Abstract
The role of aberrant methylation of fragile histidine triad (FHIT) promoters in the differentiated thyroid carcinoma (DTC) is not yet clear. Therefore, we investigated the association of the status of FHIT promoter methylation and FHIT protein expression with the clinicopathological progression of DTC, using PCR-based methylation assay and immunohistochemical technique. While no FHIT gene promoter methylation was observed in the matched non-cancerous epithelium (NCE) specimens, 24.6% of DTC samples demonstrated methylation in the FHIT promoter region. The protein expression of FHIT in NCE and DTC was 100.0% and 41.5% (P<0.01), respectively. There was a negative correlation between promoter methylation and protein expression of FHIT gene (P<0.05). Additionally, the methylation status appeared to be significantly associated with the pathological grade, tumor TNM stage, and lymph node metastasis (P<0.05), and FHIT proteins were weakly expressed in only about 20% of DTC with grade II pathological changes, TNM stage III/IV, or lymph node metastasis. Finally, the gender and tumor classification but not age marginally affected the promoter methylation and protein expression of FHIT. Our results suggest that methylation of the promoter region may play a key role in inactivation of FHIT - possibly leading to subsequent carcinogenesis and progression of DTC.
Keywords: Fragile histidine triad, differentiated thyroid carcinoma, methylation, carcinogenesis, tumor progression
Introduction
Differentiated thyroid carcinoma (DTC: papillary and follicular carcinoma) is already the most common endocrine malignancy, with reported incidence rates still rapidly increasing in recent years [1, 2]. DTC is generally indolent and is characterized by low morbidity and mortality; however, some patients are at high risk of cervical metastases and recurrence. The thyroid epithelium is the second most common tissue type, next to blood, where fusion gene products are critical for the early development of cancer [3]. Given that carcinogenesis is a multi-step process [4-7], the activation of carcinogenic pathways in DTC likewise involves many different abnormalities both genetic and epigenetic. Epigenetic alterations, particularly hypermethy-lation of promoter DNA regions with consequent gene silencing, which occur commonly in cancer [8-13] might be a common feature of DTC as well.
The short arm of human chromosome 3 is one of the most common sites of chromosomal abnormality in malignant diseases. One candidate for alterations at this location is the fragile histidine triad (FHIT) gene, located on 3p14.2, spanning the FRA3B common fragile site [14]. The FHIT gene is composed of 10 exons encompassing 1.8 Mb genome regions, of which only exons 5 to 9 are the code for protein. It encodes a short 1.1 kb mRNA transcript, and a small 16.8 kDa protein. Studies on the functional aspects of FHIT have indicated that it is a tumor suppressor gene. The FHIT gene and its protein may be involved in the regulation of proliferative and apoptotic cellular processes [15-18]. Loss of heterozygosity on chromosome 3p involving the FHIT gene is frequently associated with both benign and malignant thyroid lesions, suggest-ingthat FHIT alterations might be an early event in thyroid carcinogenesis [19, 20]. However, some reports do not support an association between FHIT loss and developmental stages, histological grade or outcome in human thyroid cancer [21, 22]. Thus the role of FHIT in thyroid cancer progression remains controversial.
DNA methylation is one of the main avenues for epigenetic modification in humans, and changes in methylation patterns play an important role in tumorigenesis [5, 23]. Methylation of the FHIT gene was found in several solid tumors - such as in esophagus, bladder, lung and breast cancers [8, 11, 13, 18, 24]. In addition, it was reported that a reduced or total loss of expression of FHIT protein resulting from gene methylation was associated with disease progression in some solid tumors [24-28]. However, the mechanism of FHIT inactivation has not yet been defined in DTC. In this study, we compared the methylation status of the FHIT gene promoter as well as the expression of its protein in 65 cases of DTC against matched non -cancerous epithelium (NCE) by using PCR-based restriction enzyme assay and immunohis-tochemical (IHC) technique, and investigated the correlation of the promoter methylation of FHIT gene with patho-clinical features of DTC.
Materials and methods
Patients and tissue specimens
Sixty-five patients underwent surgical resection for DTC between January 2005 and July 2006 in the First Affiliated Hospital of Zhengzhou University, China. The patients comprised of 19 men and 46 women with ages ranging between 15 and 75 years (mean age, 45 years). All patients had no prior chemotherapy or radiotherapy. All specimens were obtained during routine surgery. In each case, one block of tissue from the tumor and another block from non-cancerous tissue (non-cancerous epithelium, NCE) in the contralateral lobe away from the tumor was resected. The tissue samples were snap frozen in liquid nitrogen shortly after surgical removal and stored at −80 °C. A portion of each frozen tumor sample was also embedded in paraffin. Sections of each paraffin block were stained with hematoxylin and eosin (H&E) to confirm the exact tissue analyzed. Histological diagnosis of the tumors was made and confirmed individually by at least 2 experienced pathologists based on World Health Organization (WHO) criteria [29]. The clinical staging of DTC was made based on the TNM classification introduced in 2002 by the American Joint Committee on Cancer (AJCC) [30]. In this study, there were 42 cases of papillary thyroid carcinoma and 23 cases of follicular thyroid carcinoma. Amongst them, 43 had grade I carcinoma and 22 had grade II carcinoma according to histological grading. By clinical staging using TNM standards, 13 patients had stage I carcinoma, 35 had stage II, 12 had stage III, and 5 had stage IV. The presence of lymphatic metastasis in regional nodes was confirmed during surgery and was positive in 26 cancers. The specimens collected and used in this study were obtained with each patient's consent along with approval from the ethics committee of the First Affiliated Hospital of Zhengzhou University.
DNA isolation and preparation
Genomic DNA was isolated using standard techniques described previously with modification [10]. Briefly, tissue samples of 0.3-0.5 g were homogenized in liquid nitrogen and lysed in buffer containing 10 mM Tris-HCl (pH 8.0), 0.1 M EDTA, and 0.5% SDS for 20 min.
Lysates were incubated with proteinase K (final concentration 100 μg/ml) at 50 °C for 3 h, and extracted twice with phenol and twice with chloroform. The genomic DNA was precipitated with 0.2 volume of 10 M ammonium acetate and 2.5 volumes of 100% ethanol. The DNA was washed with 70% ethanol and dissolved in TE buffer. The concentration of the DNA was determined by spectrophotometry and its integrity was checked by 1.5% agarose gel electrophoresis.
PCR-based methylation assay
The methylation status of the 5'-CpG island of the FHIT gene was checked by using a PCR-based methylation assay as described by Iwase et al [31] with minor modifications [25, 32, 33]. Briefly, 1.0 micrograms of genomic DNA was digested overnight at 37 °C with 20 units of the methylation-sensitive restriction enzyme Hpall under conditions specified by the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). The sense and antisense primers used were 5'-CGG TCA CAG GAC TTT TTG-3’ and 5'-GTG GGG CGG AAG ATA CTC-3’ for the FHIT gene promoter region. The primer set amplifies a 282bp DNA fragment. The PCR reaction volume was 50 μl, containing 0.2 mM of each dNTP, 0.5 μM of each primer, 1 × reaction buffer, 2.5 mM MgCl2, 1.5 U Taq DNA polymerase (Fermentas Tamro Corp., Vantaa, Finland), and 50 ng template DNA. Conditions were as follows: 94°C for 5 min, followed by 32 cycles at 94°C for 40 Sec, 56°C (annealing temperature) for 40 Sec and 72°C for 1 min; and then followed by incubation at 72°C for 5 min.
PCR conditions were determined by cycle curve and DNA concentration curve. To rule out the possibility of false positives due to incomplete digestion and over-cycling of the PCR amplifications, the digestions of each sample and the PCR amplification were performed at least twice in independent experiments. Undigested DNA and MspI-digested DNA samples were amplified as positive and negative controls respectively. PCR products were resolved on 1.8% agarose gel. Loss of the PCR products following digestion by Hpall was assessed as demethylation (Figure 1).
Figure 1.
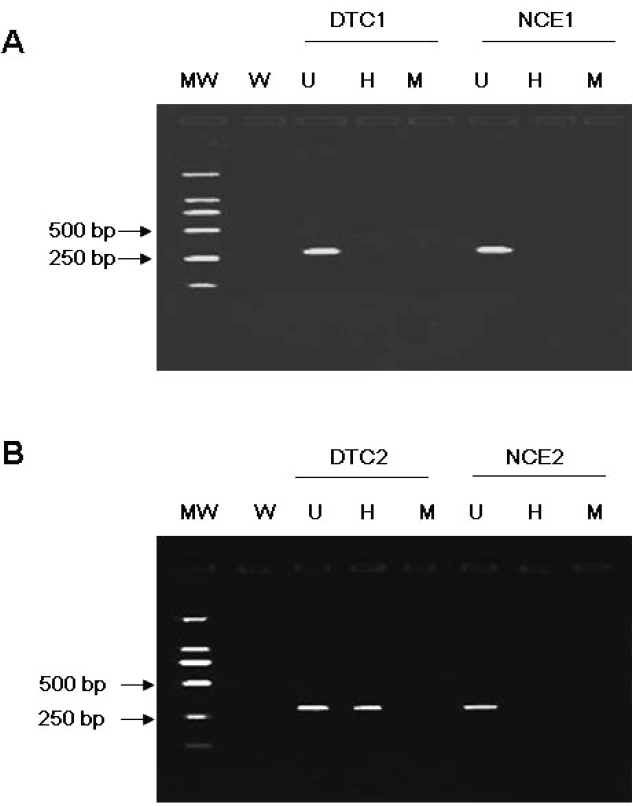
Representative results from PCR based Hpall restriction enzyme assay. A 282 bp PCR product is seen following incubation with Hpall in a methylated DTC (B: H), but not in an unmethylated DTC (A: H), as the methylated restriction site (CCGG) is resistant to digestion by the enzyme. All 65 NCE were unmethylated in the FHIT promoter region, while 16 of 65 DTC were methylated. MW: molecular weight ladder; W, water; U, undigested; H, incubated with Hpall; M, incubated with Mspl.
Immunohistochemical analysis
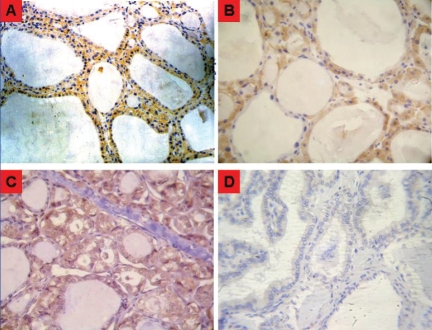
Paraffin sections of thyroid samples (4 μm) were deparaffinized in three 10-minute xylene washes and rehydrated through a series of graded alcohols. Antigen retrieval was done by incubating tissue sections in sodium citrate buffer (10 mmol/L, pH 6.0) at 95°C for 20 minutes. Endogenous peroxidase was inactivated by treatment with 3% hydrogen peroxide in methanol. Slides were incubated in 2% bovine serum albumin blocking solution, followed by incubation with a polyclonal rabbit anti-human FHIT antibody at a 1:50 dilution (Zymed Laboratories Inc., San Francisco, CA) overnight at 4°C. The bound antibody was detected by a strepta-vidin-biotin-peroxidase complex and visualized by 3,3-dinminobenzidine tetrahydrochloride supplemented with 0.01% hydrogen peroxide. Finally, the slides were lightly counterstained with Mayer's hematoxylin. All series include positive controls, and omission of the primary antibodies served as negative control. The expression of the investigated antigens was evaluated in a semi-quantitative manner [32, 34]. The immunoreaction was calculated as the percentage of positive cell * staining intensity. The percentage of positive cells was graded: < 10% as 0; 10% - 30% as 1; 31% - 60% as 2; and > 60% as 3, and the staining intensity was graded as 0, absent; 1, faint; 2, moderate; and 3, strong staining (Figure 2). Levels of immunoreaction were graded into three groups: (-): score 0; (+): score 1-4; (++): score > 4.
Figure 2.
Representative micrographs of IHC staining intensity of FHIT protein in the thyroid tissue A, Normal thyrocyte shows strong cytoplasmic staining intensity (score 3; × 200). B, Normal thyrocyte shows moderate cytoplasmic IHC staining intensity (score 2; × 400). C, Follicular thyroid carcinoma shows weak cytoplasmic IHC staining intensity (score 1; × 400). D, Papillary thyroid carcinoma shows no FHIT-specific IHC staining (score 0; × 400).
Statistical analysis
Two-tailed statistical analysis was performed using SPSS computer software (Version 13, SPSS Inc, Chicago, IL, USA). The correlation between frequency of FHIT promoter methylation and expression of FHIT gene protein or the clinicopathologic parameters was analyzed with the Pearson correlation method. A P-value of less than 0.05 was considered statistically significant.
Results
Promoter methylation and protein expression of FHIT gene in NCE and DTC
Methylation status of the FHIT promoter was assessed by PCR-based methylation assay in 65 cases of DTC and their paired NCE. FHIT gene promoter was found to be methylated in 24.5% of DTC samples (16/65), but not at all in NCE samples (Figure 1). This difference was significant between these two groups (P < 0.01). The methylation appeared to suppress expression of FHIT protein in DTC (Table 1). This is evidenced by the fact that normal thyroid tissues showed strong cytoplasmic staining by anti-FHIT, whereas DTC samples were moderately to weakly stained (Figure 2). While 100% of the NCE samples (65/65) expressed high levels of FHIT protein in the cytoplasm of epithelial cells, only 41.5% of DTC samples (27/65) expressed even just moderate or low levels of FHIT proteins (p < 0.01). Additionally, 58.5% of DTC samples (39/65) did not express any FHIT at all (Figure 3). The results suggest that hypermethy-lation of FHIT's promoter region leading to significant suppression of its protein expression occurs in some cases of DCT.
Table 1.
Promoter methylation and protein expression of FHIT gene and clinicopathologic factors in DTC
| Methylation |
Protein expression |
|||||||
|---|---|---|---|---|---|---|---|---|
| Characteristics | n | - | + | Positive rate (%) | - | + | ++ | Positive rate (%) |
| Gender | ||||||||
| Male | 19 | 16 | 3 | 15.8 | 9 | 6 | 4 | 52.6 |
| Female | 46 | 33 | 13 | 28.3 | 29 | 12 | 5 | 37.0 |
| Age | ||||||||
| <45 | 30 | 23 | 7 | 23.3 | 17 | 10 | 3 | 43.3 |
| >45 | 35 | 26 | 9 | 25.7 | 21 | 8 | 6 | 40.0 |
| Classification | ||||||||
| FTC | 23 | 15 | 8 | 34.8 | 15 | 7 | 1 | 34.8 |
| PTC | 42 | 34 | 8 | 19.0 | 23 | 11 | 8 | 45.2 |
| TNM stage | ||||||||
| I ∼ II | 48 | 41 | 7 | 14.6Δ | 25 | 14 | 9 | 47.9 |
| III ∼ IV | 17 | 8 | 9 | 52.9 | 13 | 4 | 0 | 23.5 |
| grade | ||||||||
| I | 43 | 36 | 7 | 16.3* | 20 | 15 | 8 | 53.5Δ |
| II | 22 | 13 | 9 | 40.9 | 18 | 3 | 1 | 18.2 |
| Lymph node metastasis | ||||||||
| Yes | 26 | 16 | 10 | 38.5* | 22 | 4 | 0 | 15.4Δ |
| No | 39 | 33 | 6 | 15.4 | 16 | 14 | 9 | 59.0 |
FTC: follicular thyroid carcinoma; PTC: papillary thyroid carcinoma
P<0.01;
P<0.05, as compared between groups of pathological grade, TNM stages or metastasis.
Figure 3.
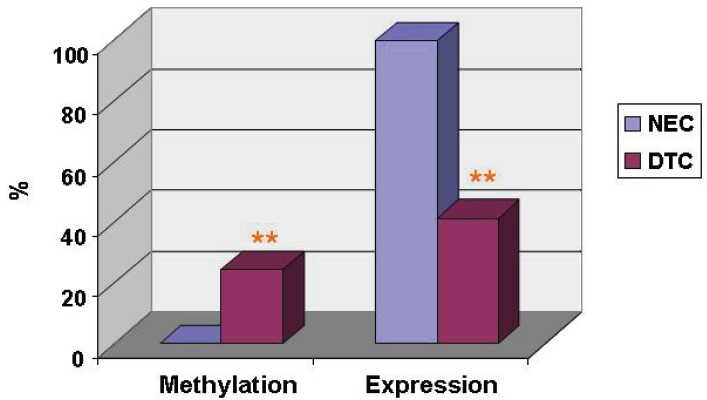
FHIT gene promoter methylation and its protein expression in DTC and NEC. FHIT gene promoter methylation and its protein expression were shown in DTC (n=65) and NEC (n=65). **, p < 0.01, as compared between DTC and NEC. Methylation: FHIT gene promoter methylation determined by PCR-based methylation analysis; Expression: FHIT proteins expression determined by IHC staining.
Inverse correlation of promoter methylation of FHIT gene with its protein expression in DTC
Theoretically, increased promoter methylation of FHIT gene could contribute to decreased FHIT protein expression in DTC. In agreement with this notion, FHIT protein expression was only detected in only 18.7% (3/16) of DTC with FHIT promoter methylation, whereas about 48.9% (24/49) of DTC with unmethylated FHIT promoter were expressed with FHIT proteins. Alternatively, FHIT promoter methylation was only detected in 11.1% (3/27) of DTC expressing FHIT proteins; while 34.2% (13/38) of DTC without expression of FHIT proteins were methylated in FHIT promoter (Table 2). Therefore, the promoter methylation of FHIT gene is inversely correlated with its proteins expression in DCT (P < 0.05). This correlation appeared not to be affected by the gender and classification of DTC, pathological changes and clinical manifestations (Table 1).
Table 2.
Correlation between FHIT gene promoter methylation and its protein expression in DTC
| Expression | Methylation |
Total | |
|---|---|---|---|
| + | - | ||
| + | 3 | 24 | 27 |
| - | 13 | 25 | 38 |
| Total | 16 | 49 | 65 |
x2=4.539, P>0.05
The correlation of promoter methylation and protein expression of FHIT gene with pathological grades of DTC
As a tumor suppressor, increased promoter methylation or decreased protein expression of FHIT gene may promote pathological changes in DTC. In agreement with this notion, the positive incidence of FHIT protein expression decreased significantly from 53.5 % (23/43) in well-differentiated cancers (grade I) DTC to 18.2 % (4/22) in poorly differentiated cancers (grade II) (P < 0.01); and consistently, the percentage of FHIT gene promoter methylation was increased from 16.3% (7 of 43) in grade I to 40.9 % (9 of 22) in grade II. (P < 0.05). Therefore, FHIT gene promoter methylation and decreased FHIT protein expression is associated with DTC progression in pathology (Figure 4A).
Figure 4.
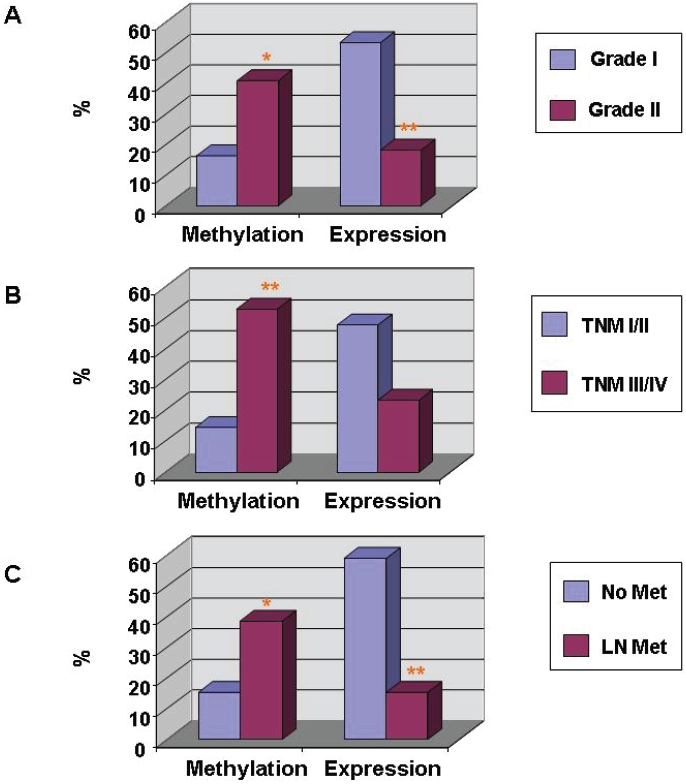
Association of FHIT gene promoter methylation and its protein expression with pathological and clinical manifestations in DTC. FHIT gene promoter methylation and its protein expression in DTC with grand I (n = 43) and grade II (n = 22) pathological changes (A), TNM stage I/II (n = 48) and TNM stages III/IV (n = 17) (B), and metastasis (n = 26) and non-metastasis (n = 39) (C) were determined by PCR-based methylation analysis and IHC staining, respectively. *, p < 0.05; ** p < 0.01, as compared between two groups.
The correlation of FHIT gene promoter methylation with manifestations of DTC
Since FHIT gene promoter methylation or reduced expression of FHIT was associated with pathological progression of DTC, we further analyzed whether these changes were associated with clinical manifestations of DTC, such as TNM stages and metastasis. As shown in Table 1 and Figure 4B & C, the presence of FHIT promoter methylation was increased from 14.6% (7 of 48) in stages I and II to 52.9% (9 of 17) in stages III and IV; and the FHIT protein expression was decreased from 47.9% (23/43) in stages I and II to 23.5% (4/22) in stages III and IV. The correlations are statistically significant (P < 0.05). The results suggest that FHIT promoter methylation or down-regulation of FHIT proteins is significantly associated with advanced clinical stages (Figure 4B). Moreover, the promoter methylation and protein expression were also associated with tumor metastasis Figure 4C). The rate of promoter methylation of the FHIT gene was 38.5% (10 of 26), and 15.4% (6 of 39), respectively, in DTC with metastasis and in DTC without metastasis (P < 0.05); and the rate of FHIT protein expression was 15.4% (4 of 26) of DTC with metastasis and in 59.0% (23 of 39) of DTC without metastasis (P < 0.05) (Table 1 & Figure 4C). Taken together, these results suggest that FHIT gene promoter methylation is associated with clinical stage and metastasis of DTC.
The effects of gender, age and classification of DTC on FHIT gene promoter methylation
The FHIT gene promoter methylation in DTC was affected by gender and classification of DTC but not gender, although the effects were not statistically significant (Table 1 & Figure 5). While the percentage of FHIT gene promoter methylation in the female patient with DTC (28.3%) was higher than that in the male (15.8%), no difference could be found between ages (23.3% at < 45 versus 25.7% at > 45) in the FHIT gene promoter methylation (P > 0.05). Moreover, the rate of FHIT gene promoter methylation in the follicular thyroid carcinoma (FTC) (34.8%) was higher than that in the papillary thyroid carcinoma (PTC) (Table 1 & Figure 5).
Figure 5.
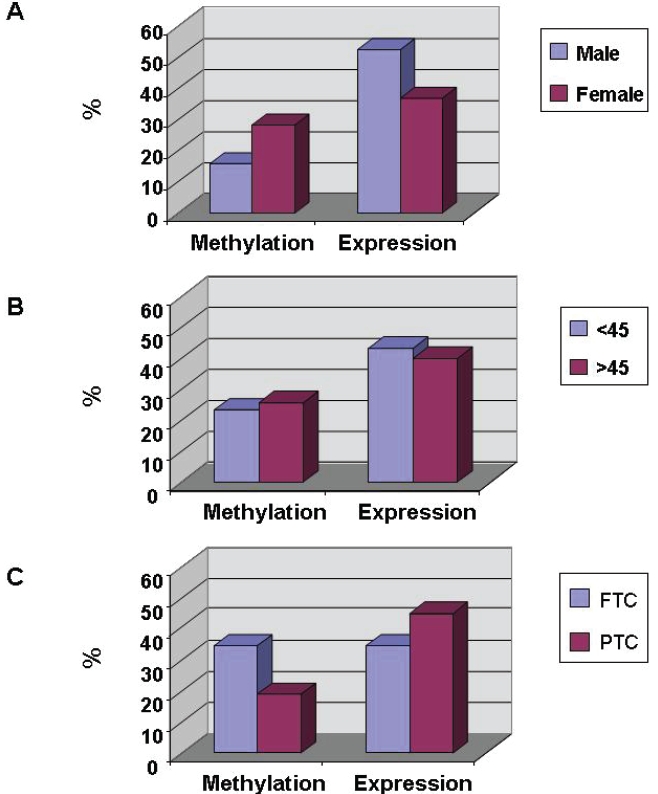
Association of FHIT gene promoter methylation and its protein expression with gender, age and tumor classification in DTC. FHIT gene promoter methylation and its protein expression in DTC of (A) male (n = 19) and female (n = 46), (B) the patients at < 45 years old (n = 30) and ≥ 45 years old (n = 35), and (C) FTC (n = 23) and PTC (n = 42) (C) were determined by PCR-based methylation analysis and IHC staining, respectively. There is no statistical significance between the groups. FTC: follicular thyroid carcinoma; PTC: papillary thyroid carcinoma.
Discussion
Defects involving the FHIT gene - such as abnormal transcripts containing deletions of one or more coding exons, intragenic homozygous deletions, or genomic DNA rearrangement - have been found frequently in many cancer cell lines, as well as in many primary tumors including that of the lung, breast, esophagus, stomach, Merkel cell, and head and neck [11, 24, 35]. FHIT's role as a possible tumor suppressor has been postulated based on the ability of FHIT to eliminate or reduce the tumorigenicity of tumor cells in nude and knockout mice [36]. Genes are often defined and identified as tumor suppressor genes if they conform to the ‘two-hits’ or “multiple hits” hypothesis which postulates that loss of both alleles of such genes is required via mutation, deletion, or rearrangement to confer a growth advantage and subsequently contribute to cancer development [4, 37, 38]. However, in contrast to many other tumor suppressor genes, inactivating point mutations are rarely observed in FHIT. While chromosomal rearrangements and deletions might be predominant in genetic alterations [39, 40], aberrant methylation causing FHIT promoter silencing was observed in several human solid cancer [41, 42]. In this study, we show that FHIT gene promoter methylation occurred in DTC but not in ECE, suggesting that aberrant epigenetic changes of FHIT may contribute to the development of DTC.
The methylation status of FHIT's promoter region in 65 cases of DTC along with their matched NCE was determined by PCR-based restriction enzyme assay. In all 65 NCE samples examined, there was no promoter methylation of the FHIT gene; whereas methylation was observed in 16 (24.5%) cases of DTC. This difference is significant (P<0.05). In esophageal squamous cell carcinomas, Tanaka et al. [43] found FHIT hypermethylation in 5 of 35 (14%) primary tumors while corresponding normal tissue showed no methylation - which is consistent with our results. Hypermethylation of the 5’ -CpG island of the FHIT gene was observed in three out of four structurally unaltered but tran-scriptionally repressed cell lines, and all methylated cell lines exhibited re-expression of the FHIT gene along with demethylation in the CpG island after treatment with the demethylating agent 5-aza-2'-deoxycytidine. These findings suggest that methylation of the 5'-CpG island of the FHIT gene is closely associated with tran-scriptional inactivation and might be involved in tumor development of the esophagus [43]. Kvasha et al. examined 22 paired samples of clear cell renal carcinoma and non-malignant renal tissue for the methylation of FHIT. Hypermethylation of FHIT was detected in 54.5% (12/22) of the clear cell renal carcinoma samples and they further demonstrated that hypermethylation of FHIT may be responsible for its inactivation in clear cell renal carcinomas [25]. Lin etal. observed that abnormal methylation of the FHIT gene was found in 26 of 55(47.2%) cases of myelodysplastic syndrome, but absent in normal controls [27]. These findings, together with ours, conclude that FHIT gene promoter is not methylated in normal cells.
We also found that incidence of FHIT promoter methylation was significant lower in cases of early stage, well-differentiated DTC, and without metastasis, as opposed to being of advanced stage, poor differentiated, and with metastasis. These findings suggests that promoter methylation of the FHIT gene might be associated with progression of DTC and a poor prognosis, consistently with our previous report that the status of aberrant DNA methylation is associated with the progression of colorectal carcinoma [10]. By Kaplan-Meier analysis, Maruyama R et al. also found that the survival of patients with methylation-positive FHIT in bladder cancers was significantly shorter than that of patients with methylation-negative tumors; and methylation-positive status was independently associated with poor survival in multivariate analyses [44].
In our current study, all NCE samples demonstrated positive immunostaining for FHIT protein, with most of them being classified as having a “strong” immunoreaction. On other hand, DTC samples had a significantly lower rate of positive expression at 41.5%, and featured variably mixed patterns of both reduced or absent and retained staining. We also noticed that FHIT expression patterns displayed significant association with lymph node metastasis and histological grade, both of which are known prognostic factors for DTC. The strong association of decreased FHIT expression with poor prognosis shown in our study suggests that epi-genetic alterations to FHIT represent a relevant molecular pathway in the carcinogenesis of DTC. Similarly to this study, we had earlier reported expression of the FHIT protein in 46.3% (19/41) of laryngeal squamous cell carcinoma (LSCC) cases [32]. The rate of positive FHIT expression in stage HI versus stage III-IV was 69.6% and 16.7% respectively; and in the lymph node metastasis group versus the group without lymph node metastasis, it was 20.0% and 61.5% respectively. We also demonstrated that FHIT might play important roles in development and metastasis of LSCC, thus opening the possibility it could be a useful marker for evaluating the biological behavior of LSCC [32]. According to these findings, it is suggested that IHC analysis of FHIT could be used to predict tumor aggressiveness in DTC.
In addition, we also observed that tumors lacking methylation of the FHIT promoter region had high levels the FHIT expression as compared to the significantly reduced levels of FHIT protein expression observed in tumors featuring methylation of the promoter. Thus, the data demonstrates a correlation between promoter methylation of FHIT and a decrease in FHIT expression, indicating that promoter methylation of FHIT may be an important mechanism for FHIT inactivation in DTC.
In summary, the FHIT gene potentially plays a vital role in the carcinogenesis and development of DTC. One possible avenue that FHIT inactivation operates through in DTC progression is via methylation of its promoter region. Thus, FHIT expression could serve as a useful biomarker in evaluating the biological behavior of FHIT and have clinical utility in devising innovative treatment strategies. A better understanding of FHIT promoter methylation and phe-notypic expression will provide new insights into DTC carcinogenesis, cancer treatment, and feasible chemopreventive measures for the future.
Acknowledgments
This work was supported by: Science Foundation for Distinguished Young Scholars of Henan Province (DTY, No. 0512000900); Medical Science Foundation for Creative Scholars of Henan Province (DTY, No. 2009WQN001). American Cancer Society (ACS) grant #IRG-112367 (JXG). Authors thank David Shi, MS-3, College of Medicine, Ohio State University, Columbus, for his editing of the manuscript
References
- 1.Giovanella L. Highly sensitive thyroglobulin measurements in differentiated thyroid carci noma management. Clinical Chemistry and Laboratory Medicine. 2008;46:1067–1073. doi: 10.1515/CCLM.2008.212. [DOI] [PubMed] [Google Scholar]
- 2.Shaha A. TNM Classification of Thyroid Carcinoma. World Journal of Surgery. 2007;31:879–887. doi: 10.1007/s00268-006-0864-0. [DOI] [PubMed] [Google Scholar]
- 3.Mizuno T, Kyoizumi S, Suzuki T, Iwamoto KS, Seyama T. Continued expression of a tissue specific activated oncogene in the early steps of radiation-induced human thyroid carcinogenesis. Oncogene. 1997;15:1455–1460. doi: 10.1038/sj.onc.1201313. [DOI] [PubMed] [Google Scholar]
- 4.Gao JX. Cancer stem cells: the lessons from precancerous stem cells. J Cell Mol Med. 2008;12:67–96. doi: 10.1111/j.1582-4934.2007.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J-X, Zhou Q. Epigenetic progenitors in tumor initiation and development. Drug Discovery Today: Disease Models In Press, Corrected Proof:
- 6.Jones PA. Epigenetics in carcinogenesis and cancer prevention. Ann N Y Acad Sci. 2003;983:213–219. doi: 10.1111/j.1749-6632.2003.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 7.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 8.Kim JS, Kim JW, Han J, Shim YM, Park J, Kim D-H. Cohypermethylation of p16 and FHIT Promoters as a Prognostic Factor of Recurrence in Surgically Resected Stage I Non-Small Cell Lung Cancer. Cancer Res. 2006;66:4049–4054. doi: 10.1158/0008-5472.CAN-05-3813. [DOI] [PubMed] [Google Scholar]
- 9.Moulin AP, Clement G, Bosman FT, Zografos L, Benhattar J. Methylation of CpG island promoters in uveal melanoma. Br J Ophthalmol. 2008;92:281–285. doi: 10.1136/bjo.2007.127035. [DOI] [PubMed] [Google Scholar]
- 10.Shen R, Tao L, Xu Y, Chang S, Brocklyn JV, Gao JX. Reversibility of aberrant global DNA and estrogen receptor-a gene methylation distinguishes the colorectal precancer from cancer. Int J Clin Exp Pathol. 2009;2:21–33. [PMC free article] [PubMed] [Google Scholar]
- 11.Eun Ju L, Bo Bin L, Jin Wook K, Young Mog S, Hoseok I, Joungho H, Eun Yoon C, Joobae P, Duk-Hwan K. Aberrant methylation of Fragile Histidine Triad gene is associated with poor prognosis in early stage esophageal squamous cell carcinoma. European journal of cancer (Oxford, England : 1990) 2006;42:972–980. doi: 10.1016/j.ejca.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Takeuchi S, Hofmann WK, Ikezoe T, van Dongen JJM, Szczepanski T, Bartram CR, Yoshino N, Taguchi H, Koeffler HP. Aberrant methylation in promoter-associated CpG islands of multiple genes in acute lymphoblastic leukemia. Leukemia Research. 2006;30:98–102. doi: 10.1016/j.leukres.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Schildhaus HU, Krockel I, Lippert H, Malfertheiner P, Roessner A, Schneider-Stock R. Promoter hypermethylation of p16INK4a, E-cadherin, O6-MGMT, DAPK and FHIT in adenocarcinomas of the esophagus, esophagogastric junction and proximal stomach. Int J Oncol. 2005;26:1493–1500. [PubMed] [Google Scholar]
- 14.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT Gene, Spanning the Chromosome 3p14.2 Fragile Site and Renal Carcinoma-Associated t(3;8) Breakpoint, Is Abnormal in Digestive Tract Cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 15.Kuroki T, Tajima Y, Furui J, Kanematsu T. Common Fragile Genes and Digestive Tract Cancers. Surgery Today. 2006;36:1–5. doi: 10.1007/s00595-005-3094-4. [DOI] [PubMed] [Google Scholar]
- 16.Huebner K, Hadaczek P, Siprashvili Z, Druck T, Croce CM. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Bio-chimica et Biophysica Acta (BBA) - Reviews on Cancer. 1997;1332:M65–M70. doi: 10.1016/s0304-419x(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 17.Kisselev LL, Justesen J, Wolfson AD, Frolova LY. Diadenosine oligophosphates (ApnA), a novel class of signalling molecules? FEBS Letters. 1998;427:157–163. doi: 10.1016/s0014-5793(98)00420-7. [DOI] [PubMed] [Google Scholar]
- 18.Huebner K, Garrison PN, Barnes LD, Croce CM. THE ROLE OF THE FHIT/FRA3B LOCUS IN CANCER. Annual Review of Genetics. 2003;32:7–31. doi: 10.1146/annurev.genet.32.1.7. [DOI] [PubMed] [Google Scholar]
- 19.Zou M, Shi Y, Farid NR, Al-Sedairy ST, Paterson MC. FHIT Gene Abnormalities in Both Benign and Malignant Thyroid Tumours. European Journal of Cancer. 1999;35:467–472. doi: 10.1016/s0959-8049(98)00370-0. [DOI] [PubMed] [Google Scholar]
- 20.Chang TJ, Tsai TC, Wu YL, Yang HM, Chi CW, Yang AH, Lee CH. Abnormal transcripts of FHIT gene in thyroid cancer. Oncol Rep. 1998;5:245–247. [PubMed] [Google Scholar]
- 21.McIver B, Grebe SK, Wang L, Hay ID, Yokomizo A, Liu W, Goellner JR, Grant CS, Smith DI, Eberhardt NL. FHIT and TSG101 in thyroid tumours: aberrant transcripts reflect rare abnormal RNA processing events of uncertain pathogenetic or clinical significance. Clin Endocrinol (Oxf) 2000;52:749–757. [PubMed] [Google Scholar]
- 22.Powell DJ, Jr, Russell JP, Li G, Kuo BA, Fidanza V Huebner K, Rothstein JL. Altered gene expression in immunogenic poorly differentiated thyroid carcinomas from RET/PTC3p53-/-mice. Oncogene. 2001;20:3235–3246. doi: 10.1038/sj.onc.1204425. [DOI] [PubMed] [Google Scholar]
- 23.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 24.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as bio-markers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 25.Kvasha S, Gordiyuk V, Kondratov A, Ugryn D, Zgonnyk YM, Rynditch AV, Vozianov AF. Hypermethylation of the 5'CpG island of the FHIT gene in clear cell renal carcinomas. Cancer Lett. 2008;265:250–257. doi: 10.1016/j.canlet.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Meng L, Wang H, Xu Q, Wang S, Wu S, Xi L, Zhao Y, Zhou J, Xu G, Lu Y, Ma D. Regulation of DNA methylation on the expression of the FHIT gene contributes to cervical carcinoma cell tumorigenesis. Oncol Rep. 2006;16:625–629. [PubMed] [Google Scholar]
- 27.Lin J, Yao D-m, Qian J, Wang Y-l, Han L-x, Jiang Y-w, Fei X, Cen J-n, Chen Z-x. Methylation status of fragile histidine triad (FHIT) gene and its clinical impact on prognosis of patients with myelodysplastic syndrome. Leukemia Research. 2008;32:1541–1545. doi: 10.1016/j.leukres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Zochbauer-Muller S, Fong KM, Maitra A, Lam S, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD. 5’ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. [PubMed] [Google Scholar]
- 29.LiVolsi VA A-SJ, Asa SL, Baloch ZW, Sobrinho-Simoes M, Wenig B, DeLellis RA, Cady B. Papillary carcinoma (World Health Organization classification of tumors) In: De Lellis RA LR, Heitz PU, Eng C, editors. Pathology and Genetics of Tumours of Endocrine Organs. Lyon, France: IARC Press; 2004. pp. 57–66. [Google Scholar]
- 30.AJCC Cancer Staging Manual: Thyroid. New York: Springer-Verlag; 2002. [Google Scholar]
- 31.Iwase H, Omoto Y, Iwata H, Toyama T, Hara Y, Ando Y, Ito Y, Fujii Y, Kobayashi S. DNA methylation analysis at distal and proximal promoter regions of the oestrogen receptor gene in breast cancers. Br J Cancer. 1999;80:1982–1986. doi: 10.1038/sj.bjc.6690631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin DT, Lu XB, Dong MM, Qiu XG, Wang QZ. [Correlation of FHIT expression to cell proliferation and metastasis of laryngeal squamous cell carcinoma] Ai Zheng. 2004;23:1338–1341. [PubMed] [Google Scholar]
- 33.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holschneider CH, Baldwin RL, Tumber K, Aoyama C, Karlan BY. The Fragile Histidine Triad Gene: A Molecular Link Between Cigarette Smoking and Cervical Cancer. Clinical Cancer Research. 2005;11:5756–5763. doi: 10.1158/1078-0432.CCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 35.Smith DI, McAvoy S, Zhu Y, Perez DS. Large common fragile site genes and cancer. Seminars in Cancer Biology. 2007;17:31–41. doi: 10.1016/j.semcancer.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Dumon KR, Ishii H, Fong LYY, Zanesi N, Fidanza V, Mancini R, Vecchione A, Baffa R, Trapasso F, During MJ, Huebner K, Croce CM. FHIT gene therapy prevents tumor development in Fhitdeficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3346–3351. doi: 10.1073/pnas.061020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- 38.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 39.Yeh KT, Chang JG, Chen YJ, Chen ST, Yu SY, Shih MC, Perng LI, Wang JC, Tsai M, Chang CP. Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in hepatocellular carcinoma. Cancer Invest. 2000;18:123–129. doi: 10.3109/07357900009038243. [DOI] [PubMed] [Google Scholar]
- 40.Lee S-H, Kim W-H, Kim H-K, Woo K-M, Nam H-S, Kim H-S, Kim J-G, Cho M-H. Altered Expression of the Fragile Histidine Triad Gene in Primary Gastric Adenocarcinomas. Biochemical and Biophysical Research Communications. 2001;284:850–855. doi: 10.1006/bbrc.2001.5038. [DOI] [PubMed] [Google Scholar]
- 41.Yanagawa N, Tamura G, Oizumi H, Kanauchi N, Endoh M, Sadahiro M, Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Foja S, Goldberg M, Schagdarsurengin U, Dammann R, Tannapfel A, Ballhausen WG. Promoter methylation and loss of coding exons of the fragile histidine triad (FHIT) gene in intrahepatic cholangiocarcinomas. Liver International. 2005;25:1202–1208. doi: 10.1111/j.1478-3231.2005.01174.x. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka H, Shimada Y, Harada H, Shinoda M, Hatooka S, Imamura M, Ishizaki K. Methylation of the 5’ CpG Island of the FHIT Gene Is Closely Associated with Transcriptional Inactivation in Esophageal Squamous Cell Carcinomas. Cancer Res. 1998;58:3429–3434. [PubMed] [Google Scholar]
- 44.Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zochbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF. Aberrant Promoter Methylation Profile of Bladder Cancer and Its Relationship to Clinicopathological Features. Cancer Res. 2001;61:8659–8663. [PubMed] [Google Scholar]