Abstract
MicroRNAs (miRNAs) are small, ~21- to 23-nucleotide (nt) non-coding RNA species that act as key regulators of gene expression along a central and well-defined cellular process known as RNA silencing, and involving the recognition and translational control of specific messenger RNA (mRNAs). Generated through the well-orchestrated and sequential processing of miRNA precursor molecules, mature miRNAs are subsequently incorporated into miRNA-containing ribonucleoprotein effector complexes to regulate mRNA translation through the recognition of specific binding sites of imperfect complementarity located mainly in the 3′ untranslated region. Predicted to regulate up to 90% of the genes in humans, miRNAs may thus control cellular processes in all cells and tissues of the human body. Likely to play a central role in health and disease, a dysfunctional miRNA-based regulation of gene expression may represent the main etiologic factor underlying diseases affecting major organs, such as the brain. In this review article, the molecular mechanisms underlying the role and function of miRNAs in the regulation of genes involved in neurological and neurodegenerative diseases will be discussed, with a focus on the fragile X syndrome, Alzheimer’s disease (AD) and prion disease.
Keywords: Brain, microRNA, RNA silencing, gene regulation, neurological diseases
1. Biogenesis and function of microRNAs
The microRNA (miRNA)-guided RNA silencing pathway is a recently discovered gene regulatory process present in eukaryotic cells and based on small, ~21- to 23-nucleotide (nt) non-coding RNAs, known as miRNAs. Our current knowledge on the biogenesis and action of miRNAs in humans has been reviewed recently (Bartel, 2009).
Encoded by the genome of nucleated cells, miRNA genes are transcribed, either by RNA polymerase II (Lee et al., 2004) or III (Borchert et al., 2006), into long RNA transcripts that fold on themselves to form primary miRNAs (pri-miRNAs). These RNA species are recognized and trimmed into miRNA precursors (pre-miRNAs) by the nuclear ribonuclease III (RNase III) Drosha (Lee et al., 2003), acting in concert with the DiGeorge syndrome Critical Region gene 8 (DGCR8) protein within the microprocessor complex (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004). After being exported to the cytoplasm via Exportin-5 (Lund et al., 2004; Yi et al., 2003), pre-miRNAs are processed by the pre-miRNA processing complex, composed of the RNAse III Dicer (Provost et al., 2002; Zhang et al., 2002) and its cofactor TAR RNA binding protein (Chendrimada et al., 2005; Haase et al., 2005), into a miRNA:miRNA* duplex. The forming ribonucleoprotein (RNP) complex is then joined by the Argonaute 2 protein, and the miRNA guide strand is selected, based on the relative stability of the duplex extremities, to form a miRNA-containing RNP (miRNP) complex (Chendrimada et al., 2005). The associated miRNAs confer to the miRNP complexes the ability to recognize cellular messenger RNAs (mRNAs) through specific binding sites generally located in the 3′ untranslated region (UTR), guiding the miRNPs for the regulation of specific mRNAs, as reviewed previously (Kim, 2005; Zamore and Haley, 2005). The targeted mRNAs will be cleaved and degraded if the complementarity between the miRNAs and their binding site is perfect, or its translation regulated if the complementarity is imperfect (Bartel, 2004). In this latter case, the repressed mRNAs are translocated to the P-bodies, after which they are either degraded or returned to the translational machinery for expression upon a specific cellular signal (Bhattacharyya et al., 2006; Liu et al., 2005). Although miRNAs are known mainly as repressors of gene expression, they have also been shown to enhance mRNA translation under specific cellular conditions (Vasudevan et al., 2007).
2. MicroRNAs in health and disease
Involving relatively few protein components, this complex and well-integrated regulatory circuit plays a key role in modulating a plethora of mRNA targets (Perron and Provost, 2008). Predicted to regulate up to 90% of the genes in humans (Miranda et al., 2006), miRNAs may control every cellular processes in all cells and tissues of the human body! Involved in the fine tuning of gene expression, a normal miRNA function is required for a tightly regulated expression of the cellular proteins (see Figure 1A), which is critical for the maintenance of health and prevention of disease, as discussed in Perron et al. (2007). In contrast, deregulated protein expression induced by a dysfunctional miRNA-based regulatory system may represent the main etiologic factor underlying diseases affecting major systems, including the central nervous system (CNS).
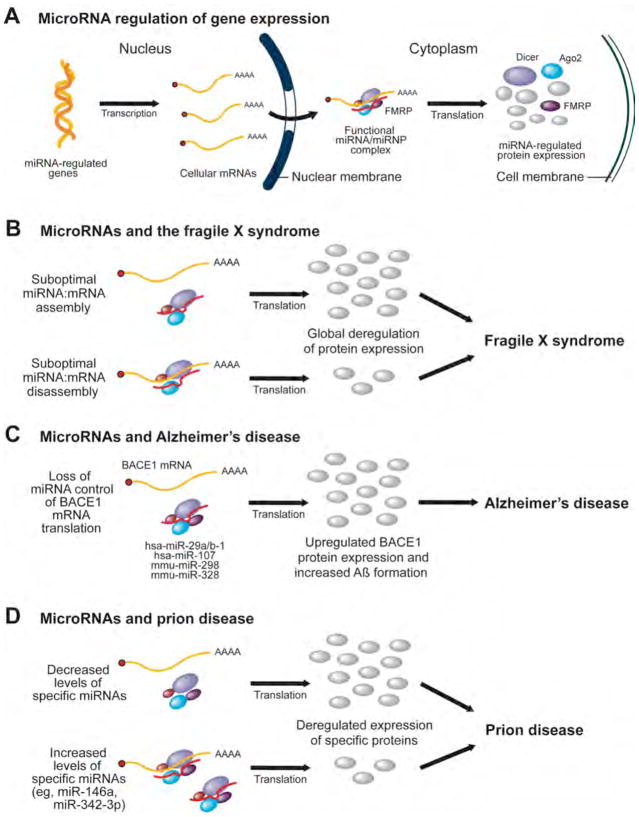
Figure 1. MicroRNAs as a molecular basis for neurological and neurodegenerative diseases, such as the fragile X syndrome, Alzheimer’s disease and prion disease.
(A) A simplified schematic illustration of the microRNA (miRNA) regulation of messenger RNA (mRNA) translation in human cells. (B) Suboptimal utilization of miRNAs, i.e. miRNA:mRNA assembly and/or disassembly, may be caused by a deregulated expression of fragile X mental retardation protein (FMRP), a protein component of the miRNA machinery, and account for some of the molecular defects in patients with the fragile X syndrome. (C) Loss of miRNA control of APP-converting enzyme (BACE1) mRNA may lead to enhanced BACE1 protein expression, with a corresponding increase in β-amyloid (Aβ) formation and deposition, which may favor the development of Alzheimer’s disease (AD). (D) Deregulated levels of specific miRNAs have been reported in brains affected by prionopathies, which may result in an altered expression of genes involved in cell death, synapse function, and neurogenesis. Ago2, Argonaute 2; hsa, homo sapiens; miRNP, miRNA-containing RNP; mmu, mus musculus.
For instance, a deregulated miRNA control of mRNA translation may occur when the function of a core protein component of the miRNA pathway is compromised, i.e. is the subject of a deletion, mutation or misexpression. In this situation, a global negative impact on miRNA biogenesis and/or function is observed, as in the case of the fragile X mental retardation protein (FMRP) in the fragile X syndrome (see section 4) (Plante and Provost, 2006) and of the behavioral and neuronal deficits associated with haploinsufficiency of the Dgcr8 gene (Stark et al., 2008).
A loss of miRNA control may also result from the deletion, mutation or misexpression of a miRNA, or that of its corresponding binding site, in a sequence of events leading to miRNA:miRNA binding site pairs becoming dysfunctional or to new miRNA:miRNA binding site functional combinations emerging. The ensuing deregulation of mRNA translation may then lead to misexpression, i.e. either downregulated or upregulated expression, of a specific protein and provoke the development of a disease (Perron et al., 2007), which may be the case of the β-amyloid precursor protein (APP)-converting enzyme (BACE) in Alzheimer’s disease (AD) (see section 5) (Boissonneault et al., 2009). Hence the relevance of using miRNAs as biomarkers and therapeutic targets/drugs in human diseases affecting major organs, such as the brain.
3. MicroRNAs in neurological and neurodegenerative diseases
Compelling evidences now link miRNAs to the control of neuronal development and differentiation, as recently reported by Decembrini et al. (2009). In that study, a set of 4 miRNAs (miR-129, miR-155, miR-214 and miR-222) was found to repress translation of mRNAs encoding for Xotx2 and Xvsx1, which directs the differentiation of late retinal progenitor cells into bipolar neurons in Xenopus. The interest in the functions of miRNAs in the CNS has also expanded to include their roles in neurodegeneration, of which little is known. Investigations have begun to reveal the influence of miRNAs on both neuronal survival and the accumulation of toxic proteins that are associated with neurodegeneration (Eacker et al., 2009). For instance, conditional Purkinje cell-specific ablation of the key miRNA-generating enzyme Dicer in mice was associated with a progressive loss of miRNAs, followed by Purkinje cell death, cerebellar degeneration and development of ataxia (Schaefer et al., 2007). In addition to provide evidence for an essential role of miRNAs in survival of differentiated neurons, these findings support the involvement of miRNAs in neurodegenerative disorders, a topic that was reviewed recently (Hebert and De Strooper, 2009).
Ongoing investigations are also providing clues as to how the accumulation of toxic proteins thought to be the cause of many neurodegenerative conditions, such as Aβ and the prion protein, cause cell death. One possible mechanism may involve interference with the miRNA regulation of specific mRNAs encoding key, pro-survival proteins (Eacker et al., 2009).
4. MicroRNAs and the fragile X syndrome
In humans, the FMR1 (fragile mental retardation 1) gene, which spans ~38 kb in the q27.3 region located at the tip of the X chromosome long arm, encodes for a protein known as FMRP. Detected in practically every tissue in humans and rodents, with high levels in the brain, testes, esophagus, lung, and kidney, FMRP is an RNA-binding protein containing two K-homology (KH) domains and an RGG box, and which is involved in regulation of mRNA translation, RNA transfer, and local modulation of synaptic mRNA translation (Khandjian, 1999). FMRP is associated with translating polyribosomes in neuronal cells (Stefani et al., 2004) and acts as a negative regulator of translation (Li et al., 2001). Loss of FMRP function, most often caused by the hypermethylation and consequent transcriptional silencing of the FMR1 gene, due to an expansion of the trinucleotide sequence CGG located in the 5′UTR, is the etiologic factor of the fragile X syndrome, which is characterized by mental retardation, autistic features and behavioral abnormalities (Penagarikano et al., 2007).
A link between miRNAs and the fragile X syndrome was proposed when the Drosophila homolog of FMRP was initially found to be associated with Dicer, Ago2 and miRNAs in vivo (Caudy et al., 2002; Ishizuka et al., 2002). Subsequently extended to human cells in culture (Jin et al., 2004), these findings raised the possibility that the fragile X syndrome could result from a defects in the miRNA machinery. These observations were intriguing, considering the ability of FMRP to recognize an abundant array of mRNAs harboring a G quartet structure (Brown et al., 2001). Developing a bioinformatic program aimed to identify mRNA targets of miRNAs, the creators of miRanda (available at: http://www.microrna.org) obtained an unexpected experimental support regarding the validity of their method. When analyzing their computational predictions in parallel with the data obtained from the micro-array analysis of mRNA transcripts bound to FMRP-containing RNP complexes, they observed a strong enrichment of predicted targets in mRNAs associated with FMRP in mammals (John et al., 2004).
Expanding previous observations made on the DNA annealing and strand exchange properties of FMRP (Gabus et al., 2004), we were able to link the nucleic acid chaperone properties of FMRP to the miRNA-guided RNA silencing pathway. Using recombinant proteins and 32P-labeled pre-miRNAs or mature miRNAs, we demonstrated that human FMRP can act as a miRNA acceptor protein for the ribonuclease Dicer and facilitate the assembly of miRNAs on specific target RNA sequences (Plante et al., 2006). Reporter gene silencing assays, performed in FMRP-deficient or wild-type murine fibroblasts, revealed the requirement of FMRP, a component of effector miRNP complexes, for optimal RNAi in vivo. Together, these results led us to propose that suboptimal utilization of miRNAs, i.e. miRNA:mRNA assembly and/or disassembly, may account for some of the molecular defects in patients with the fragile X syndrome (see Figure 1B) (Plante and Provost, 2006). Here, we need to take into consideration the fact that, in addition to repressing mRNA translation, miRNP complexes may enhance mRNA translation under specific cellular conditions, i.e. on cell cycle arrest, along a process involving the recruitement of an ortholog of FMRP (FXR1) (Vasudevan et al., 2007).
Recently, the use of a phospho-specific antibody to FMRP (P-FMRP) unveiled that P-FMRP, which is found associated with stalled untranslating polyribosomes, does not associate with Dicer or Dicer-containing complexes in a coimmunoprecipitation strategy in transiently transfected HeLa cells (Cheever and Ceman, 2009). These authors also showed that Dicer-containing complexes bind FMRP at amino acids 496–503 and that phosphorylation disrupts this association with a consequent increase in association with pre-miRNAs. Shedding new light on the molecular function of FMRP within the miRNA pathway, these results suggest that, in addition to regulating translation, phosphorylation of FMRP may regulate its involvement in the miRNA pathway by modulating association with the miRNA-generating enzyme Dicer (Cheever and Ceman, 2009).
More recently, FMRP was reported to regulate synaptic structure and function through its association with miRNAs miR-125b and miR-132 (Edbauer et al.). In fact, Edbauer and colleagues found that the mRNA encoding for the NMDA receptor subunit NR2A is a target of miR-125b and is specifically associated with FMRP in mouse brain. In hippocampal neurons, NR2A expression is negatively regulated through its 3′UTR by FMRP, miR-125b and a member of the Argonaute family of proteins, leading the authors to propose a link between these findings and the altered synaptic plasticity observed in Fragile X patients (Edbauer et al.).
5. MicroRNAs and Alzheimer’s disease
Alzheimer’s disease (AD) is a slowly progressing neurodegenerative disease that currently affects ~2% of the population in industrialized countries and whose incidence is predicted to increase dramatically over the next 40 years (http://www.alz.org) (Mattson, 2004). Affecting cholinergic neurons, AD is characterized by the accumulation of plaques formed of short β-amyloid (Aβ) peptides in the hippocampal region of the brain (Mucke, 2009). Aβ peptides are produced upon proteolytic cleavage of APP by β-secretase, also known as β-site APP-cleaving enzyme 1 (BACE1), which contributes to the formation of these plaques (Vassar et al., 1999). Accumulation of Aβ peptides may exert toxic effects and elicit an inflammatory response that may contribute to impair the function of neurons and alter the integrity of the microcircuits involved in learning, memory, and other cognitive functions.
Although a deficiency in Aβ clearance is certainly a possibility, an increased expression of the Aβ-producing enzyme BACE1 may also be an important contributor to the net accumulation of Aβ peptides and AD progression (Hebert et al., 2008). This assertion is supported by the observed dosage effect of BACE1 in inducing AD (Rovelet-Lecrux et al., 2006), suggesting that any alterations in BACE1 expression levels might contribute to the disease.
The intriguing observations suggesting a role for miRNAs in the pathogenesis of AD initially emanated from post-mortem analyses that revealed upregulation of BACE1 expression at the protein, but not at the mRNA, level in brains from patients suffering from AD, as compared to brains from unaffected patients (Holsinger et al., 2002), in accordance with a possible loss of BACE1 mRNA translational control. In an early study investigating the miRNA abundance in the hippocampal region of fetal, adult and AD brain, Lukiw (2007) observed an alteration in specific miRNA abundance in AD hippocampus, with miR-9 and miR-128 being upregulated in AD versus adult brain. This prompted the author to suggest that the misregulation of specific miRNAs in AD hippocampus may contribute to the multiple gene expression changes that accompany neural degeneration. The caveat has to be taken into account that, as brain tissues are composed of multiple neuronal, glial, vascular and other cell types, the contributions of each brain cell type to miRNA production and speciation remain to be elucidated (Lukiw, 2007).
These initial observations were followed by a string of papers addressing the relationship between miRNAs and AD, and published within an 18-month period. Investigating miRNA expression microarrays on RNA extracted from human brain tissue from the University of Kentucky AD Center Brain Bank, and confirming their data by in situ hybridization with cross-comparison to neuropathology, the group led by Peter T. Nelson documented a significant decrease in neuronal miR-107 expression even in patients with the earliest stages of AD (Wang et al., 2008). Interestingly, BACE1 mRNA levels, assessed on Affymetrix Exon Array microarrays, tended to increase as miR-107 levels decreased in the progression of AD. Computational analyses did predict the presence of several putative binding sites for miR-107 in the 3′UTR of BACE1 mRNA, and at least one of these sites was proven to be functionally regulated by miR-107 in reporter gene assays performed in transiently transfected HeLa cells (Wang et al., 2008).
Soon after, investigating changes in brain miRNA expression profiles of sporadic AD patients, Hebert and colleagues (2008) reported that several miRNAs potentially involved in the regulation of APP and BACE1 expression appeared to be decreased in diseased brain. As miR-29a, miR-29b-1, and miR-9 were found to regulate BACE1 expression in reporter gene assays performed in transiently transfected HeLa cells, the miR-29a/b-1 cluster was significantly decreased in AD patients displaying abnormally high BACE1 protein levels (Hebert et al., 2008). Providing support for a causal relationship between miR-29a/b-1 expression and AD, these findings led the authors to propose that loss of specific miRNAs can contribute to increased BACE1 and Aβ levels in sporadic AD, as illustrated in Figure 1C.
We also observed a loss of correlation between BACE1 mRNA and protein levels in the hippocampus of APPSwe/PS1 mice (Boissonneault et al., 2009), a murine model of AD. Consistent with an impairment of miRNA-mediated regulation of BACE1 mRNA translation, we were able to validate this hypothesis and demonstrate a role for two miRNAs, i.e. mmu-miR-298 and mmu-miR-328, in the regulation of BACE1 expression in transiently transfected murine neuronal N2a cells in culture. It is relevant to note that both Drosha and Dicer mRNA levels were enriched in the hippocampus and the dentate gyrus of mouse brain (Boissonneault et al., 2009), which are also the regions that are the most severely affected by Aβ deposits in AD. The concept of a link between dysfunctional miRNA regulation of BACE1 expression and AD is supported further by the detection of miR-298 and miR-328 in the hippocampus of APPSwe/PS1 mice, as well as by their decreased expression levels during aging (Boissonneault et al., 2009).
Using a different mouse model of AD, Wang and colleagues (2009) identified, among an array of 299 distinct miRNAs, a relatively highly expressed candidate miRNA, i.e. miR-34a, that was differentially expressed in the cerebral cortex of APPswe/PSΔE9 mice versus age-matched controls. miR-34a has been inversely correlated with the protein level of Bcl2 in these mice, and miR-34a expression was shown to inhibit Bcl2 mRNA translation in transiently transfected SH-SY5Y cells. As expected, neutralization of miR-34a function, through locked nucleic acid (LNA)-modified antisense oligonucleotides, increased Bcl2 protein expression in these cells, which was accompanied by a decrease of active caspase-3. Validating Bcl2 as a functional target of miR-34a, these results suggest that an abnormal expression of miR-34a may contribute to the pathogenesis of AD, at least in part, through modulation of Bcl2 expression (Wang et al., 2009).
When investigating the role of miRNAs in the etiology of AD, using established animal models of the disease, the caveat has to be taken into account that the disease is induced independently of miRNAs; most often by introducing a mutation in the APP and/or presenilin gene. Therefore, the results obtained from the use of such animals need to be interpreted with caution, as the conditions related to the disease may also influence miRNA levels and function. In that scheme, any changes in the level of specific miRNAs may represent a consequence, rather than a cause, of the disease (the chicken or the egg dilemma). Therefore, it would not be prudent to draw conclusions based solely on such data, which would need to be validated in a more relevant, different experimental context.
In addition, when comparing data related to miRNA regulation of gene expression obtained from different species (eg, human and mouse), it is important to consider that the miRNA and mRNA sequences, as well as the miRNA binding sites they harbor, may not be evolutionarily conserved. The analysis of the binding sites for miR-298 and miR-328, for example, is rather complex and difficult because, in contrast to mice, human hosts four different BACE mRNAs harboring different 3′UTR of various lengths and composition. In addition, whereas the miR-328 sequence is perfectly conserved between mouse and human, that of miR-298 is only 72% identical. This may explain, at least in part, some of the discrepancies observed in the identity of the miRNAs that effectively regulate expression of specific genes, such as BACE1.
Tracking the abundance and decay kinetics of specific miRNAs by miRNA array and Northern blot analyses of human neural cells in primary culture and in short post-mortem interval human brain tissues, Sethi and Lukiw (2009) noted a limited stability and relatively short half-life (~1–3.5 h) for specific brain-enriched miRNAs. In short post-mortem interval AD-affected temporal lobe neocortex, miRNA-9, miRNA-125b and miRNA-146a were found to be significantly upregulated, an effect that was not seen in several related neurological disorders (Sethi and Lukiw, 2009), leading the authors to propose that, unless specifically stabilized, certain brain-enriched miRNAs represent a rapidly executed signaling system employing highly transient effectors of CNS gene expression.
MiRNAs may also exert indirect effects and be involved in more complex networks of regulatory mediators of importance in the pathogenesis of AD, as depicted by the specific up-regulation of an NF-kB-sensitive miRNA, i.e. miR-146a, observed in human AD brains (Lukiw et al., 2008). This miRNA shows a high degree of complementarity to the mRNA 3′UTR of complement factor H (CFH), which is an important repressor of the inflammatory response of the brain. An inverse correlation could be established between upregulated levels of miRNA-146a and downregulation of CFH protein levels in AD brain. The use of an antisense oligonucleotide to miRNA-146a in human neural (HN) cells in primary culture confirmed the functionality of the binding sites for miR-146 in the CFH mRNA 3′UTR. These data indicate that miR-146 may exert proinflammatory effects and illustrate the potential for miRNA inhibitors as an effective therapeutic strategy against pathogenic inflammatory signaling.
In nematodes, the homolog of APP, APP-like-1 (apl-1), has been reported also to functions with and to be under the control of molecules regulating developmental progression. Caenorhabditis elegans apl-1 shows significant genetic interactions with let-7 family miRNAs, which modulate the level of the apl-1 transcription (Niwa et al., 2008), suggesting that Aβ peptide formation is under miRNA control in living organisms other than mammals.
Representing a promising approach for the treatment of AD, siRNA-mediated targeting of BACE1 mRNA has proven to be effective in down-regulating BACE1 protein levels and activity in cultured primary cortical neurons (Kao et al., 2004), as well as in a mouse model of AD (Singer et al., 2005). Interestingly, bace1-null mice do not demonstrate any developmental problems or aberrant behavioral phenotypes, making of BACE1 a potential and attractive target for RNAi-based therapies against AD (Luo et al., 2001; Roberds et al., 2001).
6. MicroRNAs and prion disease
The pathogenesis of prion diseases shares similarities with AD, as both are associated with the accumulation of fibrillar aggregates of proteins, i.e. the prion protein or β-amyloid (Aβ) respectively, that are deleterious to neuronal function, as reviewed recently (Frost and Diamond, 2009). However, prionopathies are unique among neurodegenerative diseases because they are infectious. When the nonpathogenic form of the prion protein (PrPC) comes into contact with a pathogenic prion protein conformer (PrPSc), it is induced to misfold in a process along which the conformation of a PrPSc molecule is communicated to a native PrPC protein (Pan et al., 1993). The pathogenic isoform of the cellular PrPC, or a closely related molecule, is the causative agent of transmissible spongiform encephalopathies (TSE), that comprise a group of neurodegenerative diseases including bovine spongiform encephalopathy (BSE) in cattle, Scrapie in sheep, and Creutzfeldt-Jakob disease (CJD) in humans. Although recent studies have highlighted prion-like mechanisms of propagation of protein misfolding in other neurodegenerative diseases, such as AD (Frost and Diamond, 2009), the molecular mechanisms underlying prion-induced neurodegeneration, which coincides with neuronal loss and leads to death of the host, remains to be clearly defined.
Recently, an avenue linking miRNAs with prionopathies has been proposed and explored. Microarray and reverse transcription-PCR (RT-PCR)-based miRNA profiling analyses of brains from mice infected with mouse-adapted scrapie revealed a deregulated expression for more than 15 miRNAs during prion disease. Among these, miR-342-3p, miR-320, let-7b, miR-328, miR-128, miR-139-5p and miR-146a were upregulated by more than 2.5 fold, whereas miR-338-3p and miR-337-3p were downregulated in the same proportion (Saba et al., 2008). Among these miRNAs, only one (miR-128) had previously been shown to be deregulated in neurodegenerative diseases, suggesting a disease-specific pattern of differentially expressed miRNAs associated with prion–induced neurodegeneration.
Computational analysis predicted numerous potential gene targets for the miRNAs associated with prion disease, including 119 genes previously determined to be also deregulated in mouse scrapie and whose deregulation of expression may be related to miRNAs (Saba et al., 2008). Bioinformatic and biochemical integration of miRNA and mRNA profiling identified further a correlation between miRNA expression and putative gene targets involved in intracellular protein degradation pathways and signaling pathways related to cell death, synapse function and neurogenesis (see Figure 1D) (Saba et al., 2008). Experimental validation of these miRNA and mRNA correlates may help clarify the relationship between miRNAs and prionopathies, and shed new light on the molecular basis of prion disease.
Upregulation of miR-342-3p was also observed upon microarray and RT-PCR analyses of miRNA expression in the brains of bovine spongiform encephalopathy (BSE)-infected cynomolgus macaques, used as a model for Creutzfeldt-Jakob disease (CJD) (Montag et al., 2009). More importantly, the authors could show, in a pilot study, that hsa-miR-342-3p was also upregulated in brain samples of human type 1 and type 2 sporadic CJD, suggesting that miR-342-3p might be used as a novel marker of prionopathies in animals and humans.
The Prnp gene, that encodes PrPC, is not essential, as its invalidation led to transgenic mice that are viable, apparently healthy, and resistant to challenge by the infectious prion agent, thereby offering the possibility of downregulating Prnp gene expression as a potential therapeutic approach to treat prionopathies (Tilly et al., 2003). The potential of using therapeutic RNAs to this end was verified and confirmed by a study reporting an efficient and highly specific reduction of PrPC levels in transfected cells expressing siRNAs directed against the Prnp mRNA (Tilly et al., 2003).
These observations, made in a cell culture system using rabbit kidney epithelial RK13 cells (Tilly et al., 2003), were recently transposed to transgenic mice in vivo. In this model, a small regulatory RNA designed to target the Prnp mRNA and expressed at low levels using the human Prnp promoter was found to downregulate the endogenous mouse Prnp gene expression to an extent that appears to be directly related with the transgene expression level (Gallozzi et al., 2008).
The RNAi targeting strategy can be refined further by introducing an artificial, single nucleotide mismatch into siRNAs or short-hairpin RNAs (shRNAs) directed against mutant alleles of the human Prnp gene, which appear to be associated with susceptibility to prion diseases. This modified approach was recently shown to enhance discrimination between the mutant and wild-type alleles in cultured HeLa and HEK293 cells (Ohnishi et al., 2008). These data argue for RNA interference (RNAi) and allele-specific RNAi as attractive therapeutic strategies to fight prion diseases.
7. Alzheimer’s and prion diseases share a microRNA commonality
The fact that miR-146a was found to be upregulated by independent groups, and in both AD (Lukiw et al., 2008) and prion disease (Saba et al., 2008), is noteworthy and adds further to the similarities between these two neurodegenerative diseases. Even when considering and taking into account the relatively short half-lives of some miRNAs (~1 to 3.5 h), short post-mortem interval human brain tissues from AD patients exhibited a significant up-regulation of miRNA-146a that was not seen in the same brain regions affected with amyotrophic lateral sclerosis (ALS), schizophrenia or Parkinson’s disease (Sethi and Lukiw, 2009), whose pathogenesis is closely related to AD and prion disease in that it involves misfolding and accumulation of fibrillar aggregates of proteins (Tau).
The molecular mechanism underlying the relatively specific upregulation of miR-146a commonly observed in AD and prion disease is intriguing and represents an interesting matter of speculations that raise several questions: Does the upregulation of miR-146a expression represent the chicken or the egg, i.e. is it the cause or the consequence of the diseases? Could miR-146a be used as a simple biomarker or is it actively involved in the pathogenesis of AD and prion disease? If so, is it involved in the etiology and/or in disease progression? Among the possibilities that may be worth exploring, and that would involve a transcriptional mode of regulation, is whether the accumulation of fibrillar, toxic proteins may induce an inflammatory response and activate a signaling cascade leading to an enhanced transcription of the miR-146a gene, increasing levels of miR-146a and exacerbation of the disease. A posttranscriptional mechanism modulating miR-146a expression (miRNA processing or metabolism/stability), on the other hand, would lack the observed specificity for miR-146a and appears less likely. Unless a different scenario, in which the upregulation of miR-146a expression itself would represent an etiologic basis common to both AD and prion disease, emerge. In any case, it would be most relevant to expand the list of potential mRNA targets of miR-146a, as initiated with the identification of complement factor H (CFH) by Lukiw et al. (2008), in order to get a privileged and novel insight into the pathogenesis of neurodegenerative diseases.
8. Conclusion and perspectives
Considering the ability of miRNAs to target multiple genes for expression, their potential for modulating neuroprotective mechanisms, and the deregulation of disease-specific subsets of miRNAs, the prospect of using miRNAs as therapeutics as well as biomarkers in neurological and neurodegenerative diseases is tantalizing (Saba et al., 2008). However, further investigations are required in order to improve our understanding of the biological role and importance of miRNAs in maintaining normal brain function, as well as of the etiology and molecular basis of neurological and neurodegenerative disorders, such as the fragile X syndrome, AD and prion disease, in which miRNAs may play a central role. Research advances in these areas are key to ensure the development of efficient therapeutic tools and strategies aimed at preserving, restoring or neutralizing global and/or specific miRNA function in the brain of patients at risk of developing, or suffering from, neurological and neurodegenerative diseases.
Acknowledgments
We apologize to researchers whose work could not be discussed or cited due to space limitations. P.P. is a Senior Scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). This work was supported by a Young Investigator Award from NARSAD, the World’s leading charity dedicated to mental health research, and Grant NDG-70190 from the Canadian Institutes of Health Research (CIHR) – Institute of Genetics and CIHR – Institute of Neuroscience, Mental Health and Addiction.
Abbreviations
- Aβ
β-amyloid
- AD
Alzheimer’s disease
- APP
β-amyloid precursor protein
- BACE
APP-converting enzyme
- FMRP
fragile X mental retardation protein
- miRNA
microRNA
- miRNP
miRNA-containing RNP
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real-time PCR
- RNP
ribonucleoprotein
- RT
reverse transcription
- siRNA
small interfering RNA
- UTR
untranslated region
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–24. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and MicroRNA-328 Regulate Expression of Mouse {beta}-Amyloid Precursor Protein-converting Enzyme 1. J Biol Chem. 2009;284:1971–81. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–87. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–6. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. Rna. 2009;15:362–6. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106:21179–84. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–41. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 65:373–84. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2009 doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–37. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallozzi M, Chapuis J, Le Provost F, Le Dur A, Morgenthaler C, Peyre C, Daniel-Carlier N, Pailhoux E, Vilotte M, Passet B, Herzog L, Beringue V, Costa J, Tixador P, Tilly G, Laude H, Vilotte JL. Prnp knockdown in transgenic mice using RNA interference. Transgenic Res. 2008;17:783–91. doi: 10.1007/s11248-008-9179-2. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Ann Neurol. 2002;51:783–6. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–7. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SC, Krichevsky AM, Kosik KS, Tsai LH. BACE1 suppression by RNA interference in primary cortical neurons. J Biol Chem. 2004;279:1942–9. doi: 10.1074/jbc.M309219200. [DOI] [PubMed] [Google Scholar]
- Khandjian EW. Biology of the fragile X mental retardation protein, an RNA-binding protein. Biochem Cell Biol. 1999;77:331–42. [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–83. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–22. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, Martin L, Louis JC, Yan Q, Richards WG, Citron M, Vassar R. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–2. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–9. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Montag J, Hitt R, Opitz L, Schulz-Schaeffer WJ, Hunsmann G, Motzkus D. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol Neurodegener. 2009;4:36. doi: 10.1186/1750-1326-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L. Neuroscience: Alzheimer’s disease. Nature. 2009;461:895–7. doi: 10.1038/461895a. [DOI] [PubMed] [Google Scholar]
- Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer’s amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315:418–25. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS ONE. 2008;3:e2248. doi: 10.1371/journal.pone.0002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993;90:10962–6. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–29. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Perron MP, Boissonneault V, Gobeil LA, Ouellet DL, Provost P. Regulatory RNAs: future perspectives in diagnosis, prognosis, and individualized therapy. Methods Mol Biol. 2007;361:311–26. doi: 10.1385/1-59745-208-4:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci. 2008;13:2537–47. doi: 10.2741/2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I, Provost P. Hypothesis: A Role for Fragile X Mental Retardation Protein in Mediating and Relieving MicroRNA-Guided Translational Repression? J Biomed Biotechnol. 2006;2006:16806. doi: 10.1155/JBB/2006/16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–74. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberds SL, Anderson J, Basi G, Bienkowski MJ, Branstetter DG, Chen KS, Freedman SB, Frigon NL, Games D, Hu K, Johnson-Wood K, Kappenman KE, Kawabe TT, Kola I, Kuehn R, Lee M, Liu W, Motter R, Nichols NF, Power M, Robertson DW, Schenk D, Schoor M, Shopp GM, Shuck ME, Sinha S, Svensson KA, Tatsuno G, Tintrup H, Wijsman J, Wright S, McConlogue L. BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer’s disease therapeutics. Hum Mol Genet. 2001;10:1317–24. doi: 10.1093/hmg/10.12.1317. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, Dubas F, Frebourg T, Campion D. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–6. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS ONE. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–8. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–4. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–9. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–60. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–6. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly G, Chapuis J, Vilette D, Laude H, Vilotte JL. Efficient and specific down-regulation of prion protein expression by RNAi. Biochem Biophys Res Commun. 2003;305:548–51. doi: 10.1016/s0006-291x(03)00805-2. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–41. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–23. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull. 2009;80:268–73. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. Embo J. 2002;21:5875–85. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]