Abstract
Gibberellin 2-oxidase (GA 2-oxidase) plays very important roles in plant growth and development. In this study, the AtGA2ox8 gene, derived from Arabidopsis (Arabidopsis thaliana), was transformed and over-expressed in rapeseed (Brassica napus L.) to assess the role of AtGA2ox8 in biomass accumulation and lignification in plants. The transgenic plants, identified by resistant selection, polymerase chain reaction (PCR) and reverse-transcription PCR (RT-PCR) analyses, and green fluorescence examination, showed growth retardation, flowering delay, and dwarf stature. The fresh weight and dry weight in transgenic lines were about 21% and 29% lower than those in wild type (WT), respectively, and the fresh to dry weight ratios were higher than that of WT. Quantitative measurements demonstrated that the lignin content in transgenic lines decreased by 10%–20%, and histochemical staining results also showed reduced lignification in transgenic lines. Quantitative real-time PCR analysis indicated that the transcript levels of lignin biosynthetic genes in transgenic lines were markedly decreased and were consistent with the reduced lignification. These results suggest that the reduced biomass accumulation and lignification in the AtGA2ox8 over-expression rapeseed might be due to altered lignin biosynthetic gene expression.
Keywords: Rapeseed, AtGA2ox8, Biomass, Lignification, Gibberellins
1. Introduction
Gibberellins (GAs) are endogenous plant hormones playing very important roles in plant growth and regulating many aspects of plant development, including seed germination, shoot growth, flower induction, stimulation of cell elongation and cell division, hypocotyl elongation, fruit maturation, and leaf expansion (Kende and Zeevaart, 1997; Harberd et al., 1998; Hedden and Proebsting, 1999). Only a few of the 126 presently known GAs, such as GA1, GA3, GA4, and GA7, have been shown to have biological activity (Hedden and Phillips, 2000).
GA metabolism takes place in three different cellular compartments, i.e., plastids, endoplasmic reticulum (ER), and cytosol (Hedden and Phillips, 2000; Olszewski et al., 2002; Sun and Gubler, 2004). Multiple enzymes including ent-copalyl diphosphate synthase, ent-kaurene synthase, P450 monooxygenases, and dioxygenases are involved in GA metabolism and catabolism. Two dioxygenases, GA 20-oxidase and GA 3β-hydroxygenase, catalyze the last few steps in the synthesis of bioactive GA. Another dioxygenase, GA 2-oxidase, catalyzes GA catabolism of bioactive GA or their precursors (Thomas et al., 1999; Hedden and Phillips, 2000; Helliwell et al., 2001; Olszewski et al., 2002; Zhao et al., 2007b).
The genes encoding the multi-functional GA 2-oxidase have been cloned from various species, such as runner bean (Phaseolus coccineus) (Thomas et al., 1999; Appleford et al., 2007), Arabidopsis (Arabidopsis thaliana) (Thomas et al., 1999; Schomburg et al., 2003), pea (Pisum staivum) (Lester et al., 1999; Martin et al., 1999), rice (Oryza sativa) (Sakamoto et al., 2001; 2004), poplar (Populus tremula) (Busov et al., 2003), spinach (Spinacia oleracea) (Lee and Zeevaart, 2002; 2005), and lettuce (Lactuca sativa) (Nakaminami et al., 2003). Phylogenetic analysis divides the GA 2-oxidase family into three classes according to their amino acid sequences (Lee and Zeevaart, 2005). C19-GAs (GA1 and GA4) and C19-GA precursors (GA20 and GA9) can be converted to inactive GAs by members of classes I and II, respectively (Sakamoto et al., 2004; Lee and Zeevaart, 2005; Lo et al., 2008), and C20-GAs (GA12 and GA53) can be 2β-hydroxylated by members of class III, including SoGA2ox3, AtGA2ox7, and AtGA2ox8 (Schomburg et al., 2003; Lee and Zeevaart, 2005; Lo et al., 2008). The 2β-hydroxylated C20-GA precursors cannot be converted to active GAs, resulting in a decrease in active GA levels.
The over-expression of the genes encoding AtGA2-oxidase causes a dwarf phenotype and delayed flowering in Arabidopsis (Schomburg et al., 2003; Wang et al., 2004; Zhao et al., 2007a; Rieu et al., 2008). The heterologous expression of AtGA2-oxidases also resulted in growth retardation in wheat (Hedden and Phillips, 2000), rice (Sakamoto et al., 2001), tobacco (Schomburg et al., 2003), bahiagrass (Agharkar et al., 2007), and Solanum species (Dijkstra et al., 2008). Apart from the effect of expression of AtGA2-oxidases on plant elongation, Biemelt et al. (2004) reported that biomass production was decreased in AtGA2ox1 plants, and that the transgenic plants have altered lignification. Moreover, they showed that GA have different effects on xylem and pitch cell formation.
Both the rapeseed (Brassica napus L.) and Arabidopsis are cruciferous plants. Zhao et al. (2007a) and Schomburg et al. (2003) reported that over-expression of AtGA2ox8 causes reduced bioactive GA levels in Arabidopsis. To investigate the correlation of AtGA2ox8 gene to biomass characteristics and lignin biosynthesis, we introduced it into rapeseed. Here, we show that heterologous expression of AtGA2ox8 gene in rapeseed causes growth retardation, flowering delay, dwarf stature, and reduced biomass accumulation and lignification.
2. Materials and methods
2.1. Plant materials and genetic transformation
The cultivar of rapeseed (Brassica napus L.) in this study was N529, provided by the Academy of Seed Industry of Hunan Yahua, Changsha, China. AtGA2ox8 transgenic plants were prepared by the first amplifying AtGA2ox8 (GenBank accession No. AL021960, accession number in The Arabidopsis Information Resource (TAIR) website is At4g21200) gene from Arabidopsis cDNA using a reverse-transcription polymerase chain reaction (RT-PCR) method with primers AtGA2ox8-F (5′-TCCCCCGGGATGGATCCACCATTCAACGAAATAT-3′) and AtGA2ox8-R (5′-CCGCTCGAGTTAGTAGACGTGTTAAGGAACCAGGAA-3′) (restriction sites are underlined), and then subcloning it into the SmaI and XhoI sites of pEGAD vector downstream of the CaMV35S promoter. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 by the electroporation method, as described in our previous study (Lin et al., 2009). When the rapeseed plants begin to form boll and produce flora inflorescences, the healthy flora inflorescences grown at the top of rapeseed plants were selected and immersed into a plastic bag with Agrobacterium inoculum for 30 s, and then covered with an agricultural parchment bag to maintain high humidity. One week later, the agricultural parchment bags were uncovered and the flower buds at the top of dipped inflorescences were removed. When the siliques became brown and dry, the seeds from the dry siliques were collected and stored at −20 °C for screening.
2.2. Transgenic line screening
Transgenic plants were first selected using herbicide Basta (1:800, v/v). Total genomic DNA was extracted from Basta-resistant transgenic lines using hexadecyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The detection of BastaR gene was carried out by PCR analysis with primers Bar-F (5′-CTACATCGAGACAAGCACGGT-3′) and Bar-R (5′-CTGAAGTCCAGCTGCCAGAA-3′). The PCR program was as follows: 94 °C for 5 min, and then 30 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, and a final extension of 72 °C for 5 min. The PCR products were analyzed using 1.0% (w/v) agarose gel electrophoresis.
To investigate whether the transgene is expressed in the transgenic plants, some lateral roots were excised from the Basta-resistant transgenic lines. The green fluorescent protein (GFP) fusion proteins were examined using inverted fluorescence microscope (Nikon TE2000-U, Japan).
2.3. GA3 treatment
To investigate the response of hypocotyl elongation to exogenous GA3, about 100 seeds were sown on filter paper saturated with 100 μmol/L GA3 (Shanghai Solvent) in a plate and grown under continuous white light in a temperature-controlled growth chamber at 22 °C. Hypocotyl lengths of more than twenty 6-d-old seedlings were measured manually following the method of Lin et al. (1998), and the standard deviations (SDs) of the measurements were calculated from three independent experiments with 20 samples each.
2.4. Greenhouse evaluation of growth and biomass characteristics
Transgenic plants and wild-type plants were grown in a greenhouse. The stem heights of the plants were evaluated in the flowering and maturation periods. Leaves were counted in the maturation period. Fresh weight and dry weight of the whole plant, including the roots, were determined after harvest according to the method of Biemelt et al. (2004).
The seed oil content and the glucosinolate, erucic acid, and oleic acid contents in seed oil of rapeseed were determined by near-infrared reflectance (NIR) spectroscopy after harvest (Panford and de Man, 1990).
Each value of all the experiments represents the mean±SD of 10 to 15 samples each.
2.5. Histochemical detection and quantitative determination of lignin
Histochemical detection was carried out on the hand-cut cross sections of stems from the second, eighth, and seventeenth internodes (counting from the bottom up to the top) of the plants grown in greenhouse during the flowering period. The sections were stained using phloroglucinol-HCl reagent, and then analyzed on an inverted microscope (Nikon TE2000-U, Japan) according to the methods described previously (Campbell and Ellis, 1992; Biemelt et al., 2004). Each experiment was repeated three times with at least three samples each.
The lignin contents in about 100 mg fresh weight of the apical, middle, and basal parts of the stems of both the AtGA2ox8 transgenic plants and the wild-type plants grown in the greenhouse during the flowering period were determined by acetyl bromide ultraviolet spectrophotometry (280 nm) according to the methods described previously (Fukushima and Dehority, 2000; Fukushima and Hatfield, 2001; Biemelt et al., 2004). Each value was obtained from two independent experiments with three samples each.
2.6. Semi-quantitative RT-PCR analysis
To investigate whether the AtGA2ox8 gene was over-expressed in the transgenic rapeseed plant, about 100 seeds were sown on filter paper saturated with Murashige and Skoog (MS) salt liquid medium in a plate and grown under continuous white light in a temperature-controlled growth chamber at 22 °C. Total RNA was isolated from 6-d-old seedlings using Puprep RNAeasy mini kit (Ambiogen Life Tech Ltd., China). The transcript level of AtGA2ox8 was analyzed using semi-quantitative RT-PCR as described previously (Zhao et al., 2007a). The rapeseed Actin (ACT) (European Molecular Biology Laboratory (EMBL) accession No. AF111812) gene was used as an internal control. The PCR started with a denaturation stage at 95 °C for 5 min, which was then followed by 26 (for the ACT) or 35 (for the AtGA2ox8) cycles with each cycle composed of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The RT-PCR reactions for each experiment were repeated at least three times in three independent trials. The sequences of the primers
were: AtGA2ox8-F (5′-CGGAATCAGAGGCATTAGC-3′), AtGA2ox8-R (5′-CCACCTTTGGGTTCGTCAT-3′), ACT-F (5′-TCCCTCAGCACTTTCCAACAG-3′), and ACT-R (5′-AAGGACCAGAGCATCATCACAAG-3′).
2.7. Quantitative real-time PCR analysis
To detect the mRNA level of lignin biosynthetic gene, total RNA was isolated from apical parts of stems of rapeseed during the flowering period using Puprep RNAeasy mini kit (Ambiogen Life Tech Ltd.). The amount of mRNA was analyzed using quantitative real-time PCR as described previously (Wang et al., 2009). The PCR was performed in Mx3000P (Stratagene), and started with a denaturation stage at 95 °C for 10 min, which was then followed by 40 cycles with each cycle composed of 95 °C for 30 s, 60 °C for 20 s, and 72 °C for 20 s. The PCR reactions were repeated three times from three independent experiments. The ACT gene of rapeseed was used as an internal control. The sequences of primers used in this study are listed in Table 1.
Table 1.
Primer sequences used for quantitative real-time PCR analysis
| Gene name | EMBL accession No. | Forward and reverse (5′-3′) |
| PAL1 | DQ341309 | Forward: CAAAGCGATTCACGGAGGTAA; reverse: CGCTCCTTTGAAACCGTAGTC |
| PAL2 | AY795080 | Forward: TTGGATTACGGATTCAAAGGA; reverse: CGAGATGAGTCCCAAAGAGTT |
| C4H | DQ485129 | Forward: TGACTTTAAGTATGTGCCGTTTG; reverse: GGACCTTGGCTTCATTACGAT |
| 4CL | CX190902 | Forward: TAATCCGAATCTTTACTTCCACAG; reverse: GCAACCGTCACTTTACACCTCT |
| HCT | CD832138 | Forward: CTCAAGGCTAAAGCCAAGGAG; reverse: TGACGTTGCCAAAGTAACCAG |
| CCR | CD844319 | Forward: GGTGGAAGTTTAGGTCATTAGAAGA; reverse: CCAATAGTAGACTTGAGGAGGTGAA |
| CAD | CD814354 | Forward: TTATGTCCTGGTTGGTTTCCC; reverse: CACCCTTTCAATAGCTTCGTTT |
| ACT | AF111812 | Forward: TCCCTCAGCACTTTCCAACAG; reverse: ACACTCACCACCACGAACCAG |
3. Results
3.1. Transformation and molecular characterization
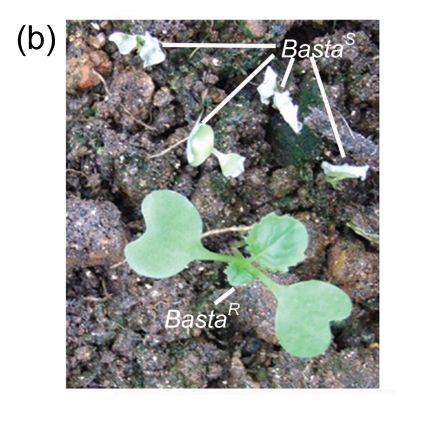
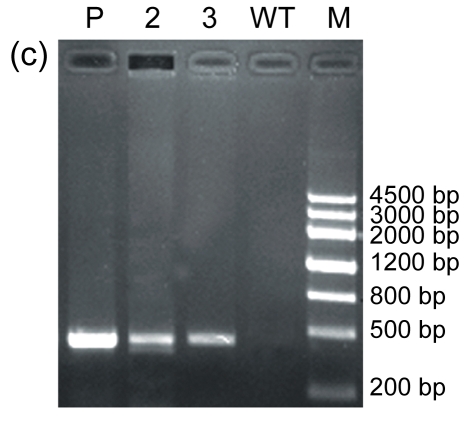
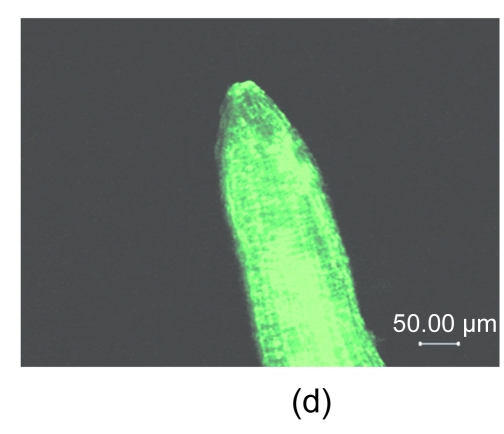
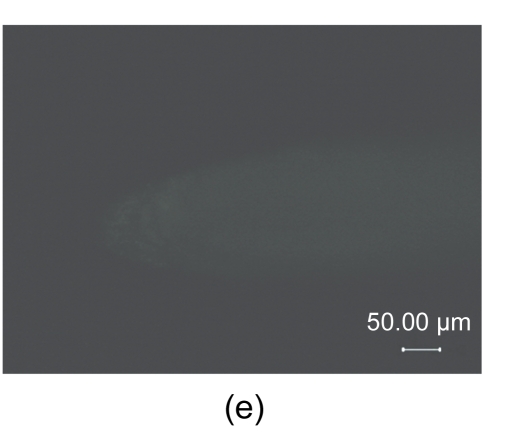
The AtGA2ox8 gene was introduced into rapeseed by Agrobacterium-mediated floral dip transformation as stated in Section 2.1. The expression cassette contains the GFP report gene, the target gene (AtGA2ox8), and the herbicide Basta-resistant (BastaR) select maker gene. AtGA2ox8 and GFP were driven by the CaMV35S promoter and would express fusion protein in the transgenic plants (Fig. 1a in p.478). Ten Basta-resistant independent lines were obtained after the first screening with herbicide Basta (Fig. 1b). The BastaR gene was detected by PCR analysis in all Basta-resistant plants but not in the wild-type plants (partly in Fig. 1c), indicating that the transgene was indeed integrated into the genomic DNA. To determine whether the transgene is normally expressed in the transgenic plants, the GFP fusion proteins were examined using inverted fluorescence microscope, and strong green fluorescence was detected in the root of transgenic lines (partly in Fig. 1d). No green fluorescence was observed in root of the wild-type plant (Fig. 1e). These data confirmed that transgenic plants were obtained. Two independent transgenic lines, 35S::GFP-AtGA2ox8-2 and 35S::GFP-AtGA2ox8-3, were then randomly selected for the further detection of the target gene expression using semi-quantitative RT-PCR analysis. The results showed that the AtGA2ox8 signals of the two independent transgenic lines were quite strong, whereas the corresponding signal of the wild type was hardly detectable (Fig. 2). These results indicated that the AtGA2ox8 gene was normally over-expressed in the two independent transgenic lines.
Fig. 1.
Transformation and transgenic rapeseed plant screening
(a) Transgene expression cassette contains the CaMV35S promoter (35S), AtGA2ox8 gene, green fluorescent protein (GFP) reporter gene, and herbicide Basta-resistant (BastaR) select marker gene, and the positions of enzyme sites and the T-DNA left border (LB) and right border (RB) are shown; (b) Transgenic rapeseed plants were screened by herbicide Basta. BastaS or BastaR symbolizes Basta-susceptible or Basta-resistance, respectively; (c) PCR analysis of BastaR gene in the transgenic rapeseed. DNA was extracted from the wild type (WT) and the Basta-resistant lines, respectively. P, 2, 3, WT, and M symbolize pEGAD plasmid, 35S::GFP-AtGA2ox8-2, 35S::GFP-AtGA2ox8-3, wild type, and marker, respectively; (d) & (e) GFP fluorescence imaging of transgenic plants with inverted fluorescence microscope. The images of the lateral root cap of the transgenic plant and WT are shown in (d) and (e), respectively
Fig. 2.
Semi-quantitative RT-PCR analysis of AtGA2ox8 gene in the transgenic rapeseed
Total RNA was isolated from 6-d-old seedlings of the wild type (WT) and the two independent transgenic lines grown on filter paper saturated with MS salt liquid medium in a plate under continuous white light in a temperature-controlled growth chamber at 22 °C. The transcript levels are shown as gel images, and the ACT is used as an internal control. Each experiment was repeated at least three times in three independent trials and similar results were obtained. Lane 1: WT; Lane 2: 35S::GFP-AtGA2ox8-2; Lane 3: 35S::GFP-AtGA2ox8-3
3.2. Effect of over-expression of AtGA2ox8 in rapeseed on plant growth
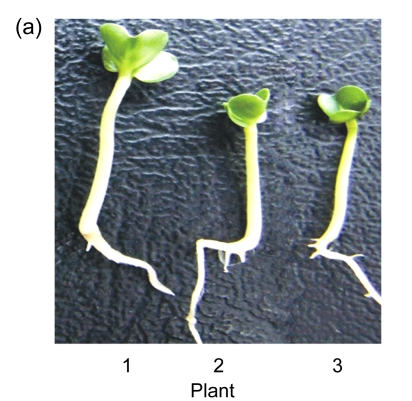
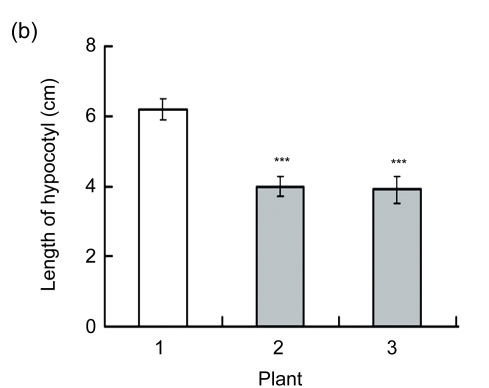
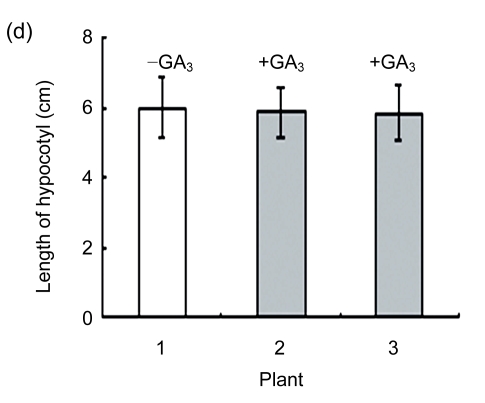
Transgenic seeds were sown in plates to investigate the hypocotyl phenotype. Compared with wild type, the over-expression of AtGA2ox8 in rapeseed inhibited the hypocotyl elongation and led to shorter hypocotyls (Figs. 3a and 3b). Furthermore, the phenotype of 35S::GFP-AtGA2ox8 could be rescued by exogenous GA3 (Figs. 3c and 3d). These results are consistent with our previous reports that the over-expression of AtGA2ox8 in Arabidopsis resulted in short hypocotyl (Zhao et al., 2007a), and the ga1 mutant impaired in the ent-copalyl diphosphate synthase gene caused short hypocotyl (Sun et al., 1992; Alabadi et al., 2004). Therefore, these observations strongly suggested that AtGA2ox8 could induce a GA-deficient phenotype in different species.
Fig. 3.
Response of AtGA2ox8 transgenic plants to GA3
(a) & (b) Six-day-old seedlings of both the wild type (WT) and the two independent transgenic lines and their hypocotyl lengths are shown, respectively; (c) & (d) GA3-rescuable phenotype of 6-d-old seedlings of the two independent transgenic lines and their hypocotyl lengths are shown, respectively. The data represent the mean values obtained from three independent experiments with 20 samples each. The error bars represent the SDs. *** P<0.001, significant difference between the WT and transgenic plants. Plant: 1. WT; 2. 35S::GFP-AtGA2ox8-2; 3. 35S::GFP-AtGA2ox8-3
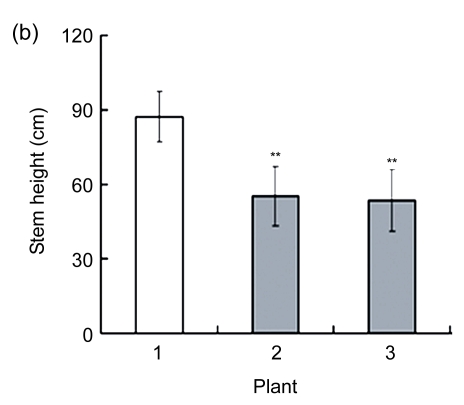
To evaluate the growth characteristics, the rapeseed seedlings were planted in a greenhouse. The phenotypes were investigated during different growth periods. The AtGA2ox8 transgenic lines exhibited growth retardation, flowering delay, and dwarf phenotype. The stem heights of the AtGA2ox8 over-expression lines were only about 70% of those of the wild-type plants (Fig. 4). Moreover, the AtGA2ox8 over-expression lines have shorter leaves (data not shown). Compared with the wild-type plants, the flowering time in the AtGA2ox8 over-expression lines was delayed for approximately one week (Fig. 4a).
Fig. 4.
Comparison of the stem heights of both the AtGA2ox8 transgenic plants and the wild type (WT) grown in a greenhouse during different growth periods
(a) & (c) Plants of the WT and the two independent transgenic lines during the flowering (a) and maturation (c) periods, respectively; (b) & (d) Stem heights of plants during the flowering (b) and maturation (d) periods, respectively. The error bars represent the SDs of 10 to 15 samples each. ** P<0.01, significant difference between the WT and transgenic plants. Plant: 1. WT; 2. 35S::GFP-AtGA2ox8-2; 3. 35S::GFP-AtGA2ox8-3
3.3. Evaluation of biomass characteristics of transgenic lines
The biomass characteristics of the transgenic plants during maturation period were also analyzed (Table 2). As far as the number of leaves is concerned, there was almost no difference between the transgenic plants and the wild type. In order to assess the impact of the transgenic plants on biomass production, the fresh weight and dry weight of the whole plants were determined. As shown in Table 2, significant differences in biomass production between the wild type and the transgenic lines were observed. Compared with the wild type, the fresh weight and dry weight in the transgenic lines decreased by about 21% and 29%, respectively. The fresh to dry weight ratios for wild type plants, 35S::GFP-AtGA2ox8-2, and 35S::GFP-AtGA2ox8-3 were 10.1, 11.2, and 11.3, respectively. Moreover, the 1000-seed weight in the transgenic plants was significantly increased, though the seed yield per plant was only slightly increased.
Table 2.
Effects of over-expression of AtGA2ox8 in rapeseed on biomass accumulation
| Plant | Number of leaves | FW (g) | DW (g) | FW/DW | Seed yield per plant (g) | 1000-seed weight (g) | Seed oil content (%) | Erucic acid content (%) | Glucosinolate content (µmol/g) | Oleic acid content (%) |
| 1 | 30.0±1.3 | 620.4±20.5 | 61.7±5.6 | 10.1 | 17.7±1.8 | 2.36±0.09 | 32.5±0.6 | 0.86±0.35 | 40.6±2.7 | 62.2±0.17 |
| 2 | 29.5±1.2 | 495.5±14.5*** | 44.2±7.6** | 11.2 | 18.8±1.8 | 2.85±0.10*** | 33.5±0.5* | 1.30±0.50 | 33.5±2.7* | 62.5±0.20 |
| 3 | 30.0±1.0 | 488.7±13.2*** | 43.5±7.3** | 11.3 | 17.8±1.4 | 2.92±0.09*** | 34.4±0.4** | 1.35±0.49 | 30.8±2.8* | 63.3±0.28 |
Results are given for two independent lines and represent the mean±SD of 10 to 15 samples each. FW: fresh weight; DW: dry weight
P<0.05 significant differences between the WT and the two mutant lines
P<0.01 significant differences between the WT and the two mutant lines
P<0.001 significant differences between the WT and the two mutant lines
Plant: 1. WT; 2. 35S::GFP-AtGA2ox8-2; 3. 35S::GFP-AtGA2ox8-3
The determination of the seed oil content and the glucosinolate, erucic acid, and oleic acid contents in seed oil revealed that the two transgenic lines produced 3% or 6% more seed oil and 15%–25% less glucosinolate in seed oil than the wild type. The erucic acid content in seed oil was higher in the transgenic lines as compared to the wild type. However, there was almost no difference in the oleic acid content between the transgenic lines and the wild type. These results demonstrated that AtGA2ox8 reduced the biomass accumulation and also affected the yield and quality of seed oil in rapeseed.
3.4. Effect of over-expression of AtGA2ox8 in rapeseed on lignification
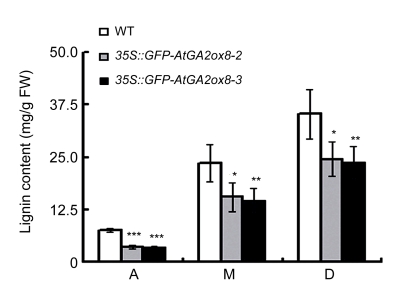
It has been reported that the change in fresh to dry weight ratio might result from the altered composition or deposition of the structure-forming substances like cellulose and/or lignin, and that higher fresh to dry weight ratio of plants usually led to a relatively low lignin content in plants (Biemelt et al., 2004). Lignification is also a characteristic feature of secondary growth and increases with maturity (Israelsson et al., 2005). To determine the different levels of lignification in the transgenic lines and the wild type, different parts of stems, i.e., apical, middle, and basal parts, were collected and their lignin contents were measured. For both the transgenic lines and the wild type, the lignin content increased gradually from the apical parts to the basal parts (Fig. 5). Lignin content comparison revealed that the two independent transgenic lines, 35S::GFP-AtGA2ox8-2 and 35S::GFP-AtGA2ox8-3, produced about 10% and 20% lower lignin contents in stems than the wild-type plants, respectively.
Fig. 5.
Lignin content in different parts of stems of both the AtGA2ox8 transgenic plants and the wild-type (WT) plants grown in a greenhouse during the flowering period
A, M, and B symbolize apical, middle, and basal parts, respectively. The error bars represent the SDs of two independent experiments with three samples each. * P<0.05, ** P<0.01, *** P<0.001, significant difference between the WT and transgenic plants. FW: fresh weight
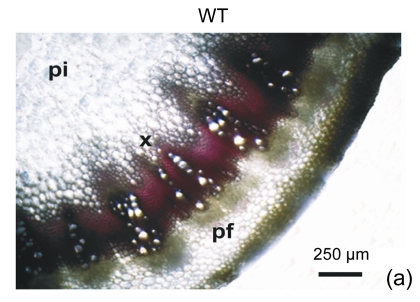
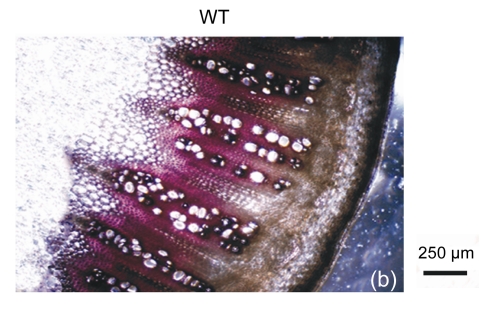
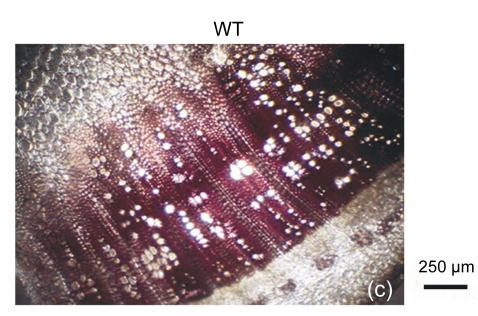
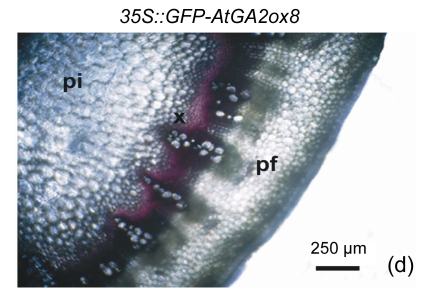
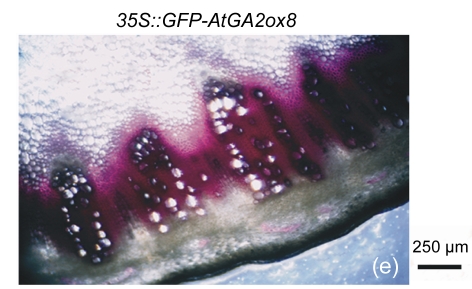
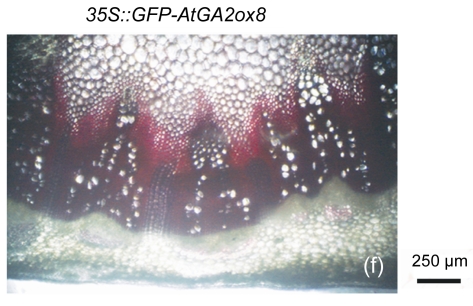
The cell walls of lignification display red or purple-red color in acidic phloroglucinol. For visually investigating the differences in lignification between the transgenic lines and the wild type, different parts of stems (the seventeenth internode, the eighth internode, and the second internode) were analyzed by histochemical detection. The biomass characteristics and lignin contents of the two transgenic lines were essentially the same. Hence, only one of the two transgenic lines, 35S::GFP-AtGA2ox8-2, was selected for the histochemical detection, and its staining pattern is shown in Fig. 6. In general, the number of lignified vessels and the amount of lignin deposition were gradually increased from the apex to the base of the stem. The AtGA2ox8 over-expression line had fewer lignified vessels than the wild-type controls, which was in good accordance with the lignin contents shown in Fig. 5. These results demonstrate that the heterologous expression of AtGA2ox8 in rapeseed can result in lignin deposition reduction.
Fig. 6.
Histochemical detection of lignin in the stem cross sections of both the AtGA2ox8 transgenic plants and the wild type (WT)
Different internodes were collected from plants during the flowering period for histochemical detection. (a), (b) & (c) Phloroglucinol-HCl stained second, eighth, and seventeenth internodes of the WT, respectively; (d), (e) & (f) Phloroglucinol-HCl stained second, eighth, and seventeenth internodes of the AtGA2ox8 transgenic plants, respectively. pf, pi, and x symbolize phloem fiber, pith, and xylem, respectively. Each experiment was repeated three times using at least three samples each with similar results
3.5. Effect of over-expression of AtGA2ox8 in rapeseed on lignin biosynthetic gene expression
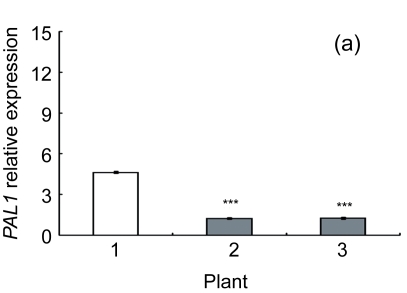
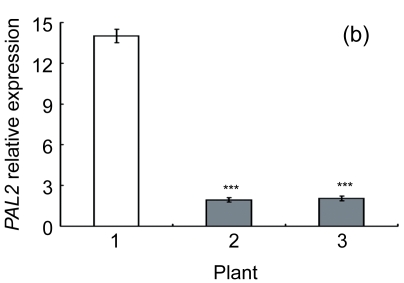
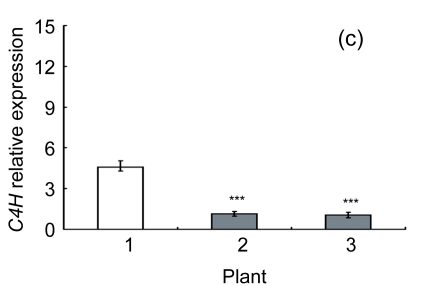
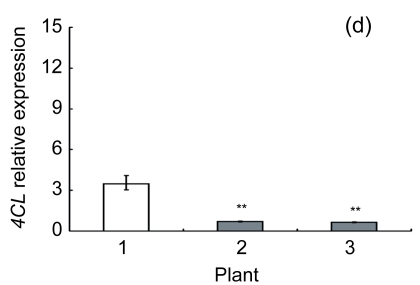
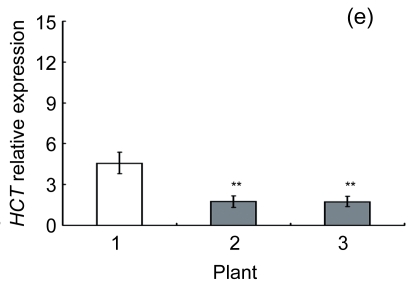
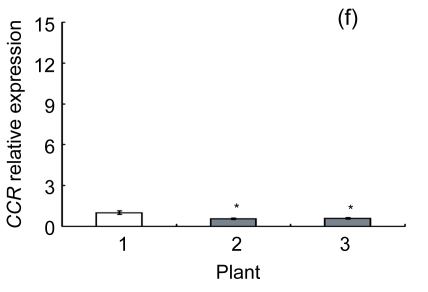
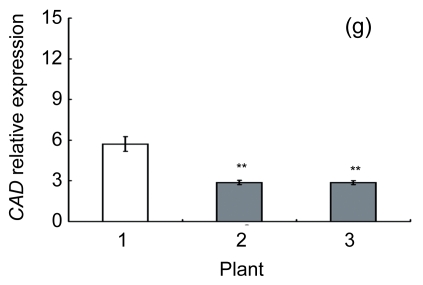
Lignin is a complex phenylpropanoid polymer, and lignin monolignols are synthesized through the phenylpropanoid pathway (Boerjan et al., 2003). To determine whether the changes of lignin content in transgenic lines could be a result of altered genes expression, a search for the cDNA coding or the expressed sequence tag (EST) sites for the enzymes in the lignin pathway was performed on the Brassica website (http://www.brassica.info/). The cDNA coding sequences of seven key enzymes in lignin synthesis, i.e., phenylalanine ammonialyase 1 (PAL1), phenylalanine ammonialyase 2 (PAL2), 4-coumarate: CoA ligase (4CL), cinnamate 4-hydroxylase (C4H), hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT), cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD), were found. The relative expression of these genes in apical parts of the stems of the AtGA2ox8 over-expression lines and the wild-type plants was analyzed by quantitative real-time PCR. The transcript levels of all the seven genes were found to have a significant difference between the wild type and the transgenic plants, and were markedly reduced in the AtGA2ox8 over-expression lines (Fig. 7). These transcript reductions coincided with the reduced lignification in the apical parts of stems of the transgenic plants (Figs. 5 and 6). All these results indicate that the changes in lignification in transgenic plants might be associated with the altered expression of lignin biosynthetic genes.
Fig. 7.
Expression analysis of lignin biosynthetic genes in both the AtGA2ox8 transgenic plants and the wild type (WT) Plants were grown in a greenhouse, and apical parts of stems were collected during the flowering period. Total RNA was isolated for quantitative real-time PCR analysis. (a) PAL1; (b) PAL2; (c) C4H; (d) 4CL; (e) HCT; (f) CCR; (g) CAD. Data represent mean values with SDs obtained from three independent assays. * P<0.05, ** P<0.01, *** P<0.001, significant difference between the WT and transgenic plants. Plant: 1. WT; 2. 35S::GFP-AtGA2ox8-2; 3. 35S::GFP-AtGA2ox8-3
4. Discussion
This study investigated the over-expression of AtGA2ox8 in rapeseed by genetic engineering technology. It was found that the over-expression of AtGA2ox8 could inhibit hypocotyl elongation in rapeseed. This inhibition effect can be removed by the application of GA3 to the AtGA2ox8 transgenic lines. AtGA2ox8 acts as GA 2-oxidases that hydroxylate carbon-2 of C20-GA precursors (e.g., GA12), but not active GAs (Schomburg et al., 2003; Lee and Zeevaart, 2005; Lo et al., 2008). Therefore, active C19-GAs such as GA3 can be employed to rescue AtGA2ox8-derived short hypocotyl phenotype.
The over-expression of AtGA2ox8 in rapeseed also resulted in dwarf phenotype with delayed flowering, shorter stems, and shorter leaves. Nevertheless, viable seeds were obtained from the transgenic lines. Moreover, the 1000-seed weight in the transgenic plants was significantly increased, though the seed yield per plant was only slightly increased. Similarly, heterologous over-expression of C20-GA2ox, including Arabidopsis GA2ox8 (Schomburg et al., 2003), spinach GA2ox3 (Lee and Zeevaart, 2005), rice GA2ox5, and GA2ox6 (Lo et al., 2008) in transgenic tobacco, resulted in reduced plant height, while allowing the production of viable seeds. The same results were also obtained from homologous over-expression of C20-GA2ox, including rice GA2ox5 and GA2ox6, and Arabidopsis GA2ox8 in transgenic plants. In contrast, the rice over-expressing C19-GA2ox, including GA2ox1, completely suppressed the formation of inflorescences (Sakamoto et al., 2001). These seemingly contradictory results may be explained as follows based on the findings in previous studies: there are so many more C20-GA precursors than the C19-GA precursors and active C19-GAs in plants that some of the C20-GA precursors could escape from inactivation by C20-GA2oxs (Sakamoto et al., 2001; Schomburg et al., 2003; Lee and Zeevaart, 2005). Hence, they can be converted to active C19-GAs by GA 20-oxidase and GA 3-oxidase (Lo et al., 2008).
To date, there has been no report about whether altered GA metabolism might change the seed oil content of rapeseed, glucosinolate and erucic acid contents in seed oil. Experimental results obtained in this work revealed that, compared to the wild-type plants, the AtGA2ox8 over-expression lines could produce significantly more seed oil content. The erucic acid content also increased in the seed oil of the transgenic plants, but agreed with the criteria for double-low rapeseed species (Olsen and Soerensen, 1980; Sivaraman et al., 2004). However, the glucosinolate content in the seed oil of the AtGA2ox8 over-expression lines was obviously lower than that of the wild type. These preliminary results suggested that AtGA2ox8 might be useful in rapeseed breeding without affecting the yield and quality of seed oil. More detailed studies are needed, however, to fully elucidate the effect of GA on the formation and quality of seed oil.
Biomass measurements demonstrated that there were some significant differences between the wild type and the transgenic lines. The fresh to dry weight ratio was higher in the transgenic plants than in the wild-type plant, which might be due to the altered deposition of lignin. The AtGA2ox8 over-expression lines produced lower lignin content than the wild-type plants. The histochemical staining results also showed that the number of lignified vessels was less in the AtGA2ox8 over-expression lines, compared to the wild-type controls. The explanations of these results are as follows: GAs stimulate xylem fiber elongation (Eriksson et al., 2000; Israelsson et al., 2005) and lignin accumulation (Biemelt et al., 2004). The AtGA2ox8 could 2β-hydroxylate the C20-GA precursors and render them unable to be converted to active GAs. The decrease in the levels of active GAs resulted in the decrease in lignin accumulation (Schomburg et al., 2003; Biemelt et al., 2004). Lignin is a complex phenylpropanoid polymer, and lignin monolignols are synthesized through the phenylpropanoid pathway (Boerjan et al., 2003). To elucidate the cause for the effect of GA on lignin accumulation, we analyzed the transcript levels of seven key enzymes in the lignin pathway and found that their transcripts were lower in the apical parts of the stems of the transgenic plants than in those of the wild type. The results demonstrate that changes in lignification in AtGA2ox8 over-expression lines might be due to the altered expression of biosynthetic genes. Similar results were obtained in AtGA2ox1 over-expression tobacco (Biemelt et al., 2004). Thus, our data suggest that GAs might affect plant lignification by regulating transcript levels of lignin biosynthetic genes.
The stems of the transgenic plants with low lignin content are conducive to rational use of resources and environment protection in the paper industry, and also more susceptible to fiber digestion by livestock (Fukushima and Dehority, 2000; Fukushima and Hatfield, 2004). Therefore, one of the major targets for intervention by genetic manipulation in our future work may be to inhibit GA biosynthesis and obtain plants with decreased lignin content. A focus will be placed on the molecular mechanism of gibberellins-regulated lignin biosynthesis.
Biography
Introducing editorial board member: Prof. Xuan-ming LIU, the corresponding author of this article, is the academic leader of a research group on plant molecular biology in Hunan University. He gained his PhD in 1995. Shortly after, he obtained a position of Professor in 1999. His study is focused on functional genomics of plant and on the molecular mechanism of phytohormone metabolism. In addition, he is interested in biochemical analysis of plant. In recent years, He dedicates to studying the function of transcription factors and receptor-like kinases (RLKs) in plant development, which was supported by different kinds of foundation, such as National High-Tech R & D Program (863), National Natural Science Foundation, National Science and Technology Major Project, and International Coordination.
Fig. A1.

Prof. Xuan-ming LIU
Footnotes
Project supported by the National High-Tech R & D Program (863) of China (No. 2007AA10Z127) and the National Natural Science Foundation of China (No. 30800080)
References
- 1.Agharkar M, Lomba P, Altpeter F, Zhang H, Kenworthy K, Lange T. Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnology Journal. 2007;5(6):791–801. doi: 10.1111/j.1467-7652.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- 2.Alabadi D, Gil J, Blazquez MA, Garcia-Martinez JL. Gibberellins repress photomorphogenesis in darkness. Plant Physiology. 2004;134(3):1050–1057. doi: 10.1104/pp.103.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleford NEJ, Wilkinson MD, Ma Q, Evans DJ, Stone MC, Pearce SP, Powers SJ, Thomas SG, Jones HD, Phillips AL, et al. Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. Journal of Experimental Botany. 2007;58(12):3213–3226. doi: 10.1093/jxb/erm166. [DOI] [PubMed] [Google Scholar]
- 4.Biemelt S, Tschiersch H, Sonnewald U. Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiology. 2004;135(1):254–265. doi: 10.1104/pp.103.036988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Biology. 2003;54(1):519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 6.Busov VB, Meilan R, Pearce DW, Ma CP, Rood SB, Strauss SH. Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiology. 2003;132(3):1283–1291. doi: 10.1104/pp.103.020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell MM, Ellis BE. Fungal elicitor-mediated responses in pine cell cultures I. Induction of phenylpropanoid metabolism. Planta. 1992;186(3):409–417. doi: 10.1007/BF00195322. [DOI] [PubMed] [Google Scholar]
- 8.Dijkstra C, Adams E, Bhattacharya A, Page AF, Anthony P, Kourmpetli S, Power JB, Lowe KC, Thomas SG, Hedden P, et al. Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Reports. 2008;27(3):463–470. doi: 10.1007/s00299-007-0471-z. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson ME, Israelsson M, Olsson O, Moritz T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nature Biotechnology. 2000;18(7):784–788. doi: 10.038/77355. [DOI] [PubMed] [Google Scholar]
- 10.Fukushima RS, Dehority BA. Feasibility of using lignin isolated from forages by solubilization in acetyl bromide as a standard for lignin analyses. Journal of Animal Science. 2000;78(12):3135–3143. doi: 10.2527/2000.78123135x. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima RS, Hatfield RD. Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. Journal of Agricultural and Food Chemistry. 2001;49(7):3133–3139. doi: 10.1021/jf010449r. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima RS, Hatfield RD. Comparison of the acetyl bromide spectrophotometric method with other analytical lignin methods for determining lignin concentration in forage samples. Journal of Agricultural and Food Chemistry. 2004;52(12):3713–3720. doi: 10.1021/jf035497l. [DOI] [PubMed] [Google Scholar]
- 13.Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE. Gibberellin: inhibitor of an inhibitor of. Bioessays. 1998;20(12):1001–1008. doi: 10.1002/SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Hedden P, Proebsting WM. Genetic analysis of gibberellin biosynthesis. Plant Physiology. 1999;119(2):365–370. doi: 10.1104/pp.119.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science. 2000;5(12):523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 16.Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(4):2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Israelsson M, Sundberg B, Moritz T. Tissue-specific localization of gibberellins and expression of gibberellin biosynthetic and signalling genes in wood-forming tissues in aspen. The Plant Journal. 2005;44(3):494–504. doi: 10.1111/j.1365-313X.2005.02547.x. [DOI] [PubMed] [Google Scholar]
- 18.Kende H, Zeevaart JAD. The five “classical” plant hormones. Plant Cell. 1997;9(7):1197–1210. doi: 10.1105/tpc.9.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DJ, Zeevaart JAD. Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiology. 2002;130(4):2085–2094. doi: 10.1104/pp.008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DJ, Zeevaart JAD. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris . Plant Physiology. 2005;138(1):243–254. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum . The Plant Journal. 1999;19(1):65–73. doi: 10.1046/j.1365-313X.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- 22.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(5):2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, Zhou B, Yang Y, Mei J, Zhao X, Guo X, Huang X, Tang D, Liu X. Piercing and vacuum infiltration of the mature embryo: a simplified method for Agrobacterium-mediated transformation of indica rice. Plant Cell Reports. 2009;28(7):1065–1074. doi: 10.1007/s00299-009-0706-2. [DOI] [PubMed] [Google Scholar]
- 24.Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20(10):2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DN, Proebsting WM, Hedden P. The SLENDER gene of pea encodes a gibberellin 2-oxidase. Plant Physiology. 1999;121(3):775–781. doi: 10.1104/pp.121.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakaminami K, Sawada Y, Suzuki M, Kenmoku H, Kawaide H, Mitsuhashi W, Sassa T, Inoue Y, Kamiya Y, Toyomasu T. Deactivation of gibberellin by 2-oxidation during germination of photoblastic lettuce seeds. Bioscience, Biotechnology, and Biochemistry. 2003;67(7):1551–1558. doi: 10.1271/bbb.67.1551. [DOI] [PubMed] [Google Scholar]
- 28.Olsen O, Soerensen H. Sinalbin and other glucosinolates in seeds of double low rape species and Brassica napus cv. Bronowski. Journal of Agricultural and Food Chemistry. 1980;28(1):43–48. doi: 10.1021/jf60227a023. [DOI] [PubMed] [Google Scholar]
- 29.Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14(Suppl. 1):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panford JA, de Man JM. Determination of oil content of seeds by NIR: influence of fatty acid composition on wavelength selection. Journal of the American Oil Chemists’ Society. 1990;67(8):473–482. doi: 10.1007/BF02540751. [DOI] [Google Scholar]
- 31.Rieu I, Eriksson S, Powers SJ, Gong F, Griffiths J, Woodley L, Benlloch R, Nilsson O, Thomas SG, Hedden P, et al. Genetic analysis reveals that C19-GA 2 oxidation is a major gibberellin inactivation pathway in Arabidopsis . Plant Cell. 2008;20(9):2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, Iwahori S, Matsuoka M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiology. 2001;125(3):1508–1516. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakamoto T, Miura K, Ihoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiology. 2004;134(4):1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15(1):151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivaraman I, Arumugam N, Sodhi YS, Gupta V, Mukhopadhyay A, Pradhan AK, Burma1 PK, Pental D. Development of high oleic and low linoleic acid transgenics in a zero erucic acid Brassica juncea L. (Indian mustard) line by antisense suppression of the fad2 gene. Molecular Breeding. 2004;13(4):365–375. doi: 10.1023/B:MOLB.0000034092.47934.d6. [DOI] [Google Scholar]
- 36.Sun T, Goodman HM, Ausubel FM. Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell. 1992;4(2):119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annual Review of Plant Biology. 2004;55(1):197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 38.Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proceedings of the National Academy of Science of the United States of America. 1999;96(8):4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell. 2004;16(5):1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang QM, Tu XJ, Deng KQ, Zeng JX, Zhao XY, Tang DY, Liu XM. A defect in zinc finger protein double B-box 1a (DBB1a) causes abnormal floral development in Arabidopsis . Journal of Plant Biology. 2009;52(6):543–549. doi: 10.1007/s12374-009-9070-6. [DOI] [Google Scholar]
- 41.Zhao XY, Yu XH, Foo E, Symons GM, Lopez J, Bendehakkalu KT, Xiang J, Weller JL, Liu XM, Reid JB, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiology. 2007;145(1):106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao XY, Yu XH, Liu XM, Lin CT. Light regulation of gibberellins metabolism in seedling development. Journal of Integrative Plant Biology. 2007;49(1):21–27. doi: 10.1111/j.1744-7909.2006.00407.x. [DOI] [Google Scholar]