Summary
MDM2 associates with ribosomal protein S7 and this interaction is required to inhibit MDM2’s E3 ligase activity leading to stabilization of MDM2 and p53. Notably, the MDM2 homologue MDMX facilitates the inhibition of MDM2 E3 ligase activity by S7. Further, ablation of S7 inhibits MDM2 and p53 accumulation induced by different stress signals in some cell types. Thus, ribosomal/nucleolar stress is likely a key integrating event in DNA damage signaling to p53. Interestingly, S7 is itself a substrate for MDM2 E3 ligase activity both in vitro and in vivo. An S7-ubiquitin fusion protein (S7-Ub) selectively inhibits Mdm2 degradation of p53 and is unaffected by MDMX. S7-Ub promotes apoptosis to a greater extent than S7 alone. This indicates that MDM2 ubiquitination of S7 is involved in sustaining the p53 response. Thus, S7 functions as both effector and affector of MDM2 to ensure a proper cellular response to different stress signals.
Introduction
MDM2 is a crucial negative regulator of the p53 tumor suppressor protein (Brooks and Gu, 2006; Poyurovsky and Prives, 2006; Toledo and Wahl, 2006). Multiple functional domains of MDM2 are involved in p53 regulation. The N-terminus of MDM2 directly binds to the transactivation domain of p53, leading to inhibiton of p53 target gene transcription. The C-terminal RING domain is required for the activity of MDM2 as an E3 ubiquitin ligase promoting ubiquitination and proteasomal degradation of p53 and itself. Additionally, the central region of MDM2 is required for its ability to degrade p53 in response to various stress signals. Multiple mechanisms including different posttranslational modifications, changes in subcellular localization and modulation of MDM2 stability are employed to rapidly uncouple the MDM2-p53 interaction and consequently stabilize and activate p53, leading to the transcriptional regulation of a variety of genes including the mdm2 gene itself (Meek and Knippschild, 2003). Thus, MDM2 forms an autoregulatory feedback loop with p53 to control both the level and activity of p53.
Besides p53, an increasing number of proteins have been shown to interact with MDM2 protein, functioning as either affectors (regulating MDM2 function) or effectors (regulated by MDM2 function) or both (Ganguli and Wasylyk, 2003; Iwakuma and Lozano, 2003). MDM2-interacting proteins can thus have controlling effects on the MDM2/p53 circuit through their association with MDM2. At the same time, in a p53 independent manner, MDM2 is able to participate in various cellular functions through its interaction with proteins such as RB, Numb, p21 and others, thereby contributing to cellular responses to different stimuli (Ganguli and Wasylyk, 2003).
Adding to the complexity of the p53/Mdm2 circuitry is the fact that the homologue of MDM2, MDMX (also called MDM4) plays an important role in facilitating MDM2 downregulation of p53 (Marine et al., 2007). While itself lacking E3 ligase activity, MDMX can both repress p53 transcriptionally and form oligomers with MDM2 that modulate its E3 ligase activity (Gu et al., 2002; Sharp et al., 1999; Stad et al., 2001) Normally cytoplasmic, upon DNA damage, MDMX is recruited to the nucleus where it is degraded by MDM2, presumably to augment the function and stability of p53 under such conditions (Kawai et al., 2003; LeBron et al., 2006; Pereg et al., 2005).
Relevant to this study, an important connection between ribosomal stress and the MDM2/p53 circuit has been revealed. Three ribosomal large subunit proteins L5, L11 and L23 were shown to interact with and regulate MDM2 activity (Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004; Lohrum et al., 2003; Marechal et al., 1994). Each, when overexpressed, can activate p53 by inhibiting MDM2-mediated p53 degradation and, when ablated by siRNA knock-down, can attenuate the p53 response to low dose actinomycin D (ActD) and 5-fluoro-uracil (5-FU) treatments. In fact, cancer-associated missense mutations (C305F, C308Y) disrupt the interaction of MDM2 with L5 and L11 and these mutant MDM2 proteins are impaired in undergoing nuclear export, proteasomal degradation and promoting p53 degradation (Lindstrom et al., 2007a). Furthermore, exogenous L11 stimulates MDMX polyubiquitination by MDM2 while knockdown of L11 by siRNA reduces the ability of ActD to downregulate MDMX (Gilkes et al., 2006).
Ribosomal proteins are not the only nucleolar-associated proteins that have been shown to interact with MDM2. Nucleophosmin (Colombo et al., 2002; Kurki et al., 2004) and nucleolin (Saxena et al., 2006) have also been reported to bind and regulate MDM2 functions. Further, one of the mechanisms by which the nucleolar-associated p19ARF protein can counteract the repressive effect of MDM2 is to relocalize it to nucleoli (Lohrum et al., 2000; Weber et al., 1999).
Here, we have identified ribosomal small subunit protein 7 (S7) as an MDM2 interacting protein that is required for regulating the stability of MDM2. There are several features of our observations with S7 that go beyond previous findings with other ribosomal proteins. First, its requirement for counteracting MDM2 is very broad, since virtually every stress inducer we have tested requires S7. Second, there is an important and selective role for MDMX in this process. Third, S7 is itself a substrate of MDM2 and S7 ubiquitination may serve to extend the p53 stress response and facilitate cell death. Our results strengthen the hypothesis that interference with ribosomal and nucleolar integrity is the key integrator of the p53-MDM2 stress response. During our studies another group reported that S7 binds MDM2 and described the consequences of their interaction (Chen et al., 2007). Similarities and differences between their results and ours are addressed in the Discussion.
Results
S7 Interacts with the Central Region of MDM2
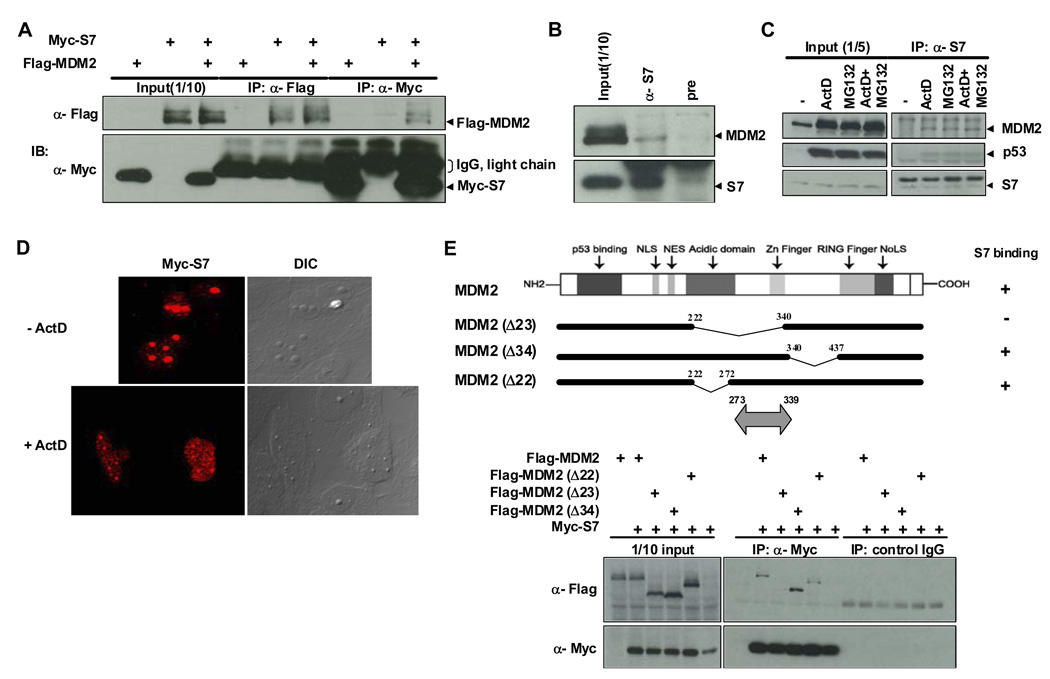
Using a construct expressing MDM2 (amino acids 4–491) as bait, a high-throughput yeast two-hybrid screen was carried out to identify MDM2-interacting proteins. Multiple clones identified from three different cDNA libraries encoded the ribosomal protein S7. The interaction between MDM2 and S7 was confirmed by co-immunoprecipitation of the two proteins from extracts of H1299 (Figure 1A) or U2OS (data not shown) cells transfected with constructs expressing S7 and MDM2. The association of endogenous S7 and MDM2 was also demonstrated using cell lysates from HCT116 cells (Figure 1B). Levels of MDM2 detected in α-S7 immunoprecipitates were increased when cells were treated with a low dose of ActD or with the proteasome inhibitor MG132. Combining ActD and MG132 did not further enhance the interaction between S7 and MDM2 (Figure 1C). Although the increased association of S7 and MDM2 is likely due to the increased cellular MDM2 levels caused by these treatments, it is also possible that relocalization of S7 or MDM2 in response to these agents is involved in this response. Indeed, MDM2 associates with nucleoli in response to MG132 treatment (Klibanov et al., 2001; Latonen et al., 2003) (data not shown). In addition, while ectopic S7 accumulated in nucleoli in unstressed U2OS cells, a low dose of ActD (Figure 1D) or daunorubicin (data not shown) led to redistribution of a significant proportion of S7 into the nucleoplasm (Figure 1D). p53 was also found in α-S7 immunoprecipitates, suggesting that it formed a complex with S7 and MDM2.
Figure 1. Binding between MDM2 and Ribosomal Protein S7 and Mapping of the MDM2 Domain for S7 Binding.
(A) Interaction between ectopic MDM2 and S7. H1299 cells were transfected with plasmids expressing Flag-MDM2 (1.5 µg), Myc-S7 (1.5 µg), or both (1.5 µg each) as indicated. Twenty-four hours after transfection, whole-cell lysates were subjected to immunoprecipitation (IP) with anti-Flag or anti-Myc antibodies followed by immunoblotting (IB) with indicated antibodies. IgG light chains cross-reacting with anti-Myc antibody are also indicated. (B) Co-association of endogenous MDM2 and S7. Whole-cell lysates prepared from HCT116 cells were subjected to IP with either anti-S7 or pre-immune serum, followed by IB with anti-MDM2 and anti-S7 antibodies. (C) Increased binding between endogenous MDM2 and S7 in response to ActD or MG132 treatment. HCT116 cells were treated with ActD (5 nM), MG132 (20 µM) or both for 6 hours before harvesting. IP and IB were carried out as in (B). (D) Relocalization of ectopic S7 in response to ActD treatment. U2OS cells were transfected with a Myc-S7 (1.5 µg). Twenty hours after transfection, the cells were treated with ActD (5 nM) for 7 hours, and then subjected to immunofluorescent staining. (E) Mapping of the MDM2 domain for S7 binding. H1299 cells were transfected with plasmids (1.5 µg each) expressing different MDM2 deletion mutants as indicated. IP with anti-Myc antibody or control mouse IgG was carried out as in (A) and followed by IB with indicated antibodies. Schematic representation of MDM2 mutants used for transfection is shown on top with the S7 binding region indicated as double headed arrow at bottom.
Further, using a series of Flag-MDM2 mutants co-expressed with Myc-S7 in H1299 cells, a sequence within amino acids 273–339 of MDM2 was found to be required for S7 binding (Figure 1E and Figure S1). This region is within the central portion of MDM2 that is required for its degradation of p53 and also overlaps sequences shown previously to interact with other ribosomal proteins L5, L11 and L23 (Bhat et al., 2004; Dai and Lu, 2004; Dai et al., 2004; Jin et al., 2004; Lohrum et al., 2003; Marechal et al., 1994).
MDMX is Selectively Required for S7 Inhibition of MDM2 Ubiquitination
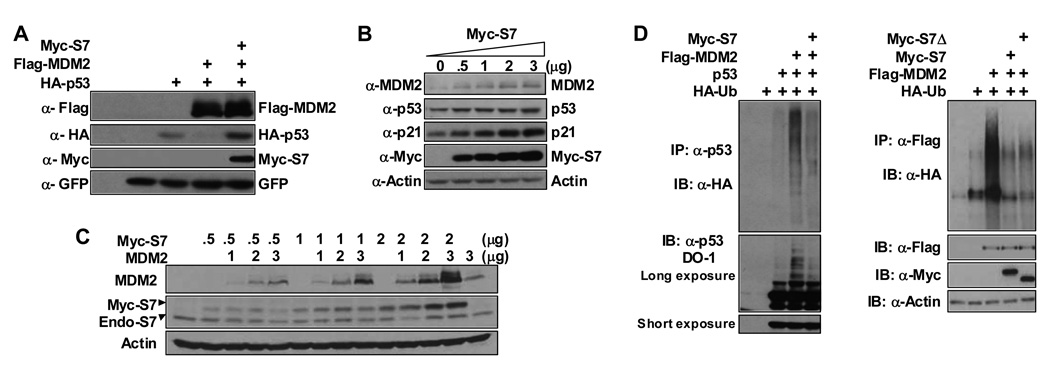
To elucidate the impact of S7 on MDM2 we co-expressed S7 with MDM2 and p53 in U2OS cells. MDM2-mediated degradation of p53 was greatly inhibited by S7 (Figure 2A). More significantly, the levels of endogenous p53, MDM2 and p21 in U2OS cells were elevated following S7 expression (Figure 2B). Consistent with this observation overexpression of S7 led to cell cycle arrest in U2OS cells (Figure S2). The increase in MDM2 was not solely due to augmentation of its expression resulting from increased p53 protein because expression of S7 also resulted in stabilization of MDM2 in H1299 cells that lack p53 (Figure 2C). The ability of S7 to inhibit MDM2-induced p53 degradation is most likely related its ability to inhibit both MDM2-mediated p53 ubiquitination and MDM2 autoubiquitination in vivo (Figure 2D). Under some circumstances the ability of S7 to relocalize MDM2 to nucleoli (in a relatively small proportion of the cells) may also contribute to its inhibitory effects (Figure S3).
Figure 2. Ribosomal Protein S7 Stabilizes p53 and Inhibits MDM2-mediated p53 Polyubiquitination.
(A) U2OS cells were transfected with HA-p53 (0.35 µg), Flag-MDM2 (1.5 µg), and Myc-S7 (1.5 µg) constructs as indicated. Cell lysates were used for IB with indicated antibodies. GFP construct was added for transfection and loading control. (B) Ectopic expression of S7 stabilizes endogenous MDM2 and p53. U2OS cells were transfected with increasing amounts of Myc-S7 (0.5, 1, 2, and 3 µg) as indicated. Cell lysates were subjected to IB with indicated antibodies. (C) Ectopic expression of S7 stabilizes exogenous MDM2 independently of p53. H1299 cells were transfected with increasing amount of Myc-S7 (0.5, 1, 2 µg) or pcDNA3-MDM2 (1, 2, 3 µg) as indicated. Cell lysates were used for IB with indicated antibodies. (D) Ectopic expression of S7 inhibits MDM2-mediated p53 polyubiquitination and MDM2 autoubiquitination. H1299 cells were transfected with combinations of Myc-S7 (1.2 µg), Myc-S7Δ (1.2 µg), and Flag-MDM2 (1.2 µg) plasmids with (left panel) or without (right panel) p53 (0.3 µg) plasmid in the presence of an HA-Ub (1.2 µg) plasmid as indicated. The cells were treated with MG132 (20 µM) for 4 hours before harvesting. Whole-cell lysates were subjected to IP with anti-p53 (FL393-G) antibody followed by IB with anti-HA antibody to detect ubiquitinated p53 (left panel) or IP with anti-Flag antibody followed by IB with anti-HA antibody to detect ubiquitinated MDM2 (right panel).
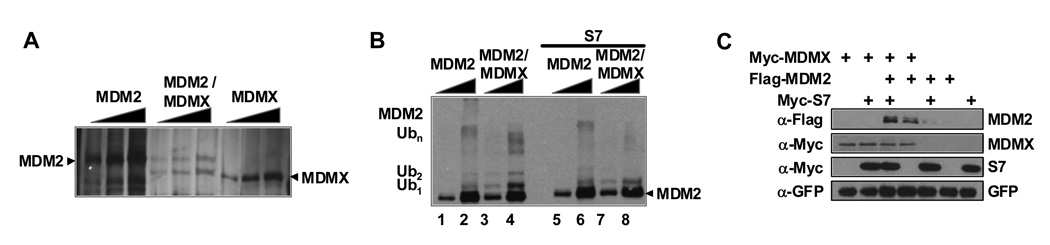
To gain further insight into S7 inhibition of the E3 ligase activity of MDM2, an in vitro ubiquitination assay was carried out using purified S7, MDM2 and p53 proteins. S7 was found to inhibit MDM2-mediated p53 ubiquitination but had no impact on MDM2 autoubiquitination in vitro (Figure 3B compare lanes 1 and 2 with lanes 5 and 6; and see Figure 6A below). We considered the possibility that MDMX might be required for S7 to block MDM2 autoubiquitination. In fact, when we compared the autoubiquitination of MDM2 alone with that of a purified preformed MDM2/MDMX complex (see Figure 3A) S7 was only able to inhibit the heteromeric complex (Figure 3B compare lanes 3 and 4 with lanes 7 and 8). Furthermore, co-expression of MDMX and S7 led to markedly greater stabilization of MDM2 than either of these two proteins alone (Figure 3C).
Figure 3. S7 Inhibits MDM2 E3 Ligase Activity in a Manner that Involves MDMX.
(A) Purification of MDM2/MDMX complex. Increasing amounts (0.05, 0.1 and 0.2 µg) of Flag-MDM2, HA-MDMX or MDM2/MDMX complex purified from insect cells were resolved on SDS-PAGE gel and visualized by silver staining. Amounts of the complex used in subsequent experiments were adjusted so that total amounts of MDM2 protein were equivalent. (B) S7 inhibits MDM2 E3 activity only when it is present in an MDM2/MDMX complex. Ubiquitination reaction mixtures containing either 0.1 or 0.3 µg of MDM2 alone or the MDM2/MDMX complex with or without 0.5 µg S7 protein as indicated, along with ~300 pmol ubiquitin, UbcH5c and rabbit E1 were incubated at 37°C for 40 min. Reactions were terminated by addition of SDS-sample buffer and mixtures with equivalent amounts of MDM2 were resolved by SDS-PAGE. Ubiquitin conjugates were visualized by Western blotting with anti-MDM2 antibody. Migration of mono- (Ub1) di- (Ub2), and poly- (Ubn) ubiquitinated MDM2 species are indicated at left. (C) S7 and MDMX increase MDM2 stabilization. U2OS cells were transfected with the indicated plasmids (1.2 µg each). Cells were harvested after 24 hours and cell lysates were subjected to IB with indicated antibodies. GFP construct was added for transfection and loading control.
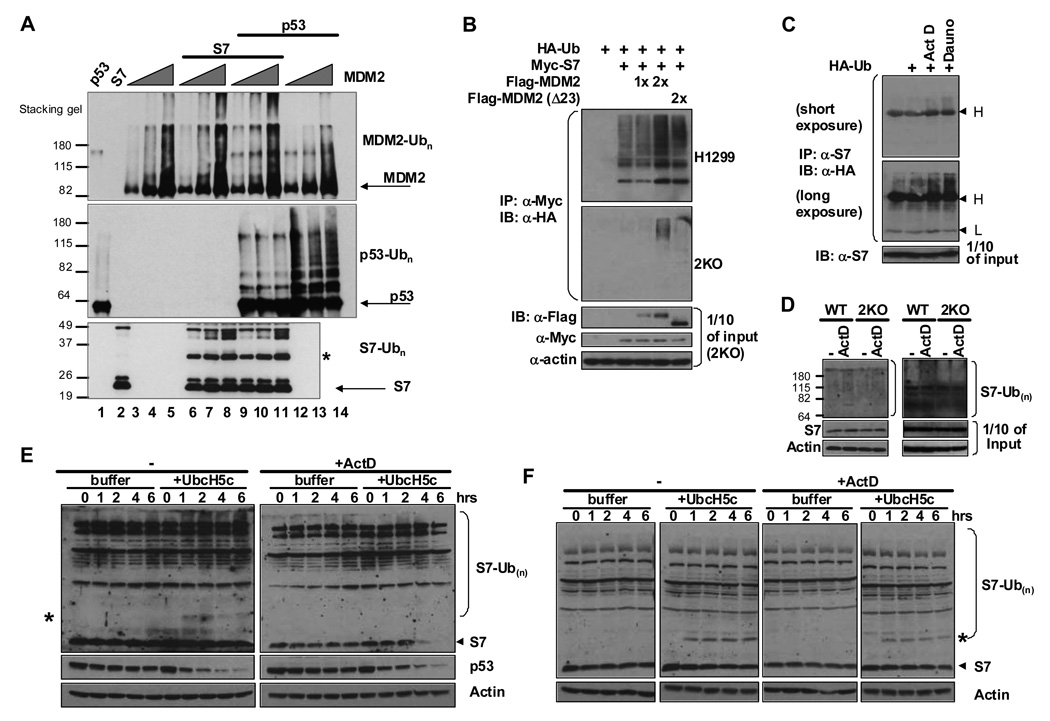
Figure 6. S7 Is a Substrate for Ubiquitin Ligase Activity of MDM2 and Inhibits MDM2-mediated p53 Ubiquitination In Vitro.
(A) MDM2 ubiquitin ligase activity in vitro toward itself, p53 and S7. In vitro ubiquitination assays were performed in the presence of MDM2 alone, MDM2 and p53, MDM2 and S7, or all three. Reaction mixtures contained increasing amounts of Flag-MDM2 (0.5, 1, 1.5 µg) per reaction (lanes 3–14), 500 ng of p53 (lanes 1, 9–14), 1 µg of S7 (lanes 2, 6–11), ubiquitin, E1 and UbcH5c. Equivalent amounts of each reaction mixture were resolved by SDS-PAGE. MDM2, p53 and S7-ubiquitin conjugates were detected by IB with indicated antibodies. The asterisk indicates the position of mono-ubiquitinated S7. (B) S7 ubiquitination is increased by MDM2 in vivo. H1299 (upper panel) or 2KO (p53 −/− mdm2 −/−; lower panels) cells were transfected with combinations of Myc-S7 (0.6 µg), Flag-MDM2 (0.6 or 1.2 µg) or Flag-MDM2 (Δ23) (1.2 µg) plasmids in the presence of an HA-Ub (1.2 µg) plasmid as indicated for 24 hours. The cells were treated with MG132 (20 µM) for 4 hours before harvesting. Whole-cell lysates were subjected to IP with anti-Myc antibody followed by IB with anti-HA antibody to detect ubiquitinated S7. Ten percent of input cell lysates from 2KO cells were subjected to IB with indicated antibodies. (C) Endogenous S7 ubiquitination is increased in response to stress signals. U2OS cells were transfected with HA-Ub (2 µg) for 24 hours and treated with ActD (5 nM) or Dauno (0.22 µM) for 6 hours before harvesting. IP was carried out with anti-S7 antibody followed by IB with anti-HA antibody to detect ubiquitinated endogenous S7. IgG heavy and light chains cross-reacting with anti-HA antibody are indicated at right. (D) MDM2 is involved in ubiquitination of endogenous S7 in response to ActD. Wild-type and 2KO MEFs were transfected with His-Ub (2 µg) for 24 hours and treated with ActD (5 nM) for 6 hours before harvesting. A Ni-NTA pull down assay was carried out and eluted proteins were analyzed by Western blotting with anti-S7 antibody. (E–F) MDM2 plays a role in ubiquitination and degradation of endogenous expressed S7 in response to ActD. Twenty-four hours after plating, Wild-type MEF cells (E) and 2KO cells (F) were treated with ActD (5 nM ) for 6 hours before harvesting. In vitro degradation assays were performed and aliquots were removed from reaction mixtures at the indicated time points and subjected to IB with indicated antibodies. Mono-ubiquitinated S7 species are indicated with asterisks.
Down-regulation of S7 Inhibits MDM2 and p53 Stabilization after Multiple Sources of Genotoxic Stress
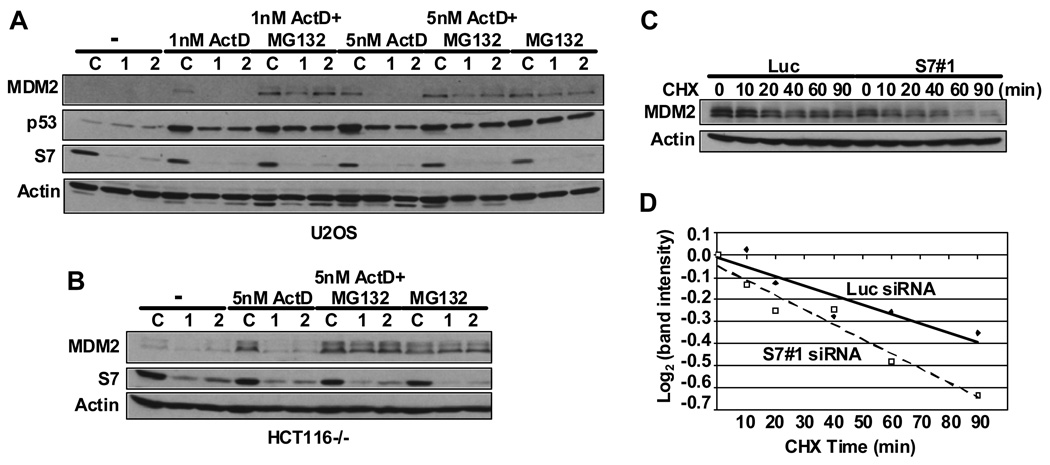
We next evaluated how MDM2 is regulated by endogenously expressed S7. To approach this we examined the effect of down-regulation of S7 on the function of MDM2 and p53 using two different siRNAs (Figure 4). In both U2OS and SAOS-2 cells even though S7 could be lowered to more than half of its original level there was no discernable effect on cell proliferation under the conditions used i.e. with up to 200 nM siRNA and 48 hours after siRNA treatment (data not shown). Furthermore, ablation of S7 in unstressed U2OS cells did not lead to increased levels of p53 and MDM2 (Figure 4A). It was shown previously that low doses of ActD or 5-FU trigger ribosomal stress and consequent activation of p53 (Bhat et al., 2004; Dai et al., 2004; Gilkes et al., 2006; Lindstrom et al., 2007b; Sun et al., 2007). When we introduced S7 siRNAs into U2OS cells, ActD-induced p53 stabilization was attenuated and the levels of MDM2 were significantly decreased (Figure 4A). Incubating cells with MG132 abrogated this effect, showing that S7 modulates proteasome-mediated p53 and MDM2 degradation. In HCT116 p53−/− cells S7 reduction by siRNA also resulted in destabilized MDM2 after ActD treatment indicating that the lower levels of MDM2 were due to its increased rate of degradation (Figure 4B). This was further supported by a cycloheximide half-life assay in U2OS cells which showed that S7 ablation significantly increased MDM2’s turnover (Figure 4C and D). These results indicate that normal levels of S7 are critical to MDM2 stability under conditions of ribosomal stress.
Figure 4. siRNA Ablation of S7 Inhibits Actinomycin D-induced MDM2 and p53 Stabilization.
(A) Down-regulation of S7 attenuates ActD-induced MDM2 and p53 stabilization. U2OS cells were transfected with 0.025 µM control siRNA or two different S7 (1 and 2) siRNAs. Forty-two hours after transfection, cells were treated with ActD (1 nM or 5 nM), or MG132 (20 µM), or both as indicated. Six hours later, cell lysates were subject to IB with indicated antibodies. (B) Down-regulation of S7 attenuates ActD-induced MDM2 stabilization in the absence of p53. HCT116 p53−/− cells were transfected with siRNAs and treated with ActD or MG132 as in (A). MDM2, S7 and Actin were detected by Western blot analysis. (C) Reducing S7 levels accelerates MDM2 degradation after ActD treatment. U2OS cells were transfected with control or S7 (S7#1) siRNA and treated with 5 nM ActD as in (A). Then, cyclohexamide (CHX, 100 µg/ml) was added and cells were harvested at the indicated times. Cell lysates were subjected to IB with indicated antibodies. (D) Quantification of the western blot data in (C) was carried out using image J software .
It should be mentioned that these observations were not obtained in all cell lines tested. In some cell lines S7 ablation by siRNA led to p53 dependent and independent cell cycle arrest (Figure S4 and data not shown). We do not know the basis for cell-specific responses to reduced S7 but speculate that in some cells the ensuing nucleolar stress may lead to relocalization of proteins that are capable of counteracting MDM2 and thereby stabilizing p53 even in the absence of S7, while in others (eg. U2OS and SAOS-2) such proteins either do not accumulate to sufficiently high levels or other mechanisms are involved.
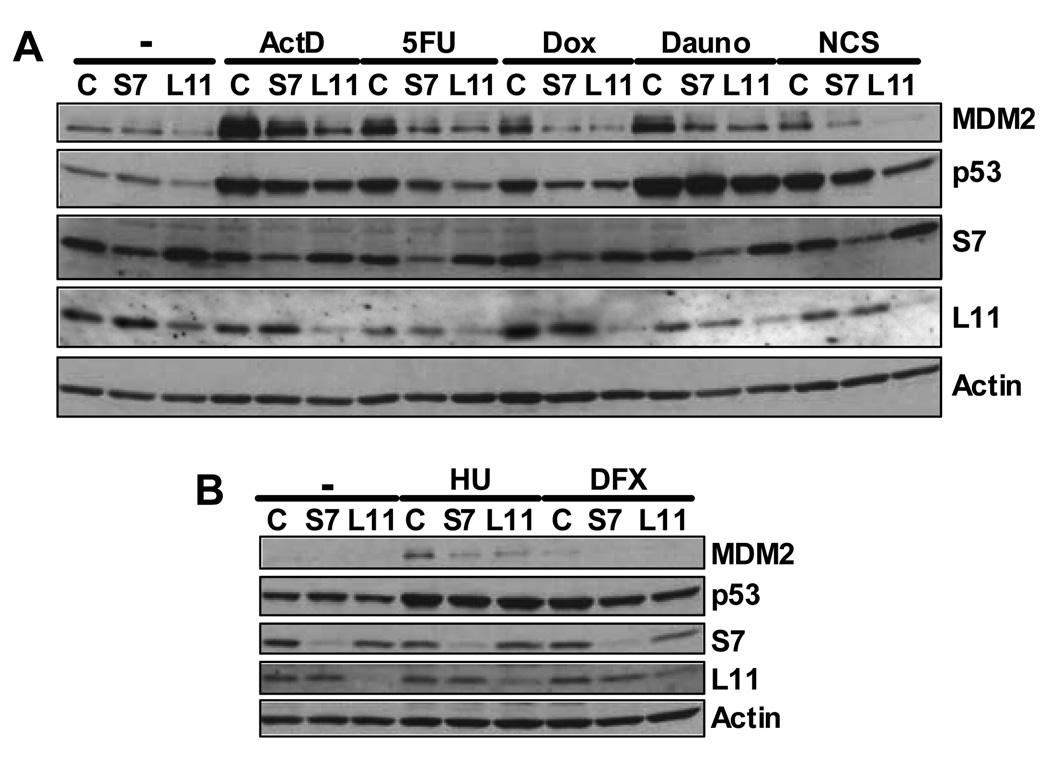
We anticipated that agents specifically known to induce ribosomal stress would require S7 to inhibit MDM2 and stabilize p53. It was therefore surprising that DNA damaging agents that elicit a variety of different genotoxic assaults including doxorubicin, daunorubicin, hydoxylurea, deferoxamine mesylate, and neocarzinostatin gave essentially the same response of reducing MDM2 in response to ablation of S7 (Figure 5A and B). Stress-induced p53 stabilization was also attenuated by the various agents (to different extents) after S7 ablation. Note that these agents work by various distinct mechanisms and cause arrest at different stages of the cell cycle.
Figure 5. S7 or L11 Ablation Attenuates MDM2 Stabilization after Multiple Stress Inducing Agents.
U2OS cells were transfected with 0.025 µM control , S7 (S7#1), or L11 siRNA. Six hours before harvesting, cells were treated with ActD (5 nM), 5-FU (50 µM), doxorubicin (DOX, 0.03 µM) and daunorubicin (Dauno, 0.22 µM) (A) or hydroxyurea (HU, 1.5 mM) and deferoxamine mesylate (DFX, 250 µM) (B). Neocarzinostatin (NCS, 640 ng/ml) was added 2 hours before harvesting (A). Forty-eight hours after transfection, cell lysates were subjected to IB with indicated antibodies.
We also ablated L11 using a previously reported siRNA (Bhat et al., 2004). As previously shown, downregulation of L11 led to decreased levels of p53 in U2OS cells even under unstressed condition and attenuated the impact of low dose ActD- or 5-FU-induced p53 stabilization. Interestingly, like S7 ablation, L11 depletion also led to the inhibition of stress-induced MDM2 stabilization for all the other drugs that were tested (Figure 5 A and B). This indicates that S7 and L11 do not compensate for each other in stabilization of MDM2. Based on these results we propose that modulation of MDM2 function by MDM2-interacting ribosomal proteins is a common if not universal mechanism employed by cells in response to multiple stress stimuli.
S7 is Ubiquitinated by MDM2 and Ubiquitinated S7 Differentially Impacts p53 and MDM2 Stabilization
A large-scale analysis of the human ubiquitin-related proteome by mass spectrometric analysis revealed that many ribosomal proteins including S7 are ubiquitinated (Matsumoto et al., 2005). Based on its apparently strong interaction with MDM2 we tested whether S7 can serve as a substrate for the E3 ligase activity of MDM2. Indeed S7 was ubiquitinated by MDM2 in a dose-dependent manner in vitro (Figure 6A). Remarkably, under the conditions used, while S7 could inhibit p53 ubiquitination, p53 had no impact on the ability of MDM2 to ubiquitinate S7. In line with this, ubiquitination of ectopic S7 was also increased by co-expressed MDM2 in vivo both in H1299 and mouse 2KO (p53−/−, mdm2−/−) cells (Figure 6B). S7 ubiquitination by MDM2 requires the interaction between S7 and MDM2 as no ubiquitinated S7 species were detected in 2KO cells with a deletion mutant MDM2 (Δ23) that cannot bind S7 (Figure 6B). Moreover, ubiquitination of endogenous S7 was increased when U2OS cells were treated with either ActD or daunorubicin (Figure 6C). Similar results were obtained for HCT116 cells treated with ActD (data not shown). Since only polyubiquitinated S7 species were detected when cells were treated with these agents (Figure 6C) it is likely that polyubiquitination of S7 occurs rapidly and there is very little steady state mono-ubiquitinated S7.
To examine the involvement of endogenous MDM2 in the ubiquitination and degradation of endogenous S7 we employed two different approaches using embryonic fibroblasts (MEFs) from wild-type and Mdm2/p53 double null (2KO) mice. In the first case we transfected His-tagged ubiquitin into the MEFs and performed Ni-NTA pull-downs from cell extracts followed by immunoblotting for S7. We detected a subtle but reproducible increase in endogenous poly-ubiquitinated S7 species after ActD treatment in wild-type but not in 2KO cells (Figure 6D shows two separate experiments). Second, we subjected lysates of untreated or ActD treated wild-type and 2KO MEFs to an in vitro degradation assay (Rape and Kirschner, 2004) in which extracts were supplemented with ubiquitin and UbcH5c known to be the E2 that works with MDM2 to degrade p53 (Saville et al., 2004) (Figure 6E and F). We reasoned that if MDM2’s E3 ligase activity is indeed involved in S7 ubiquitination, the addition of ubiquitin and specific E2 proteins (UbcH5c in this case) to cell extracts would very likely to drive S7 towards ubiquitin-mediated degradation. As shown in Figure 6E, extracts of ActD treated wild-type MEF cells were capable of degrading endogenous S7 while extracts of untreated cells were unable to do so. Note that both wild-type cell extracts were able to degrade endogenous p53 in a UbcH5c-dependent manner. Extracts of 2KO cells were not able to degrade S7 significantly even when cells were treated with ActD (Figure 6F). Taken together, our data indicate that endogenous MDM2 plays a role in ubiqutination and degradation of endogenous S7. However, since a polypeptide species corresponding to mono-ubiquitinated S7 (indicated by an asterisk) was detected upon addition of the E2 even in 2KO cells this indicates that MDM2 is not the only E3 ubiquitin ligase for S7. Note that cell lysates were centrifuged as part of the procedure leading to depletion of the rough-ER associated ribosomes, so only a fraction of the total ribosomes are present in the reaction mixtures. ActD treatment disrupts nucleoli and this may yield a lot more available free S7 for degradation.
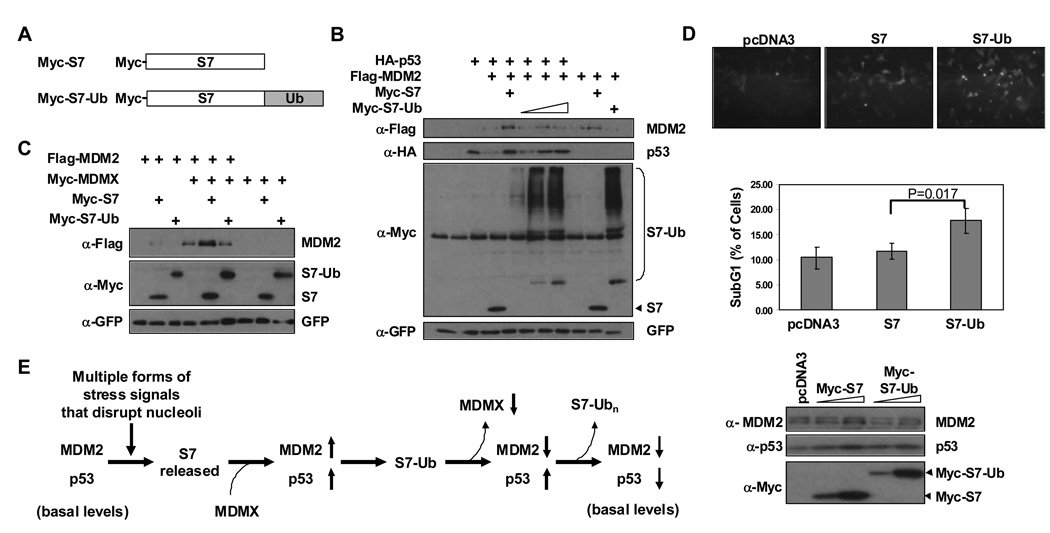
To gain further insight into the significance of S7 ubiquitination by MDM2 we followed an approach previously demonstrated to be instructive for understanding the impact of p53 ubiquitination in which a p53-ubiquitin fusion protein was utilized (Carter et al., 2007; Li et al., 2003; Sasaki et al., 2007). An S7-ubiquitin fusion protein (S7-Ub) was generated in which a single ubiquitin is joined to the C-terminus of S7. When S7-Ub was introduced into H1299 cells a fraction of it was retained in the cytoplasm and was extensively polyubiquitinated (Figure S5 A and B). The portion of S7-Ub that localized to nuclei was largely un-polyubiquitinated. Importantly, when the impacts of S7 and S7-Ub on p53 and MDM2 protein levels were compared, while both versions of S7 could stabilize p53, only S7 but not S7-Ub was capable of also stabilizing MDM2 (Figure 7B). Further, MDMX was unable to co-operate with S7-Ub to stabilize MDM2 (Figure 7C). Correspondingly, upon ectopic expression of S7 or S7-Ub in U2OS cells at similar levels, S7-Ub induced more apoptosis than S7 (Figure 7D). Based on these observations we propose that MDM2 ubiquitination of S7 is a mechanism for extending the stabilization of p53 after conditions of ribosomal stress.
Figure 7. An S7-ubiquitin Fusion Protein Stabilizes p53 but not MDM2 and Promotes Apoptosis.
(A) Schematic representation of Myc-S7 and its derivative (Myc-S7-Ub) that has one copy of ubiquitin fused to the C-terminus. (B) S7-Ub stabilizes p53 more efficiently than MDM2. U2OS cells were transfected with the indicated combinations of HA-p53 (0.28 µg), Flag-MDM2 (1.6 µg), Myc-S7 (0.8 µg) or increasing amounts of Myc-S7-Ub (0.8, 1.6, 2.2 µg). Cell lysates were subjected to IB with indicated antibodies. GFP construct was added for transfection and loading control. (C) S7-Ub cannot cooperate with MDMX to stabilize MDM2. U2OS cells were transfected with the indicated combinations of Flag-MDM2 (0.6 µg), Myc-MDMX (0.6 µg), Myc-S7 (0.6 µg), or Myc-S7-Ub (2.2 µg). Cell lysates were subjected to IB with indicated antibodies. GFP construct was added for transfection and loading control. (D) S7-Ub induces more apoptosis than S7. U2OS cells were transfected with Myc-S7 (0.6 µg) or Myc-S7- Ub (2.2 µg) together with an F-GFP plasmid (0.1 µg). Thirty hours after transfection, cells were examined by immunofluorescence microscopy for floating cells (top panel) and then harvested for both FACS analysis (middle panel) and Western blot analysis (bottom panel). Apoptotic cell index and standard deviations were obtained from three independent experiments. P-value was calculated using Student’s t-Test with two-tailed distribution. Immunofluoresence and western blot images are representative data from these independent experiments. (E) A Model for S7 Regulation of the MDM2-p53 Circuit. Stress signals lead to ribosomal biogenesis perturbation and partial nucleolar disruption. S7 is released from nucleoli where it can interact with MDM2 leading to inhibition of MDM2-E3 ligase activity towards p53. MDM2 autoubiquitination is inhibited by S7 in an MDMX dependent manner. In addition, S7 serves as an E3 ligase substrate for MDM2 and since ubiquitinated S7 cannot stabilize MDM2, this may promote a feed-forward mechanism to maintain p53 mediated cellular response. Reduced levels of S7 would eventually allow p53 to return to basal levels.
Discussion
Our studies have revealed a complex relationship between the ribosomal protein S7 and MDM2 that both extends previous observations with other ribosomal proteins and provides new insight into the role that nucleolar stress plays in stabilizing p53: (1) S7 levels in some cells are correlated with the stability of MDM2. When overexpressed, S7 stabilizes MDM2, while depletion of S7 leads to rapid MDM2 and p53 turnover after numerous types of genotoxic stress. In other cell types ablation of S7 leads to p53 dependent and independent cell cycle arrest. (2) MDMX plays a critical and interesting role in allowing MDM2 to be stabilized by S7. (3) S7 and another well studied ribosomal protein that interacts with MDM2, L11, are not mutually redundant in their impact on MDM2. (4) S7 itself is an excellent substrate for MDM2’s E3 ligase activity and S7 ubiquitination may serve to maintain the p53 response to DNA damage. Our findings not only characterize another ribosomal protein that intercepts the MDM2/p53 circuit, they implicate ribosomal stress as a critical integrating factor in regulating this circuit. Indeed, not only treatment with ActD (Figure 1D), but also Daunorubicin (data not shown) causes redistribution of ectopic S7 from nucleolus to the nucleoplasm. We envisage a pathway whereby, upon disruption of nucleoli caused by numerous stress signaling pathways, S7 is released into the nucleoplasm and then interacts with and represses the E3 activity of MDM2 in a manner that requires MDMX (Figure 7E). S7 is itself an MDM2 substrate and our data suggest that ubiquitinated S7 selectively destabilizes MDM2 thereby enhancing the p53 stress response until S7 levels subside (likely due to eventual polyubiquitination of S7) at which point p53 levels would return to ground state.
While our work was in progress it was reported that S7 binds MDM2 and inhibits MDM2-mediated p53 ubiquitination and degradation leading to cell death, while S7 ablation by a pool of siRNAs inhibits apoptosis in unstressed U2OS cells (Chen et al., 2007). In opposition to their results, using two individual S7 siRNAs we observed virtually no effect on levels of basal p53 and MDM2 in normally cycling U2OS cells, although the stabilization of both proteins was compromised following various forms of genotoxic stress. We also found that the population of U2OS cells undergoing apoptosis under normal growth conditions (or cells transfected with control siRNA) was very small (4% or less) and S7 siRNAs had no discernable effect on this low rate. This is consistent with our observation that ectopically expressed S7 has very little impact on apoptosis in these cells. Perhaps using a pool of siRNAs at a relatively high concentration as reported by Chen et al. can explain the differences between their and our data. Whatever the basis for the differences, both studies show that S7 is a new and interesting regulator of MDM2.
Going forward we pose three questions that can be approached in future studies regarding the regulation of the p53 stress response by S7 and other ribosomal proteins.
Why do ribosomal proteins and nucleoli regulate MDM2 function after genotoxic stress?
Numerous studies have implicated disruption of ribosomal biogenesis and nucleoli in the eukaryotic stress response (Boisvert et al., 2007; Mayer and Grummt, 2005). Laser ablation experiments have provided evidence that loss of nucleolar integrity is obligatory for activation of p53 after several forms of DNA damage (Rubbi and Milner, 2003). Disruption of nucleoli results in dispersion of nucleolar proteins into the nucleoplasm including reported MDM2-interacting ribosomal proteins (Andersen et al., 2005). It is thus somewhat surprising that loss of a single one (in our experiments either S7 or L11) markedly affects MDM2 levels, suggesting that S7 and L11 are not functionally redundant and may have unique modes by which they regulate MDM2. By contrast, L11 appears to be required for p53 mediated cell cycle arrest caused by depletion of S6 in A549 cells (Fumagalli et al., 2009). Alternately, at least in the cell types we used, these two ribosomal proteins may function similarly but levels of either one alone are inadequate for stabilizing MDM2 after nucleolar stress and release of both (and perhaps other nucleolar proteins as well) are required to reach a threshold necessary to inactivate MDM2. Different cell cycle stages or the duration and strength of a stress signal might also involve unique ribosomal proteins for regulation of MDM2. We speculate that the ribosomal/nucleolar stress response is so critical that there is considerable “overkill” to ensure that MDM2 is functionally compromised. The centrality and pleotropic nature of the p53 stress response is served extremely well by harnessing the availability of the very abundant and dynamic ribosomal proteins which are released from disrupted nucleoli.
What is the role of MDMX in inhibition of MDM2 by S7?
MDMX plays a critical and complex role in the regulation of MDM2 and p53 (Marine et al., 2007). Here we show that MDMX is involved in the inhibitory effects of S7 towards MDM2. It was reported that p53 activation by ribosomal stress requires degradation of MDMX in an MDM2-dependent fashion and that exogenous L11 (but not L5 and L23) stimulates MDMX polyubiquitination by MDM2 while L11 ablation reduces the downregulation of MDMX after ActD treatment (Gilkes et al., 2006). While we have not seen a similar impact of S7 on MDMX degradation (data not shown), our experiments reveal another role for MDMX which is to facilitate the ability of S7 to inhibit the E3 activity of MDM2. As MDMX is actively transported to nuclei after DNA damage to form MDM2/MDMX hetero-oligomers leading to its eventual destruction (Kawai et al., 2003; LeBron et al., 2006; Pereg et al., 2005), our results suggest that MDMX may have an important role in the nucleus prior to its destruction which would be to allow S7 to inhibit the MDM2 E3 ligase activity. MdmXmay work in this regard by providing a better MDM2-S7 interacting surface that allows the inhibitory effect of S7 on MDM2 autoubiquitination (and MDM2-mediated p53 ubiquitination). This may also be one of the mechanisms employed by cells to protect MDM2 from the inhibitory effect of S7 under unstressed conditions.
Why is S7 ubiquitinated by MDM2?
We were prompted to test whether S7 can be an MDM2 E3 ligase substrate by the fact that it binds to and competes with p53 for MDM2. We do not yet know the site(s) at which S7 is ubiquitinated by MDM2 nor do we have any direct evidence that S7 is targeted for proteasomal degradation by MDM2 although our data suggests that the S7-Ub fusion protein we generated differs from unubiquitinated S7 in three important ways. First, S7-Ub is unable to stabilize MDM2 or cooperate with MDMX although it retains the ability to stabilize p53. Second, unlike S7 alone, S7-Ub is capable of augmenting apoptosis. Third, S7-Ub is extensively polyubiquitinated especially in the cytoplasm (Figure S5). The selective inhibitory attenuation of MDM2 function by S7-Ub and S7-Ub’s increased apoptotic potential supports a feed-forward mechanism employed by MDM2/S7 to extend the period of p53 activation. Although S7-Ub may not replicate the behavior of S7 ubiquitinated by MDM2, our results with this construct are tantalizing and provide the basis for a model in which ubiquitinated S7 serves to extend p53 stabilization until it eventually becomes degraded (Figure 7E).
It is also possible that ubiquitination of S7 serves a different function than causing its proteasomal degradation. Yeast ribosomal protein L28 (L27a in human) was found to be polyubiquitinated in a cell cycle regulated manner. Its ubiquitination was suggested to be involved in translation regulation rather than degradation (Spence et al., 2000). Ubiquitination of proteins can affect their cellular location as well as their ability to function in various complexes and activities (Mukhopadhyay and Riezman, 2007).
While this work was in progress Oren and colleagues showed that another ribosomal protein L26, that stimulates translation of p53 mRNA after DNA damage (Takagi et al., 2005), can be ubiquitinated by MDM2 as a new means of holding p53 in check (Ofir-Rosenfeld et al., 2008). It remains to be determined which ribosomal proteins previously shown to interact with Mdm2 can also serve as its E3 ligase substrates.
Experimental Procedures
Plasmids and Cell Culture
Flag-MDM2, HA-p53 and HA-Ubiquitin were described previously (Poyurovsky et al., 2003). His-Ubiquitin (pcBH2Ub) was a kind gift from Dr. R. Baer (Columbia University). Myc-MDMX was constructed by subcloning MDMX fragment from Myc6-MDMX construct (Dr. A. Yoshimura, Kurume University, Japan) into pCMV-Myc vector between BamHI and XhoI restriction sites. To construct HA-MDMX baculovirus, an HA tag was inserted upstream of MDMX by PCR and HA-MDMX fragment was cloned into pFestBac HTa plasmid (Invitrogen) between RsrII and KpnI restriction sites. Flag-MDM2 baculovirus was a kind gift from Dr. S. Grossman (University of Massachusetts Medical School). Both baculoviruses were prepared according to the Bac-to-Bac baculovirus expression system (Invitrogen). Detailed procedures and the primers used for making constructs Myc-S7, Flag-MDM2 (Δ22), Flag-MDM2 (Δ23), Flag-MDM2 (Δ24), Flag-MDM2 (Δ34), Myc-S7Δ (Δ98–118) and S7-His6 are described in Supplemental Data.
U2OS, H1299, HCT116 (p53+/+ and p53−/−) (Dr. B. Vogelstein, Johns Hopkins Medical Institutions), or wild type and 2KO (p53−/−, mdm2−/−) mouse embryo fibroblast cells (Dr. G. Lozano, Anderson Cancer Center, Houston) were grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum at 37°C.
Antibodies and Drugs
Commercially obtained antibodies included: p53 (FL393-G, Santa Cruz), Flag (M2, Sigma), c-Myc (9E10, Santa Cruz), c-Myc (rabbit polyclonal, Sigma), HA (HA.11, Covance), GFP (B-2, Santa Cruz), p21 (C-19, Santa Cruz), and Actin (Sigma). Mouse monoclonal antibodies against human p53 (DO-1, 1801) and MDM2 (3G5, 4B11, 5B10, SMP14, 2A10) were used as supernatants from hybridoma cultures. A rabbit polyclonal antibody to rat S7 that was used in most of the Western blot analyses was generated using purified rat S7 protein as antigen (Lutsch et al., 1990). Another rabbit polyclonal antibody to human S7 was generated using S7-His6 protein as the antigen and produced by ProteinTech (Chicago, IL, USA). It was used for immunoprecipitation and some of the Western blot analyses. Rabbit polyclonal antibody to human L11 was a kind gift from Dr. H. Lu (Indiana University). Drugs used in this study are as follows: hydroxyurea, daunorubicin, doxorubicin, and deferoxamine mesylate from Sigma; Actinomycin D, MG132, and cyclohexlomide from Calbiochem; neocarzinostatin from Kayaku Co.(Tokyo, Japan).
Protein Purification
PK-Ubiquitin and His-UbcH5c were prepared as previously described (Poyurovsky et al., 2007). Insect Sf9 cells were infected with recombinant viruses encoding either Flag-MDM2, HA-MDMX, or both. Cells were lysed by sonication and soluble extracts bound to M2-Agarose (Sigma) in buffer A (50 mM Tris.Cl pH 8.0, 150 mM NaCl, 10% glycerol and protease inhibitors). Following extensive washing with buffer A containing 300 mM NaCl proteins were eluted with 1 mg/ml Flag-peptide (Sigma) in buffer A. For complex purification Flag-peptide eluted material was bound to HA-agarose (Roche) in buffer A and eluted with 1 mg/ml HA-peptide at 30°C. S7-His6 was expressed in BL21 (DE3) E. coli cells with 1 mM IPTG induction. Cell pellets were washed once with buffer B (50 mM Tris.Cl pH 8.0, 150 mM KCl, 0.5 mM PMSF), resuspended in buffer B containing 1 M LiCl, and then disrupted by sonication. Cleared cell lysates were incubated with Ni-NTA resin (Qiagen) for 2 hours. Following extensive washing the bound protein was eluted with buffer B containing 1 M LiCl and 250 mM imidozale. Samples were then dialyzed against 1 × PBS with 10% glycerol and stored at −80°C.
Transfection and Western Blot Analysis
Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. pcDNA3 plasmid was added to ensure equal amounts of total DNA used for transfecting cells. Total cell lysates were prepared and separated on SDS polyacrylamide gels, transferred to nitrocellulose and immunoblotted with the indicated antibodies as described previously (Gottifredi et al., 2001).
Ubiquitination and Degradation Assays
In vivo ubiquitination assays were carried out using either an HA-Ub or a His-Ub construct. For experiments using HA-Ub, cleared cell lysates were subjected to immunoprecipitation followed by Western blotting with anti-HA antibody. When His-Ub was used, a Ni-NTA pull down assay was carried out as previously described (Xirodimas et al., 2001). Eluted proteins were analyzed by Western blotting with anti-S7 antibody.
In vitro ubiquitination assays were carried out as previously described using purified proteins (Poyurovsky et al., 2007).
In vitro degradation assays were performed as previously described (Rape and Kirschner, 2004) with extracts of wild-type MEF cells or 2KO cells with or without ActD treatment (6 hours) using purified PK-Ubiquitin and His-UbcH5c protein. Aliquots were removed at indicated time points and subjected to Western blotting. Detailed procedures are described in Supplemental Data.
siRNA Interference
Individual pre-designed duplex small interfering RNA (siRNA) oligonucleotides targeting human S7 were purchased from Dharmacon (Lafayette, CO). The siRNA target sequences were #1 GGGCAAGGATGTTAATTTT; #2 CTAAGGAAATTGAAGTTGG. Reported siRNA oligonucleotides targeting human L11 (Bhat et al., 2004) and luciferase (Urist et al., 2004) were obtained from Invitrogen (Carlsbad, CA). Transfection was performed using Dharmafect 1 (Dharmacon, Lafayette, CO) according to the manufacturer's instructions.
Immunofluorescent Staining
U2OS cells grown on coverslips were transfected with Myc-S7 (1.5 µg). Twenty hours after transfection, cells were either untreated or treated with ActD (5 nM) for 7 hours. Immunofluorescent staining was carried out as described previously (Karni-Schmidt et al., 2007). Rabbit polyclonal anti-Myc antibody and secondary Alexa fluor 594 donkey anti-rabbit antibody (Molecular Probes, Eugene, OR) were used to detect Myc-S7. Images were analysed by confocal laser scanning microscopy (Olympus model 1X81) using Fluoview software (Canter Valley, PA). DIC imaging was used in parallel to directly visualize cells.
Supplementary Material
Acknowledgements
We thank Ella Freulich for expert technical assistance and members of the Prives’ laboratory for helpful suggestions. This work was supported by NIH grant CA58316.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yan Zhu, Department of Biological Sciences, Columbia University, New York 10027, USA.
Masha V. Poyurovsky, Department of Biological Sciences, Columbia University, New York 10027, USA
Yingchun Li, Department of Biological Sciences, Columbia University, New York 10027, USA.
Lynn Biderman, Department of Biological Sciences, Columbia University, New York 10027, USA.
Joachim Stahl, Max Delbruck Center for Molecular Medicine, Berlin 13125, Germany..
Xavier Jacq, Hybrigenics, Paris 75014, France.
Carol Prives, Department of Biological Sciences, Columbia University, New York 10027, USA.
References
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. The EMBO journal. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Molecular cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nature cell biology. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of biological chemistry. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and cellular biology. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1:1027–1035. [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. The EMBO journal. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. The Journal of biological chemistry. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Molecular and cellular biology. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, Bouvet P, Sheetz M, Fersht A, Prives C. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene. 2007;26:3878–3891. doi: 10.1038/sj.onc.1210162. [DOI] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. The Journal of biological chemistry. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- Klibanov SA, O'Hagan HM, Ljungman M. Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. Journal of cell science. 2001;114:1867–1873. doi: 10.1242/jcs.114.10.1867. [DOI] [PubMed] [Google Scholar]
- Kurki S, Peltonen K, Latonen L, Kiviharju TM, Ojala PM, Meek D, Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- Latonen L, Kurki S, Pitkanen K, Laiho M. p53 and MDM2 are regulated by PI-3-kinases on multiple levels under stress induced by UV radiation and proteasome dysfunction. Cellular signalling. 2003;15:95–102. doi: 10.1016/s0898-6568(02)00044-x. [DOI] [PubMed] [Google Scholar]
- LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. The EMBO journal. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. Vol. 302. New York, N.Y: 2003. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2; pp. 1972–1975. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Deisenroth C, Zhang Y. Cell cycle. Vol. 6. Georgetown, Tex: 2007a. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions; pp. 434–437. [DOI] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Molecular and cellular biology. 2007b;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nature cell biology. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- Lutsch G, Stahl J, Kargel HJ, Noll F, Bielka H. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. European journal of cell biology. 1990;51:140–150. [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Molecular and cellular biology. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. Journal of cell science. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. Cell cycle. Vol. 4. Georgetown, Tex: 2005. Cellular stress and nucleolar function; pp. 1036–1038. [DOI] [PubMed] [Google Scholar]
- Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res. 2003;1:1017–1026. [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Science. Vol. 315. New York, N.Y: 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling; pp. 201–205. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Molecular cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, Shiloh Y. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Jacq X, Ma C, Karni-Schmidt O, Parker PJ, Chalfie M, Manley JL, Prives C. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Molecular cell. 2003;12:875–887. doi: 10.1016/s1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The MDM2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. The EMBO journal. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes & development. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. The EMBO journal. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Nie L, Maki CG. MDM2 binding induces a conformational change in p53 that is opposed by heat-shock protein 90 and precedes p53 proteasomal degradation. The Journal of biological chemistry. 2007;282:14626–14634. doi: 10.1074/jbc.M610514200. [DOI] [PubMed] [Google Scholar]
- Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. The Journal of biological chemistry. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- Saxena A, Rorie CJ, Dimitrova D, Daniely Y, Borowiec JA. Nucleolin inhibits Hdm2 by multiple pathways leading to p53 stabilization. Oncogene. 2006;25:7274–7288. doi: 10.1038/sj.onc.1209714. [DOI] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. The Journal of biological chemistry. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, Saville MK, Jochemsen AG. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO reports. 2001;2:1029–1034. doi: 10.1093/embo-reports/kve227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. The Journal of biological chemistry. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature reviews. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes & development. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nature cell biology. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.