Abstract
The Williams–Beuren syndrome (WBS) region at 7q11.23 is subject to several genomic rearrangements, one of which, the WBSinv-1 variant, is an inversion polymorphism. The WBSinv-1 chromosome has been shown to occur frequently in parents of individuals with WBS, implying that it predisposes the region to the WBS deletion. Here we investigate two WBS families with multiple affected children, and show that in one family, both siblings have a deletion on a WBSinv-1 chromosome background that arose due to interchromosomal recombination. These results suggest that the two WBS deletions in this family were independent events, and that there is likely a significant increase in the risk of deletion of the WBS region associated with the WBSinv-1 chromosome. The rarity of multiplex WBS families would suggest that the overall risk of having a child with WBS is still relatively low; however, families with an existing member with WBS may choose to opt for WBSinv-1 testing and genetic counseling.
Introduction
Williams–Beuren syndrome (WBS) is a multisystem developmental disorder with an incidence of between 1/7,500 and 1/20,000 (Greenberg 1990; Stromme et al. 2002) caused by the hemizygous deletion of chromosome 7q11.23 (Ewart et al. 1993). A 1.55-Mb commonly deleted interval is present in more than 95% of individuals with clinically diagnosed WBS and the mechanism of deletion is unequal meiotic recombination between directly aligned, related segments of DNA sequence flanking the region (Urban et al. 1996; Dutly and Schinzel 1996; Bayés et al. 2003). These large DNA blocks share more than 95% DNA nucleotide similarity, with up to 99.5% identity in some regions (Bayés et al. 2003). Since these chromosome 7 low-copy repeats (LCRs) are apparently present in all individuals it was predicted that the occurrence of non-allelic homologous recombination was a largely random event. Consequently the predicted risk in any particular family of a chromosomal rearrangement occurring that would result in deletion of the WBS region would be expected to be identical.
We previously reported the WBSinv-1 variant of the WBS region in one third of transmitting parents of individuals with WBS (Osborne et al. 2001). This inversion spanned around 1.9 Mb of DNA, including the commonly deleted interval, and was predicted to stem from recombination between blocks of DNA within the LCRs that are aligned in an inverted orientation. The genomic variant is found in approximately 5% of non-WBS controls (Hobart et al. 2004) but in 25–33% of transmitting WBS parents (Osborne et al 2001; Bayés et al. 2003; Hobart et al. 2004), suggesting its presence predisposes the chromosome to subsequent meiotic rearrangements, specifically deletion of the WBS region. If this were the case, the deletion of the WBS region would not simply be a stochastic event but instead carriers of the WBSinv-1 would have an elevated risk of having a child with a deletion, resulting occasionally (depending upon the increase in risk) in recurrence within a family.
We identified two families in which two siblings each had a clinical and molecular diagnosis of typical WBS and investigated whether the recurrence of WBS could be due to a WBSinv-1 in the transmitting parent.
Materials and methods
Families with WBS
The affected siblings from one family (S), which had previously been described in the literature prior to the discovery of the WBSinv-1, were hypothesized to be due to maternal germline mosaicism (Kara-Mostefa et al. 1999). The second family (H) was newly ascertained by us (K.W.G. and L.N.) and consisted of two siblings. The younger sister presented with failure to thrive, developmental delay and seizures and was diagnosed at age 15 months. Subsequently her brother, aged 5 years 5 months, was also shown to carry the typical deletion. Both patients have typical physical and behavioral characteristics. These studies were approved by the University of Toronto Research Ethics Board and informed consent was obtained from each participant. Unfortunately, a blood sample from the father in family H was not available for study.
Polymorphic marker analysis
All members of family H and family S were genotyped at 13 polymorphic loci spanning 7q11.23 (listed in Table 2). The PCR reactions were performed using 20 ng of genomic DNA template, 200 μM dNTPs, 10 × PCR buffer II (Applied Biosystems), 25 mM MgCl2 and 1 U AmpliTaq DNA polymerase (Applied Biosystems). Cycles were performed as follows: 94°C for 5 min; 25–28 cycles of 94°C for 30 s, an appropriate annealing temperature for 45 s, and 72°C for 30 s; and a final extension of 72°C for 15 min. Annealing temperatures for each primer pair were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). One-microliter volumes of each PCR reaction were suspended in a 10-μl mixture of 5 μl GeneScan 500HD[liz] size standard in 980 μl Hi–Di formamide (Applied Biosystems). The samples were run on an ABI3730xL genetic analyzer (Applied Biosystems) using the POP7 polymer and dye set G5. Results were analyzed using the software GeneMapper, version 3.5.
Table 2.
Genotyping analysis of family H with two WBS siblings
| DNA SAMPLE | MARKER | ALLELE 1 | ALLELE 2 | COMMENTS | |
|---|---|---|---|---|---|
| Size in bp | |||||
| Mother | D7S1766 | 135 | 135 | Paternal allele present* | |
| WBS Sibling 1 | 139 | 135 | |||
| WBS Sibling 2 | 139 | 135 | |||
| Mother | D7S2415 | 131 | 137 | Paternal allele present* | |
| WBS Sibling 1 | 129 | 137 | |||
| WBS Sibling 2 | 129 | 131 | |||
| Mother | D7S653 | 209 | 215 | Uninformative† | |
| WBS Sibling 1 | 209 | 215 | |||
| WBS Sibling 2 | 209 | 209 | |||
| Mother | D7S672 | 146 | 158 | Uninformative† | |
| WBS Sibling 1 | 146 | 158 | |||
| WBS Sibling 2 | 146 | 146 | |||
| Common WBS deletion | Mother | D7S2476 | 149 | 162 | Single maternal allele |
| WBS Sibling 1 | 162 | ||||
| WBS Sibling 2 | 149 | ||||
| Mother | D7S613 | 117 | 117 | Single maternal allele | |
| WBS Sibling 1 | 117 | ||||
| WBS Sibling 2 | 117 | ||||
| Mother | D7S2472 | 87 | 87 | Single maternal allele | |
| WBS Sibling 1 | 87 | ||||
| WBS Sibling 2 | 87 | ||||
| Mother | D7S3194 | 199 | 207 | Single maternal allele | |
| WBS Sibling 1 | 199 | ||||
| WBS Sibling 2 | 207 | ||||
| Mother | D7S3195 | 409 | 409 | Single maternal allele | |
| WBS Sibling 1 | 409 | ||||
| WBS Sibling 2 | 409 | ||||
| Mother | D7S3196 | 226 | 226 | Single maternal allele | |
| WBS Sibling 1 | 226 | ||||
| WBS Sibling 2 | 226 | ||||
| Mother | D7S1870 | 132 | 134 | Single maternal allele | |
| WBS Sibling 1 | 132 | ||||
| WBS Sibling 2 | 134 | ||||
| Mother | D7S675 | 210 | 212 | Paternal allele present* | |
| WBS Sibling 1 | 208 | 212 | |||
| WBS Sibling 2 | 208 | 210 | |||
| Mother | D7S669 | 182 | 182 | Paternal allele present* | |
| WBS Sibling 1 | 174 | 182 | |||
| WBS Sibling 2 | 174 | 182 | |||
Alleles that are not present in the mother, and must therefore have been inherited from the father are italicized
The presence of a paternally inherited allele could not be proven and these were termed uninformative
Fluorescence in situ hybridization
Deletion detection was performed by fluorescence in situ hybridization (FISH) of metaphase spreads using biotin-labeling kits (Oncor) according to the protocols provided by the manufacturer. WBSinv-1 inversion testing was carried out on both blood and transformed lymphoblastoid cell lines from each family member, according to previously described protocols (Osborne et al. 2001). Chromosome spreads for interphase FISH analysis were prepared from peripheral blood lymphocytes using standard methodologies. Three-color FISH was performed with two probes located within the commonly deleted region (RP5-1186P10 at the GTF2IRD1 locus and CTA-208H19 at the FZD9 locus) and one probe located telomeric to the WBS deleted region CTB-139P11 at the HIP1 locus (Osborne et al. 2001). Purified BAC DNAs were labeled by nick-translation (Roche), and unincorporated nucleotides were removed by ethanol precipitation. The three probes were labeled separately with digoxigenin, biotin, and a 3:2 mix of digoxigenin:biotin. Hybridization was performed using standard protocols and was detected with anti-digoxigenin-rhodamine (red), avidin-FITC (green) or both (yellow) (Roche). Nuclei were counterstained with DAPI (Sigma).
SSN analysis
The PCR primers, restriction enzyme digests and product sizes are detailed in Table 1. PCR reactions (25 μl) were set up with 50 ng of genomic DNA, 10 pmols of each primer, and 0.2 U of Taq polymerase (Ecogen) in the manufacturer’s buffer. Cycles were performed as follows: 94°C for 5 min; 25–28 cycles of 94°C for 40 s, 60°C for 30 s, and 72°C for 40 s; and a final extension of 72°C for 10 min. The amplimers were digested with restriction enzymes according to the manufacturer’s instructions (New England Biolabs), and the products size-fractionated on 3% agarose gels.
Table 1.
SSN amplimers
| SSN | Location | Primers | Restriction enzyme and sizes (bp) |
|---|---|---|---|
| SSN10 | GTF2IRD2 Intron 4 |
Forward: 5′-TGCAAGGTCGTAATTCTCAGG-3′ Reverse: 5′-TGTCTTTCCATAGGCATGAAGA-3′ |
MspI Bt: 42; 201; 753 Bc+Bm: 42; 954 |
| SSN11 | GTF2IRD2 Intron 3 |
Forward: 5′-TTGTAAAATGGTGTTTATTTTAGG-3′ Reverse: 5′-GCCCCACAAACTTGGATCTG-3′ |
Tru9I Bt: 20; 51; 242; 156 Bc+Bm: 20; 51; 100; 142; 156 |
A digital image of the gel was captured at varying exposure times, to ensure that the bands were not saturated. Then, intensities of bands corresponding to blocks Bc, Bm, and Bt were quantified by use of the Volume Tool from the Quantity One software package (Bio-Rad). Relative intensities were calculated by means of a dosage quotient for Bt relative to [Bc+Bm] and a final ratio was calculated by relating the dosage obtained for each WBS sibling to the mean dosage value of their mother. Each experiment was performed at least twice with a different number of PCR cycles. A WBS family whose parents do not carry an WBSinv-1 inversion, and WBS proband with a transmitting parent who does carry WBSinv-1 were used as controls in the same assay.
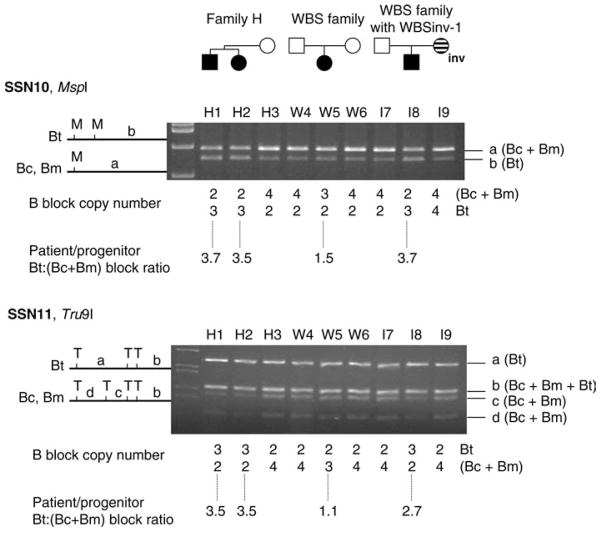
By calculating the patient/progenitor block B ratio, patient Bt:(Bc+Bm)/progenitor Bt:(Bc+Bm), we can determine whether the deletion in a WBS proband occurred on a normal or a WBSinv-1 chromosome background. We have previously demonstrated that patient/progenitor ratios are significantly different for a WBS proband when the deletion occurs on a normal chromosome, as opposed to when it occurs on a WBSinv-1 chromosome (Bayés et al. 2003). For SSN10, the patient/proband ratio is 1.33 (mean 1.43; max 1.75; min 1.09; SD 0.23) for a deletion on a normal chromosome, and 3 (mean 3.83; max. 4.55; min. 2.95; SD 0.53) for a deletion on a WBSinv-1 chromosome. For SSN11, the patient/proband ratio is 1.33 (mean 1.22; max. 1.68; min. 1.0; SD 0.21) for a deletion on a normal chromosome, and 3 (mean 3.44; max. 4.62; min. 2.62; SD 0.61) for a deletion on a WBSinv-1 chromosome.
Results
Polymorphic marker analysis
To determine which parental chromosome 7 had undergone non-allelic homologous recombination, all family members for whom DNA was available were genotyped at 13 polymorphic loci spanning 7q11.23.
In family S, the affected siblings carried a deletion that occurred on a chromosome inherited from their mother, as originally reported (data not shown). In family H, the siblings displayed only maternal alleles throughout the commonly deleted WBS region, although they had inherited non-maternal alleles outside this region, demonstrating that their deletion had occurred on a paternally inherited chromosome (Table 2). Both siblings were also found to have the same paternal alleles for both proximal and distal markers, suggesting that they both inherited the same 7q11.23 chromosome region from their father.
FISH analysis to detect WBSinv-1
Affected individuals (but not parents or unaffected siblings) from both families had a deletion of 7q11.23 confirmed by metaphase FISH analysis. We then performed three-colour interphase FISH analysis to identify inversions of the WBS region. Initially, no inversions were readily observed in any of the available samples from families, however, because in family H we were unable to test the transmitting parent, we chose an alternative method of detecting the presence of WBSinv-1 in this family.
Detection of WBSinv-1 using site-specific nucleotide analysis
This method utilizes dosage analysis of site-specific nucleotide differences (SSNs) between the centromeric, medial and telomeric blocks of repeat. We have previously shown that the copy number of block-specific SSNs in individuals with WBS is different when the deletion has occurred on a normal versus an WBSinv-1 chromosome (Bayés et al. 2003). Therefore, by analyzing the relative dosage of sequences within the B-block in an individual with a WBS deletion we can infer the inversion status of the parental chromosome on which the deletion occurred.
Two SSNs within the B-blocks from the centromeric (Bc), medial (Bm) and telomeric (Bt) LCRs were used in this study. Both SSN10 and SSN11 distinguish the Bt copy with respect to the other two copies, Bc and Bm. The SSN10 is characterized by a nucleotide change in Bt that creates an MspI site not present in Bc or Bm, whilst SSN11 is characterized by nucleotide change in Bt that destroys a Tru9I site present in Bm. We have shown that the interchromosomal exchange, which generates a deletion in an inversion carrier takes place within the last 38 kb of blocks Bm and Bt (Bayés et al. 2003). This event results in a gain of a Bt-type block within the region covered by SSN10 and SSN11 if the rearranged WBS chromosome originated in an inversion carrier. This gain of a Bt-type block can be identified by calculating the patient/progenitor ratio, as described in Materials and methods.
Analysis of family H using SSN analysis and dosage comparisons showed patient/progenitor ratios of 3, indicating a de novo gain of a telomeric-type block B and deletion of a medial-type block B: a pattern consistent with a typical 1.55-Mb deletion due to interchromosomal exchange between misaligned chromosomes in an inversion carrier (Fig. 1; Bayés et al. 2003). This pattern was also seen in the WBS proband where the deletion was known to have occurred on a WBSinv-1 chromosome, but not in the WBS proband where neither parent carried the WBSinv-1 variant. In family H, since the mother was shown to not carry the WBSinv-1 polymorphism, and both siblings displayed only maternally inherited alleles throughout the common WBS deletion region, it can be concluded that the WBSinv-1 chromosome that underwent interchromosomal exchange was inherited from the father.
Fig. 1.
SSN analysis of WBS families. SSN analysis of samples from family H, a WBS family without WBSinv-1, and a WBS family where the WBS deletion arose on a WBSinv-1 progenitor chromosome. Relative block dosages calculated from band intensities are depicted below each lane. Representative results for the SSN10 and SSN11 assays, which allow the detection of the WBSinv-1 in patients with WBS, are shown. Details of these assays are presented in Table 1 and in the main text. SSN10: As represented in the scheme, a nucleotide change in block Bt introduces an MspI site that is not present in blocks Bc and Bm. The WBS family proband (W5) shows two digested copies of fragment b (block Bt) versus three undigested copies of fragment a (blocks Bc and Bm), as shown below the gel. The WBSinv-1 proband, however (I8), shows a reduction of the intensity of fragment a (Bc+Bm) to two copies and an increase of the intensity of fragment b (Bt) to three copies. A ratio of the patient to progenitor B blocks is calculated from these copy numbers using the following equation: patient Bt:(Bc+Bm) ratio/progenitor Bt:(Bc+Bm) ratio, as previously described in Bayes et al. 2003. These ratios are shown at the bottom of the figure. A patient/progenitor block B ratio of approximately 3 indicates that in the WBSinv-1 proband I8, the WBS chromosome arose in a progenitor heterozygous for the inversion, I9. SSN11: As represented in the scheme, a nucleotide change in block Bt destroys a Tru9I site that is present in blocks Bc and Bm. The WBS family proband (W5) shows three digested copies of fragments corresponding to blocks Bc and Bm, versus two undigested copies of fragments corresponding to block Bt. The WBSinv-1 proband, however (I8), shows a reduction of the intensity of fragments c and d (Bc+Bm) and an increase of the intensity of fragment a (Bt). The patient/progenitor block B ratio of approximately 3 indicates that in the WBSinv-1 family proband I8, the WBS chromosome arose in a progenitor heterozygous for the inversion, I9. Dosage calculations in family H show that the two WBS siblings (H1 and H2) have patient/progenitor block B ratios of 3 for both SSN10 and SSN11, indicating that their deletions both arose in a progenitor heterozygous for WBSinv-1
Discussion
Our finding that two siblings in family H carry a deletion of the WBS region that has occurred on the background of an inverted chromosome inherited from their father, supports the hypothesis that the WBSinv-1 variant confers an increased risk of deletion during meiotic recombination. It is likely that the inversion of inv(7)(q11.23) on one chromosome 7 causes misalignment of the WBS region between sister chromatids, resulting in deletion and/or duplication of the region if non-homologous recombination takes place.
We have recently identified two deletion breakpoint hotspots within the final 38-kb region of block B that are associated exclusively with deletions in which the transmitting progenitor is an inversion carrier. One of these hotspots is associated with maternal crossovers, and one with paternal crossovers (Rivera et al. 2004). Direct sequencing of a PCR product spanning the deletion breakpoint in the family H siblings revealed, as expected, that both had deletion breakpoints within the 950-bp hotspot associated with paternal crossovers.
While we could not definitively prove that the deletions were independent events by sequence analysis, such deletion events could only occur in a progenitor carrying an inversion by means of interchromosomal recombination during meiosis I (Bayés et al. 2003). This effectively excludes the occurrence of a premeiotic event leading to gonadal mosaicism in the father. We did not, however, find the WBSinv-1 in family S, but the occurrence of multiply affected siblings in this case can be explained by maternal gonadal mosaicism for the deletion which could occur through premeiotic intrachromosomal recombination in the mother (Kara-Mostefa et al. 1999). Although these are currently the only two families with WBS siblings reported, both likely arise due to the existence of a progenitor genomic rearrangement (maternal mosaic deletion in family S and paternal inversion variant in family H).
An accurate estimate of the risk of having a child with WBS associated with the presence of the WBSinv-1 variant being present in a parents is currently not known and would require the sampling of its frequency in a large number of chromosomes from the general population. There are efforts to complete such a study and its value will have added impact since an initial survey estimates the population frequency of the WBSinv-1 variant at 5% (Hobart et al. 2004). With the relatively high frequency of the WBSinv-1 variant, the incidence of WBS might be expected to be somewhat higher than the current estimate of 1 in 7,500 (Stromme et al. 2002). It may be that carrier status for WBSinv-1 is associated with a higher incidence of spontaneous fetal loss, but such potential associations can only be investigated in a large, population-based study.
Although WBS is not a common disorder, the perceived elevation of risk associated with this genomic polymorphism should not be underestimated. Even a several-fold increase in risk for WBSinv-1 carriers might be sufficient for many families who already have a child or a close relative with WBS to opt for inversion testing, and subsequent prenatal deletion testing if either parent were found to carry the inversion. Indeed, in our experience a number of WBS families are already searching for this information for these precise reasons (S.W.S., L.R.O. and L.A.P.-J.). Ultimately, the parents would need to be counseled for the risk of WBS-associated chromosome rearrangements occurring due to their genetic composition, weighed against odds of other potential negative outcomes brought on by administering the genetic testing. Our findings in this report provide the first molecular characterization of multiply affected WBS families most likely resulting because of predisposing genetic factors.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), Sick Kids Foundation, the Spanish Ministries of Health and Science and Technology and Genome Canada/Ontario Genomics Institute and Genome Spain. S.W.S. is a CIHR Investigator and an International Scholar of the Howard Hughes Medical Institute and L.R.O. is a CIHR Scholar. We thank the patients and their families for their participation.
Contributor Information
Stephen W. Scherer, Genetics and Genomic Biology Program, Sick Kids Hospital, Toronto, ON, Canada, M5G 1X8, Department of Molecular and Medical Genetics, University of Toronto, Toronto, Ontario, Canada
Karen W. Gripp, Division of Medical Genetics, Alfred I. duPont Hospital for Children, Wilmington, Delaware, USA
Jaume Lucena, Unitat de Genetica, Departament de Ciencies Experimentals i de la Salut, Universitat Pompeu Fabra, 08003, Barcelona, Spain.
Linda Nicholson, Division of Medical Genetics, Alfred I. duPont Hospital for Children, Wilmington, Delaware, USA.
Jean-Paul Bonnefont, Department of Genetics, Hopital Necker-Enfants Malades, Assistance Publique-Hopitaux de Paris, Paris, France.
Luis A. Pérez-Jurado, Unitat de Genetica, Departament de Ciencies Experimentals i de la Salut, Universitat Pompeu Fabra, 08003, Barcelona, Spain
Lucy R. Osborne, Department of Molecular and Medical Genetics, University of Toronto, Toronto, Ontario, Canada, Department of Medicine, University of Toronto, 7238 Medical Sciences Building, 1 King’s College Circle, Toronto, ON, M5S 1A8, Canada, Tel.: +1-416-9465804, Fax: +1-416-9788765
References
- Bayés M, Magano LF, Rivera N, Flores R, Pérez Jurado LA. Mutational mechanisms of Williams–Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutly F, Schinzel A. Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams–Beuren syndrome. Hum Mol Genet. 1996;5:1893–1898. doi: 10.1093/hmg/5.12.1893. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Greenberg F. Williams syndrome professional symposium. Am J Med Genet. 1990;6(Suppl):85–88. [Google Scholar]
- Hobart HH, Gregg RG, Mervis CB, Robinson BF, Kimberley KW, Rios CM, Morris CA. Heterozygotes for the microinversion of the Williams–Beuren syndrome region have an increased risk for affected offspring. Am Soc Hum Genet; 54th Annual Meeting; Toronto. 2004. Abstr. 891. [Google Scholar]
- Kara-Mostefa A, Raoul O, Lyonnet S, Amiel J, Munnich A, Vekemans M, Magnier S, Ossareh B, Bonnefont JP. Recurrent Williams–Beuren syndrome in a sibship suggestive of maternal germ-line mosaicism. Am J Hum Genet. 1999;64:1475–1478. doi: 10.1086/302362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW. A 1.5 million-base pair inversion polymorphism in families with Williams–Beuren syndrome. Nat Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera N, Lucena J, Bayés M, Pérez-Jurado LA. Sex-preferential hotspots for non-allelic homologous recombination within segmental duplications in Williams–Beuren syndrome deletions. Am Soc Hum Genet; 54th Annual Meeting; Toronto. 2004. Abstr. 849. [Google Scholar]
- Stromme P, Bjornstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- Urban Z, Helms C, Fekete G, Csiszar K, Bonnet D, Munnich A, Donis-Keller H, Boyd C. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover. Am J Hum Genet. 1996;59:958–962. [PMC free article] [PubMed] [Google Scholar]