from polyclonal anti-Z serum recognized the GST-Z protein and other copurified peptide (ca. 29 kDa), whiles the control, GST alone, did not generate signal. Nevertheless, apparently this peptide is not a substrate of Factor Xa. In summary, the copurified peptides had high affinity for glutathione resin, so they probably contained GST derived amino acids, and one of them was recognized by the anti-Z serum, so it had Z derived amino acids. To confirm these facts the N-terminus of the copurified peptides, and some peptides obtained by limited proteolysis with chymotrypsin were sequenced (Figure 3(g)). This assay allowed the identification of structured domains, which were less sensitive to proteolysis, and could be better candidates for crystallographic studies. We found that one of the copurified peptides, the one of approximately 25 kDa, was stable even at high chymotrypsin concentrations. The N-terminus of this peptide shared the sequence with the first six amino acids of GST, indicating that degradation occurred by the carboxyl terminus of the recombinant protein. The same results were obtained after analyzing the N-terminus of the approximately 29 kDa copurified protein, which was also stable after the chymotrypsin treatment. Thus, it was possible that these low molecular weight peptides contained fragments of Z protein at its C-terminus. Two of them were sequenced at the N-terminus because, according to the molecular weight estimated by the electrophoretical migration, the Z protein sequence would be included (indicated with II and III in the Figure 3(g)). We could not establish the C-terminal sequence of the two copurified peptides, so we still do not know the specific site of cleavage of GST-Z in order to determine a stable domain. However, the approximate stable polypeptide corresponds to most of GST, which is not of our interest for crystallographic studies.
A subsequent approach to obtain Z protein in a native conformation could take advantage of its interactions with host proteins, such as the promyelocytic leukemia protein (PML) and others; it might be possible to overexpress Z and its interaction partner fused to a suitable tag for affinity purification. This would allow the copurification of the complex by affinity chromatography to later obtain native Z.
For Lassa arenavirus it was demonstrated that expression of Z in mammalian cell lines was sufficient for budding of “pseudo-virions” or Z-containing membranous particles [46]. Apparently, glycine myristoylation at position 2 of Z sequence is critical for this phenomenon [47–49]. This glycine residue is completely conserved in Z protein from all members of Arenaviridae (Figure 1). This suggests that all Z homologues within the Arenaviridae family are myristoylated at this position. Currently, all reports of pseudo-virion budding employed mammalian cell lines. At the present it is not known whether insect cell lines are capable of recognizing this post-translational modification signal present in Z, so during this work we started testing this possibility.
When insect cell lines were used for Z expression, high levels of expression were not achieved (Figure 4(b)). For this system, Z was His-Tagged so it could be purified by IMAC and then untagged by TEV protease cleavage. Interestingly, Z was detected in the cellular fraction by means of Western blotting with α-Tio-Z-V5-His IgGs, but it was not possible to detect it using an anti α-His Tag antibody (Figures 4(c) and 4(d), resp.). The absence of His-Tag signal could indicate the removal of the His-Tag from the protein, by a yet unidentified cellular process, or possible protein degradation at this particular site. Besides protein Western blot, a purification of the protein by IMAC was attempted, but no retention of protein from a cellular protein extract was observed (data not shown). Interestingly, when the cell culture supernatant fraction was analyzed using a Proteinase K protection assay, the results indicated that Z was included within small lipid vesicles, although other confirmatory assays such as immuno electron microscopy will be required in order to unambiguously confirm this.
The next step will be to further investigate the Z-membrane vesicles obtained from these cell lines. If N-myristoylation of Z in insect cells is proven to be effective, it will be necessary to modify the constructions for purification purposes. The employment of insect cell lines would represent a much safer methodology to obtain virus like particles [50], which have potential use as vaccine against arenaviruses for which successful treatment has not yet been established.
Acknowledgments
The authors are very grateful to Dra. Ana María Ambrosio and Dra. María del Carmen Saavedra who prepared Candid#1 virions; Dra. Graciela Glikman for help with antibodies production; Dr. Stenio Fragoso and Dra. Claudia Nunes dos Santos from IBMP (Paraná, Brasil), who provided the modified pGEX-B vector; and Dr. Félix Rey (Unité de Virologie Structurale, Pasteur Institute) for help with expression and purification experiments. This work was supported by PICT 38138 from ANPCYT and PIP 5813 from CONICET (Argentina). S.E.G., B.I.S., M.F.B, R.P.R., and C.S.B. have a research fellowship from CONICET, J.A.I. holds a research fellowship from ANPCYT, P.D.G. and M.E. Lozano are members of the research career of CONICET. S. E. Goñi and C. S. Borio contributed equally to this work.
References
- 1.Salvato MS, Clegg JCS, Buchmeier MJ, et al. Family arenaviridae. In: Van Regenmortel MHV, Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press; 2005. [Google Scholar]
- 2.Auperin DD, Romanowski V, Galinski M, Bishop DHL. Sequencing studies of Pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. Journal of Virology. 1984;52(3):897–904. doi: 10.1128/jvi.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghiringhelli PD, Rivera-Pomar RV, Lozano ME, Grau O, Romanowski V. Molecular organization of Junin virus S RNA: complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. Journal of General Virology. 1991;72(9):2129–2141. doi: 10.1099/0022-1317-72-9-2129. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier MJ, de la Torre JC, Peters CJ. Arenaviridae: the virus and their replication. In: Knipe M, Howley PM, editors. Fields Virology. 5th edition. Vol. 2. Philadelphia, Pa, USA: Lippincott Williams & Wilkins; 2007. pp. 1791–1827. [Google Scholar]
- 5.Jácamo R, López N, Wilda M, Franze-Fernández MT. Tacaribe virus Z protein interacts with the L polymerase protein to inhibit viral RNA synthesis. Journal of Virology. 2003;77(19):10383–10393. doi: 10.1128/JVI.77.19.10383-10393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iapalucci S, López N, Rey O, Zakin MM, Cohen GN, Franze-Fernández MT. The 5’ region of Tacaribe virus L RNA encodes a protein with a potential metal binding domain. Virology. 1989;173(1):357–361. doi: 10.1016/0042-6822(89)90257-2. [DOI] [PubMed] [Google Scholar]
- 7.Charrel RN, Feldmann H, Fulhorst CF, Khelifa R, Chesse RD, Lamballerie XD. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochemical and Biophysical Research Communications. 2002;296(5):1118–1124. doi: 10.1016/s0006-291x(02)02053-3. [DOI] [PubMed] [Google Scholar]
- 8.Salvato MS, Schweighofer KJ, Burns J, Shimomaye EM. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Research. 1992;22(3):185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- 9.Borden KLB, Campbell Dwyer EJ, Salvato MS. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. Journal of Virology. 1998;72(1):758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden KLB, Campbelldwyer EJ, Carlile GW, Djavani M, Salvato MS. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. Journal of Virology. 1998;72(5):3819–3826. doi: 10.1128/jvi.72.5.3819-3826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell Dwyer EJ, Lai H, MacDonald RC, Salvato MS, Borden KLB. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. Journal of Virology. 2000;74(7):3293–3300. doi: 10.1128/jvi.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kentsis A, Dwyer EC, Perez JM, et al. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. Journal of Molecular Biology. 2001;312(4):609–623. doi: 10.1006/jmbi.2001.5003. [DOI] [PubMed] [Google Scholar]
- 13.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. Journal of Virology. 1993;67(2):807–812. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López N, Jácamo R, Franze-fernández MT. Transcription and RNA replication of tacaribe virus genome and antigenome analogs require n and l proteins: Z protein is an inhibitor of these processes. Journal of Virology. 2001;75(24):12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(22):12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed EO. Viral late domains. Journal of Virology. 2002;76(10):4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends in Cell Biology. 2002;12(12):569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Serrano J, Pérez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. Journal of Virology. 2004;78(11):5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344(1):55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 20.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. Cellular factors required for Lassa virus budding. Journal of Virology. 2006;80(8):4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goñi SE, Iserte JA, Ambrosio AM, Romanowski V, Ghiringhelli PD, Lozano ME. Genomic features of attenuated Junín virus vaccine strain candidate. Virus Genes. 2006;32(1):37–41. doi: 10.1007/s11262-005-5843-2. [DOI] [PubMed] [Google Scholar]
- 22.Parodi AS, Greenway DJ, Rugiero HR, et al. Sobre la etiología del brote epidémico en Junín. Día Médico. 1958;30:2300–2302. [PubMed] [Google Scholar]
- 23.Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae) In Vitro. 1977;13(4):213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 24.Granados RR, Guoxun L, Derksen ACG, McKenna KA. A new insect cell line from Trichoplusia ni (BTI-Tn-5B1-4) susceptible to Trichoplusia ni single enveloped nuclear polyhedrosis virus. Journal of Invertebrate Pathology. 1994;64(3):260–266. [Google Scholar]
- 25.Luria SE, Burrous JW. Hybridization between Escherichia coli and Shigella. The Journal of Bacteriology. 1957;74:461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. 1st edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Volpon L, Osborne MJ, Borden KL. NMR assignment of the arenaviral protein Z from Lassa fever virus. Biomolecular NMR Assignments. 2008;2(1):81–84. doi: 10.1007/s12104-008-9090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villegas GA, Argüelles MH, Castello AA, Mas NJ, Glikmann G. A rapid method to produce high yields of purified rotavirus particles. Journal of Virological Methods. 2002;104(1):9–19. doi: 10.1016/s0166-0934(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 30.Argüelles MH, Orellana ML, Castello AA, et al. Measles virus-specific antibody levels in individuals in Argentina who received a one-dose vaccine. Journal of Clinical Microbiology. 2006;44(8):2733–2738. doi: 10.1128/JCM.00980-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Research. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hulo N, Bairoch A, Bulliard V, et al. The 20 years of PROSITE. Nucleic Acids Research. 2008;36(1):D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurer-Stroh S, Eisenhaber F. Myristoylation of viral and bacterial proteins. Trends in Microbiology. 2004;12(4):178–185. doi: 10.1016/j.tim.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. Journal of Virology. 2009;83(14):7029–7039. doi: 10.1128/JVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nature Structural Biology. 2002;9(11):812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nature Structural and Molecular Biology. 2007;14(3):194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urata S, Yasuda J, De La Torre JC. The Z protein of the new world arenavirus tacaribe virus has bona fide budding activity that does not depend on known late domain motifs. Journal of Virology. 2009;83(23):12651–12655. doi: 10.1128/JVI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. Journal of Virology. 2005;79(5):2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demain AL, Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnology Advances. 2009;27(3):297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Kentsis A, Gordon RE, Borden KLB. Self-assembly properties of a model RING domain. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(2):667–672. doi: 10.1073/pnas.012317299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi K, Kojima C. pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expression and Purification. 2008;62(1):120–127. doi: 10.1016/j.pep.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Strecker T, Eichler R, Ter Meulen J, et al. Lassa virus Z protein is a matrix protein sufficient for the release of virus-like particles. Journal of Virology. 2003;77(19):10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strecker T, Maisa A, Daffis S, Eichler R, Lenz O, Garten W. The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virology Journal. 2006;3, article 93 doi: 10.1186/1743-422X-3-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, De La Torre JC. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. Journal of Virology. 2007;81(17):9451–9460. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez M, Greenwald DL, De La Torre JC. Myristoylation of the RING finger Z protein is essential for arenavirus budding. Journal of Virology. 2004;78(20):11443–11448. doi: 10.1128/JVI.78.20.11443-11448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warfield KL, Posten NA, Swenson DL, et al. Filovirus-like particles produced in insect cells: immunogenicity and protection in rodents. Journal of Infectious Diseases. 2007;196(2):S421–S429. doi: 10.1086/520612. [DOI] [PubMed] [Google Scholar]