Summary
MicroRNAs (miRNAs) are ~21 to 24-nucleotide RNAs that mediate repression of messenger RNA (mRNA) translation through recognition of specific miRNA binding sites usually located in the 3′ nontranslated region. Designed to simulate miRNAs, small interfering RNAs represent a powerful genetic approach to potently inhibit gene expression by mediating cleavage of the intended mRNA target. This strategy has been applied successfully to suppress replication of several viruses, including human immunodeficiency virus type 1 (HIV-1). However, recent evidences indicate that viral RNAs may themselves be processed, to some extent, by the endogenous miRNA biosynthetic machinery in mammalian cells, extending previous observations in plants. The resulting viral miRNAs may exert regulatory effects towards host and/or viral genes that may influence viral replication and modulate the course of infection. Viral miRNA generation and/or action may be limited by counteraction through inhibitory viral RNAs and/or proteins. This review article will focus on the relationship between HIV-1 and miRNA-guided RNA silencing, and discuss the different aspects of their interaction. As we learn more about the mechanism and importance of small RNA-based antiviral systems, a more intricate picture of the interaction between HIV-1 and a proven antiviral defense mechanism in lower eukaryotes is emerging.
Keywords: HIV-1, RNA silencing, microRNA, small interfering RNA, gene expression
A new family of small regulatory RNAs
MicroRNAs (miRNAs) are short ~21 to 24-nucleotides (nt) RNA species expressed in most eukaryotes examined to date, such as fungi, plants and animals, and known as key regulators of gene expression (Kim, 2005; Zamore and Haley, 2005). According to the latest update of miRBase, the repository of miRNA data on the web, more than 326 different miRNA sequences giving rise to 319 unique mature miRNAs have been identified in human, 234 of which have been experimentally validated (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006). A recent study suggested that the number of miRNAs, which may regulate as much as 30 to 50% of the human genes (Lewis et al., 2005; Xie et al., 2005), may reach up to 1000 (Berezikov et al., 2005)!
The endogenous miRNA-based RNA silencing machinery
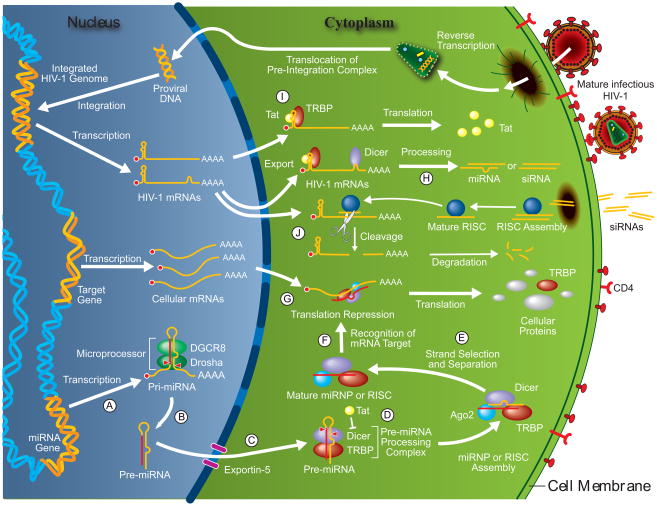
Encoded by the genome of most eukaryotes examined so far, miRNA genes are transcribed by RNA polymerase (pol) II into stem-loop structured primary miRNAs (pri-miRNAs) (see Figure 1-A). As illustrated in Figure 1-B, these pri-miRNAs are then trimmed into miRNA precursors (pre-miRNAs) by the nuclear ribonuclease (RNase) III Drosha (Lee et al., 2003), acting in concert with the DiGeorge syndrome critical region 8 (DGCR8) protein within the microprocessor complex (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004). The resulting ~60–70-nt pre-miRNAs are then exported via Exportin-5 (Bohnsack et al., 2004; Brownawell and Macara, 2002; Lund et al., 2004; Yi et al., 2003) to the cytoplasm (see Figure 1-C), where they are cleaved at the base of the loop by the RNase III Dicer to generate miRNA:miRNA* duplexes (Bernstein et al., 2001; Provost et al., 2002; Zhang et al., 2002; Zhang et al., 2004). Dicer was recently shown to operate together with the transactivating response RNA-binding protein (TRBP) (Gatignol et al., 1991) within a pre-miRNA processing complex (Chendrimada et al., 2005; Haase et al., 2005) (see Figure 1-D). Following a strand selection and separation step (see Figure 1-E), which is based on the thermodynamic stability of the RNA duplex (Schwarz et al., 2003), the miRNA strand (~21 to 24-nt) with the least stable 5′ end pairing is incorporated into effector miRNA-containing ribonucleoprotein (miRNP) complexes, containing Argonaute 2 (Ago2), TRBP and Dicer (Chendrimada et al., 2005), and guiding them towards specific messenger RNAs (mRNAs) (see Figure 1-F). The opposite miRNA* strand is encountered much less frequently and is presumably degraded (Matranga et al., 2005). miRNA assembly on specific mRNA sequences may be facilitated by the fragile X mental retardation protein, which can accept and use miRNAs derived from Dicer (Plante et al., 2006). The targeted mRNA will be primarily subjected to translational repression (see Figure 1-G), although mRNAs containing partial miRNA complementary sites may also be targeted for degradation in vivo (Bagga et al., 2005). These regulatory events may occur at specific cytoplasmic foci referred to as processing bodies (P-bodies) (Liu et al., 2005; Teixeira et al., 2005), or GW182-containing bodies (GW-bodies) (Eystathioy et al., 2002), that are enriched in proteins involved in mRNA degradation (Eystathioy et al., 2003), such as Ago2 (Liu et al., 2005).
Figure 1.
Schematic representation of the possible interactions between HIV-1 and the host miRNA-guided RNA silencing pathway. miRNA genes are transcribed into primary miRNAs (pri-miRNAs) (A), which are trimmed into miRNA precursors (pre-miRNAs) by the microprocessor complex (B). Pre-miRNAs are then exported via Exportin-5 to the cytoplasm (C), where they are cleaved by the pre-miRNA processing complex to generate miRNA:miRNA* duplexes (D). Following a strand selection and separation step (E), the mature miRNA is incorporated into effector miRNA-containing ribonucleoprotein (miRNP) complexes to mediate recognition (F) and translational repression (G) of specific mRNA targets. Secondary structures in HIV-1 mRNAs may themselves be recognized and processed by Dicer into viral miRNAs or siRNAs (H). TAR is the target for the viral transactivating protein Tat (I), which has been suggested to inhibit Dicer activity (D). Conversely, cellular transactivating response RNA-binding protein (TRBP), which cooperates synergistically with Tat function (I), was recently reported to assist Dicer function in RNA silencing (D). Finally, siRNAs targeting HIV-1 and introduced in an infected cell can be accommodated by the endogenous machinery to form a mature RNA-induced silencing complex (RISC) that will cleave HIV-1 mRNAs (J).
Biological roles of miRNAs
miRNAs have been shown to control various processes, such as cell proliferation and apoptosis in flies, and hematopoietic cell differentiation in mice (Bartel, 2004). Their biological role is linked to their ability to repress translation of specific mRNAs. This is accomplished through recognition of specific miRNA binding sites usually located in the mRNA 3′ nontranslated region (NTR), thereby inhibiting translation initiation (Pillai et al., 2005). Because recognition by miRNAs is mainly based on imperfect sequence complementarity, the identification of their physiological mRNA targets is difficult to predict and is rather arduous. Characterization of a few experimentally validated miRNA:mRNA interactions (eg, let-7 and lin-41) (Vella et al., 2004), allowed to establish a context in which this interaction is favored. For example, the critical miRNA:mRNA pairing region, referred to as the “miRNA seed”, involves nt 2 to 8 of the miRNA in the 5′ to 3′ orientation. Pairing of the miRNA 3′ region appears to be less important, but may compensate a weaker binding of the 5′ region (Vella et al., 2004). A better understanding of mRNA recognition by miRNAs helped develop bioinformatic approaches that have proven to be instrumental for identifying potential miRNA targets and initiating characterization of miRNA function.
A role for small RNAs in antiviral host defenses
In addition to fulfilling important gene regulatory functions in their eukaryotic hosts, small RNAs may also help defend against invasion of the host genome by RNAs of foreign origin, such as viruses. Initial evidences for such a role came from observations made by plant biologists. Indeed, while investigating the natural antiviral defense mechanism known as posttranscriptional gene silencing (PTGS), Hamilton and Baulcombe detected the presence of antisense viral RNA of ~25-nt in virus-infected plants by Northern blot (1999). The authors noted that these small RNAs, which were later found to originate from viral double-stranded RNA (dsRNA) processing by Dicer or DICER-LIKE 1 (DCL1 in Arabidopsis) (Reinhart et al., 2002), were long enough to convey sequence specificity and suggested their probable role in limiting virus infection in plants. The antiviral function of small RNAs and their biosynthetic machinery in plants has recently been extended to insects (Li et al., 2002), nematodes (Lu et al., 2005; Wilkins et al., 2005) and mammals.
Biology of HIV-1
The genome of HIV-1 is composed of two identical single stranded 9.6 kb RNA molecules. The viral genetic material must be reverse transcribed into DNA and integrated into the host genome before directing viral gene expression. HIV-1 relies heavily on the cellular transcription and translation machineries for the synthesis of viral genomic RNA and proteins. The full-length RNA is transcribed by RNA pol II and serves both as genomic RNA and as a template for expression of the structural proteins Gag and Gag-Pol. It is also spliced into multiple 4 kb and 2 kb mRNA species encoding the envelope (Env) proteins as well as several non-structural proteins. All these RNA species adopt complex and dynamic secondary structures that may resemble miRNA precursors and thus could potentially be targeted by the host miRNA-guided RNA silencing machinery (see Figure 1-H). Although the high variability of the HIV-1 genome gives rise to a multitude of RNA folding possibilities, a number of structures are very well conserved because of their essential function in the virus life cycle. Among these are the dimerization site, Trans-Activation Responsive (TAR) region and Rev-Responsive Element (RRE). These elements all have in common dsRNA structures (dimerization or stem-loop), which may potentially be processed into miRNAs. Whether these structures are recognized and processed by the Drosha·DGCR8 complex remains to be determined.
HIV-1 dimerization site
In virus particles, the genome consists of two identical molecules of RNA that are non-covalently linked near their 5′ ends. The dimerization process involves a series of conformational changes of the untranslated leader region in which a first structure referred as the kissing-loop complex is rearranged into a more extended molecular duplex (Huthoff and Berkhout, 2002). These conformations possess a number of dimer RNA molecules and stem-loop structures that could potentially be recognized by Drosha and/or Dicer, whereby the latter preferentially cleaves dsRNAs at their termini (Zhang et al., 2002), to generate miRNAs.
HIV-1 TAR
The TAR region is a 59-nt stem-bulge-loop structure located at the 5′ end of all HIV-1 transcripts found in the nucleus and cytoplasm, which is essential for efficient viral transcription. TAR is the target for the viral transactivating protein Tat that is known to act at the RNA level to enhance virus gene expression by more than 100 fold (see Figure 1-I). Upon binding to TAR, Tat recruits the positive transcription-elongation factor b (P-TEFb), a complex made of cyclin T1 and the cyclin dependent kinase 9 (CDK9), to the initiation complex. The CDK9 then phosphorylates the RNA pol II carboxy-terminal domain, which promotes the formation of an efficient elongation transcription complex (Bannwarth and Gatignol, 2005). Apart from this nuclear-based function, TAR is also important after RNA export to the cytoplasm since it inhibits translation by two mechanisms, i.e. through a direct block of translation initiation by its secondary structure and by activation of the dsRNA binding protein kinase R (PKR) which in turn phosphorylates the eukaryotic initiation factor 2 alpha (eIF2α) leading to an arrest of translation initiation. Both of these negative effects are alleviated by TRBP, which inhibits PKR (Park et al., 1994) and releases the translational block due to the TAR structure (Dorin et al., 2003).
HIV-1 RRE
The RRE domain is a large RNA structure present in all 9 kb and 4 kb RNAs, located within the Env intron. Through its interaction with the Rev protein, RRE is responsible for the nuclear export of these unspliced or singly spliced RNAs. In the absence of Rev, these RNAs are sequestered in the nucleus and only the multiply spliced 2 kb RNA encoding the regulatory proteins Tat, Rev and Nef are exported in the cytoplasm and translated. The interaction between Rev and RRE promotes the transition between this early phase of the viral life cycle to the late phase where structural proteins are produced (Pollard and Malim, 1998). The RRE is a 351 nt complex structure that comprises several stem-loop structures on which Rev assembles as a multimeric complex. This structure may resemble pri-miRNAs, which are often composed of multiple stem-loop structures, and represent a very good candidate for the source of viral miRNA. Of real interest for that matter is the observation that like TAR, RRE interacts with TRBP (Park et al., 1994).
siRNAs directed against HIV-1
Small interfering RNAs (siRNAs) are synthetic 21-nt RNA duplexes that have been designed to mimic the endogenous miRNAs or DICER-generated siRNAs. Their efficiency in downregulating expression of specific genes in cultured mammalian cells (Elbashir et al., 2001) established the basis for the use of siRNAs or RNA interference (RNAi) technology in therapeutics. This strategy was exploited successfully to inhibit the replication of several viruses, including HIV-1 (see Figure 1-J and Supplementary Table S1). Approaches based on siRNA targeting of host genes have been also used to restrict HIV-1 production (Martinez et al., 2002).
Although RNAi-based antiviral therapies are promising, HIV-1 have been shown to escape RNAi induced by a specific siRNA. In these cases, the emergence of mutants was observed, either showing nucleotide substitutions or deletions within the targeted sequence (Das et al., 2004), or evolving an alternative structure in its RNA genome occluding the siRNA binding site (Westerhout et al., 2005). A single substitution in the targeted sequence is sometimes sufficient to abolish the antiviral activity of siRNAs (Sabariegos et al., 2006). Such problem may be circumvented by targeting the most conserved sequences at multiple locations in the HIV-1 genome (Leonard and Schaffer, 2005).
Viral miRNAs
The observed sequence-specific HIV-1 RNA degradation induced by siRNAs implies that certain HIV-1 RNA sequences are accessible to the RNA silencing machinery in vivo. This concept, which is applicable to other viruses, is supported by the demonstrated improvement of RNA-induced silencing complex (RISC)-mediated target RNA cleavage when the target site access is increased (Brown et al., 2005). This raises the following question: Is the RNA silencing machinery capable of processing HIV-1 RNA, or other viral RNAs, into miRNAs or siRNAs naturally, thereby playing a role in antiviral defense against viruses?
Several laboratories, mostly working in collaboration, have started to address that issue from different angles. A research group led by Thomas Tuschl was the first to investigate the role of RNA silencing in human cells. Recording the small RNA profile of a Burkitt’s lymphoma cell line latently infected with Epstein-Barr virus (EBV), they found that this large DNA virus expresses several miRNAs (Pfeffer et al., 2004). Bioinformatic analysis of the genomic sequences flanking the cloned RNAs, which were detectable by Northern blot, unveiled fold-back structures characteristics of miRNA precursors. As for plant virus-derived siRNAs (Molnar et al., 2005), EBV miRNAs may originate predominantly from Dicer processing of highly structured single-stranded RNA. However, despite the recent identification of 18 new EBV miRNAs (Grundhoff et al., 2006), their biological roles can only be deduced from computational mRNA target predictions (Pfeffer et al., 2004) and remains to be fully appreciated.
Using similar approaches, the same group investigated several other viruses and identified miRNAs encoded in the Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8), mouse gammaherpesvirus 68 and human cytomegalovirus (also called HHV5) (Pfeffer et al., 2005). However, viral miRNAs derived from HIV-1 were neither predicted (using an algorithm identifying genomic regions that may assume a secondary structure similar to that of pri- or pre-miRNAs) nor found among 260 cloned miRNA sequences derived from HeLa cells stably expressing CD4 and CXCR4, and infected by HIV-1, isolate Bru (LAV-1) (Pfeffer et al., 2005). These findings suggested that HIV-1 may effectively hide its highly structured RNA from RNase III cleavage.
HIV-1-derived miRNAs
However, this assertion is being challenged, as concurrent studies about HIV-1 miRNAs have been reported. Using a computational method designed to uncover well-ordered folding patterns in nucleotide sequences, five candidate pre-miRNAs encoded by different regions of the HIV-1 genome were flagged (Bennasser et al., 2004). Omoto and colleagues (2004) reported on a miRNA (miR-N367) derived from the nef region, an accessory gene partially overlapping with the 3′ long terminal repeat (LTR). This HIV-1 miRNA could be detected by Northern blot analysis of total RNA prepared from MT-4 T cells persistently infected with HIV-1 IIIB and cloned from a ~25-nt RNA sub-population. Overexpression of miR-N367, which shows perfect complementarity with nef, seemed to suppress HIV-1 LTR-driven transcription in reporter gene assays performed in Jurkat T cells (Omoto and Fujii, 2005), suggesting that this nef-derived miRNA could act as a negative regulator of HIV-1 transcription. The biogenesis and action of this particular miRNA require further investigations.
More recently, a study reported that the HIV-1 RNA genome also encodes an siRNA derived from the env gene (Bennasser et al., 2005). The authors observed that two RNA strands forming a perfect 19-bp duplex, and joined by an extended 198-nt loop, could be converted into siRNAs upon incubation with recombinant Dicer in vitro. A probe specific for the viral siRNA detected a ~24-nt signal not seen in mock-infected cells by Northern blot analysis (Bennasser et al., 2005). Overexpression of this viral siRNA effectively reduced Env mRNA levels and viral replication, whereas its neutralization with complementary 2′O-methyl oligonucleotides led to a dose-dependent increase in HIV-1 replication in human cells (Bennasser et al., 2005). These results suggest that an HIV-1-derived siRNA can reduce virus production.
Whether or not HIV-1 miRNAs are effectively produced in infected cells and fulfill important biological roles warrants further experimental validation and confirmation. It may be that they are restricted to specific strains. Or, perhaps HIV-1 miRNAs may have escaped detection by small RNA cloning strategies, since methylation of the 2′ hydroxyl of the terminal ribose significantly reduces the cloning efficiency of silencing-associated small RNAs (Ebhardt et al., 2005). This would explain some of the discrepancies observed between laboratories using different techniques to identify viral miRNAs.
Biosynthetic mechanism of HIV-1 miRNAs
HIV-1 miRNAs may also be expressed at levels barely detectable using the techniques currently available. If their existence is proven unequivocally, their biosynthesis would merit due mechanistic considerations. For instance, are these viral miRNAs processed by the successive action of Drosha and Dicer like cellular miRNAs, by Dicer only or, as in plants, by an RNAi pathway adapted to viruses? How does the cleavage of an RNA substrate flanked by genomic single-stranded RNA sequences occur, knowing the preference of Dicer for RNA duplexes bearing terminal 2-nt 3′overhangs (Zhang et al., 2002)? How does the presence of an abnormaly extended loop in env siRNA precursor influence its processing? Is the expression level of viral miRNAs related to the relatively inefficient processing of poor HIV-1 dsRNA substrates by RNases III and/or to the limited access to a structure embedded within the HIV-1 RNA? Although regions of the HIV-1 genome possess structures relatively close to that of pri- or pre-miRNAs, the fact that they are complexed with many cellular and viral proteins may also affect their recognition and processing by Drosha and/or Dicer.
HIV-1, RNA silencing and RNA editing
The susceptibility of viral RNAs to RNases III may also be modified by structural changes produced by adenosine deaminases that act on RNA (ADARs) (Knight and Bass, 2002; Tonkin and Bass, 2003). The predominant form of RNA editing in human converts adenosine (A) into inosine (I) within largely double-stranded cellular and viral RNAs [reviewed in (Bass, 2002)]. A-to-I RNA editing may thus alter base pairing of a dsRNA substrate and reduce its susceptibility to Dicer cleavage, preventing it from initiating RNAi (Knight and Bass, 2002). Several viral genomes or transcripts show sequence changes consistent with such modification, including HIV-1. Indeed, TAR was previously reported to be a substrate for ADAR in Xenopus Oocytes and edited in a process dependent on Tat (Sharmeen et al., 1991). Although it has been speculated that editing of viral RNAs may somehow lead to viral persistence (Bass, 2002), the possible implications of this modification in HIV-1 pathogenesis remains unknown.
Biological significance of HIV-1 miRNAs
It may be too early to discuss about the potential biological function of miRNAs derived from HIV-1. However, the possibility that they directly influence viral pathogenesis and persistence in human cells is appealing. Cellular mRNAs that could potentially be regulated by these viral miRNAs have been tentatively identified (Bennasser et al., 2004). Investigation of the human and viral genes and processes possibly regulated by HIV-1 miRNAs, which have been the subject of speculations, awaits their prior experimental confirmation and validation.
Interaction between HIV-1 Tat and the miRNA-based RNA silencing machinery
In addition to protein-RNA and RNA-RNA interactions, recent studies have revealed an intriguing link between protein components involved in HIV-1 pathogenesis and RNA silencing, such as the virally-encoded Tat protein and the cellular TRBP.
Overexpression of Tat in mammalian cells was shown to attenuate silencing of reporter genes induced by short hairpin RNAs (shRNAs), but not siRNA (Bennasser et al., 2005). Knowing that the former elicits RNAi upon Dicer processing, the authors investigated and determined that Tat could inhibit Dicer activity in vitro (see Figure 1-D). However, prior to qualifying HIV-1 Tat as a proven inhibitor of Dicer function, it would be prudent (i) to determine if the Dicer inhibitory effect of Tat can be extended in vivo and occurs at physiological expression levels, (ii) to confirm that the observed inhibitory effects of Tat are specific and not due to random binding to dsRNAs, (iii) to verify if RNAi proceeds normally in the context of HIV-1 infection, and (iii) to assess whether Dicer function is indeed inhibited by HIV-1 in infected cells.
TRBP functions in RNA silencing
TRBP was originally discovered as a cellular protein that cooperates synergistically with Tat function and enhances transactivation of the HIV-1 5′ LTR (see Figure 1-I) (Gatignol et al., 1991). Intriguingly, recent studies indicated a role for TRBP in miRNA-guided RNA silencing, more specifically, in assisting Dicer function within a pre-miRNA processing complex (see Figure 1-D) (Chendrimada et al., 2005; Haase et al., 2005). Immunoprecipitation approaches, followed by mass spectrometry analysis of bound proteins, identified TRBP as a Dicer-interacting protein (Chendrimada et al., 2005; Haase et al., 2005). The Dicer-binding region on TRBP could be delineated to its third C-terminal dsRNA-binding domain (dsRBD) by deletional analysis in the yeast two-hybrid system (Haase et al., 2005). Depletion of TRBP was found to negatively affect pre-miRNA processing using cell extracts in vitro (Haase et al., 2005). Similar findings were obtained in Drosophila S2 cells, where the TRBP homolog Loquacious (Loqs) was found to interact with Dicer-1 and to be required for pre-miRNA processing into mature miRNA (Forstemann et al., 2005; Saito et al., 2005). However, no pronounced effects of TRBP (Haase et al., 2005) or Loqs (Forstemann et al., 2005; Saito et al., 2005) depletion were observed on mature miRNA levels in vivo, although contrasting observations were also reported (Chendrimada et al., 2005). The apparent discrepancy between the observed effect of TRBP on pre-miRNA processing in cells and cell extracts may be readily explained by incomplete depletion of the protein, allowing for the manifestation of processing deficiency in vitro but not in vivo (Haase et al., 2005). Whether incubated in the absence or presence of TRBP, recombinant Dicer was equally effective in processing pre-miRNAs in vitro (Chendrimada et al., 2005). The stimulatory effects of TRBP that become apparent in cell extracts suggests the possible implication of another cellular factor in vivo.
TRBP also may form a ternary complex with Dicer and Ago2 (Chendrimada et al., 2005), thereby coupling the initiation and effector steps of RNA silencing. This may explain the importance of TRBP in mRNA cleavage mediated by miRNAs (Haase et al., 2005) or siRNAs (Chendrimada et al., 2005; Haase et al., 2005) acting downstream of Dicer. The function of TRBP within the effector complex remains to be elucidated.
A dual role for TRBP - Implications for HIV-1
TRBP may thus exert a dual role in HIV-1 pathogenesis and RNA silencing, as recently discussed (Gatignol et al., 2005). The requirement of TRBP to achieve a higher virus production may have forced the virus to evolve under selective pressure from the RNA silencing machinery. There is a possibility that TAR and RRE RNA structures could also compete with Dicer for TRBP binding, and thus inhibit RNA silencing (Gatignol et al., 2005). The delicate balance thereby created may have conferred to HIV-1 the ability to replicate in TRBP-expressing cells and be responsible, to some extent, for the low virus load and persistence in HIV-1-infected individuals. Pharmacological interventions aimed at dissociating TRBP functions may represent a relevant therapeutic area to combat the HIV-1 pandemic.
Perspectives for HIV-1
A number of studies published recently have provided key insights into the increasingly complex interaction between HIV-1 and host RNA silencing machineries. It has been known for some time that HIV-1 induces drastic changes in gene expression programming of infected cells. With the recent idea that HIV-1 may encode miRNAs, the identification and validation of the complete HIV-1 miRNA array as well as their cellular and viral mRNA targets, which pose a considerable challenge, may significantly improve our understanding of HIV-1 pathogenesis. In particular, it may help determine to what extent the perturbed gene expression profiles in HIV-1-infected cells (Chun et al., 2003; Corbeil et al., 2001; van ’t Wout et al., 2003) can be related to virus-derived miRNAs and how it ultimately influences viral replication as well as the efficiency of host defenses. This raises the attractive hypothesis that HIV-1 replication may result, at least in part, from a delicate balance between the structural requirements to support HIV-1 replication versus the potential beneficial role of HIV-1 miRNAs in blocking host gene expression.
More recently, candidate HIV-1 genes that could be controlled by host miRNAs have been predicted in view of thermodynamically favorable miRNA:target pairing (Hariharan et al., 2005). In addition, changes in miRNA expression profiles, where a large pool of miRNAs is downregulated, have been observed in human HeLa cells transfected with the infectious molecular clone pNL4-3 (Yeung et al., 2005). This suggests that the virus may counteract the biosynthesis and action of cellular miRNAs. To what extent do these changes contribute to HIV-1 replication remains to be appreciated.
Applicability to other viruses
The various aspects of the interaction between HIV-1 and the RNA silencing machinery may also be applicable to other viruses of global importance for human health. In turn, mechanisms described for other viruses may ultimately be transposed to HIV-1. A few examples that may be relevant include (i) herpesvirus Kaposi’s sarcoma-associated herpesvirus (KSHV), which encodes as much as 11 distinct miRNAs that may play critical roles in establishment and/or maintenance of KHSV latent infection (Cai et al., 2005), (ii) simian virus 40 (SV40), which encodes miRNAs that regulate viral gene expression and reduce susceptibility to cytotoxic T cells (Sullivan et al., 2005), (iii) a cellular miRNA, miR-32, that was recently shown to restrict the accumulation of the retrovirus primate foamy virus type 1 (PFV-1) (Lecellier et al., 2005), and, in contrast, (iv) an abundant miRNA specifically expressed in the human liver, miR-122, that has been shown to assist hepatitis C virus (HCV) replication through a genetic interaction with the 5′ noncoding region of the viral genome (Jopling et al., 2005).
Conclusion
Further investigation on the relationship between HIV-1 and RNA silencing pathways may unveil key aspects of viral pathogenesis, provide new insights into the persistence of the virus in infected patients and offer novel basis for anti-HIV-1 therapies. Undoubtedly, the intersection of the virus and RNA silencing research avenues offers new perspectives on host-pathogen interactions and promises to be much more frequented, enhancing the chances to discover additional close encounters.
Supplementary Material
Acknowledgments
We express our gratitude to Gilles Chabot for graphic design. P.P. is a New Investigator of the Canadian Institutes of Health Research (CIHR). M.J.T. is the recipient of the Canada Research Chair in Human Immuno-Retrovirology (senior level). This work was financially supported by grant EOP-64706 from Health Canada/CIHR (P.P. and M.J.T.).
References
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3(1):61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22(5):607–19. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1(1):43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10(2):185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol. 2005;12(5):469–70. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J Cell Biol. 2002;156(1):53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102(15):5570–5. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Justement JS, Lempicki RA, Yang J, Dennis G, Jr, Hallahan CW, Sanford C, Pandya P, Liu S, McLaughlin M, Ehler LA, Moir S, Fauci AS. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl Acad Sci U S A. 2003;100(4):1908–13. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeil J, Sheeter D, Genini D, Rought S, Leoni L, Du P, Ferguson M, Masys DR, Welsh JB, Fink JL, Sasik R, Huang D, Drenkow J, Richman DD, Gingeras T. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 2001;11(7):1198–204. doi: 10.1101/gr.180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78(5):2601–5. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dorin D, Bonnet MC, Bannwarth S, Gatignol A, Meurs EF, Vaquero C. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J Biol Chem. 2003;278(7):4440–8. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc Natl Acad Sci U S A. 2005;102(38):13398–403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13(4):1338–51. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. Rna. 2003;9(10):1171–3. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3(7):e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251(5001):1597–600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gatignol A, Laine S, Clerzius G. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology. 2005;2:65. doi: 10.1186/1742-4690-2-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. Rna. 2006 doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6(10):961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337(4):1214–8. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41(33):10439–45. doi: 10.1021/bi025993n. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10(4):809–17. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14(23):2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308(5721):557–60. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Leonard JN, Schaffer DV. Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape. J Virol. 2005;79(3):1645–54. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–21. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–3. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Martinez MA, Gutierrez A, Armand-Ugon M, Blanco J, Parera M, Gomez J, Clotet B, Este JA. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. Aids. 2002;16(18):2385–90. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-Strand Cleavage Facilitates Assembly of siRNA into Ago2-Containing RNAi Enzyme Complexes. Cell. 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79(12):7812–8. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86(Pt 3):751–5. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1(1):44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci U S A. 1994;91(11):4713–7. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Plante I, Davidovic L, Ouellet DL, Gobeil L-A, Tremblay S, Khandjian EW, Provost P. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J Biomed Biotech. 2006 doi: 10.1155/JBB/2006/64347. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21(21):5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–26. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabariegos R, Gimenez-Barcons M, Tapia N, Clotet B, Martinez MA. Sequence Homology Required by Human Immunodeficiency Virus Type 1 To Escape from Short Interfering RNAs. J Virol. 2006;80(2):571–7. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3(7):e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sharmeen L, Bass B, Sonenberg N, Weintraub H, Groudine M. Tat-dependent adenosine-to-inosine modification of wild-type transactivation response RNA. Proc Natl Acad Sci U S A. 1991;88(18):8096–100. doi: 10.1073/pnas.88.18.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–6. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. Rna. 2005;11(4):371–82. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302(5651):1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Wout AB, Lehrman GK, Mikheeva SA, O’Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003;77(2):1392–402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA:target interaction. Chem Biol. 2004;11(12):1619–23. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33(2):796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M, Machaca K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436(7053):1044–7. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Bennasser Y, Myers TG, Jiang G, Benkirane M, Jeang KT. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology. 2005;2:81. doi: 10.1186/1742-4690-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21(21):5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.