Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy
The yeast Ubp3–Bre5 deubiquitination complex interacts with Cdc48 and Ufd3. These proteins are required for ribophagy and likely cooperate to recognize and deubiquitinate ribophagy substrates prior to their vacuolar degradation.
Keywords: ubiquitin, deubiquitination, proteasome, ribophagy
Abstract
Ubiquitin-dependent processes can be antagonized by substrate-specific deubiquitination enzymes involved in many cellular functions. In this study, we show that the yeast Ubp3–Bre5 deubiquitination complex interacts with both the chaperone-like Cdc48, a major actor of the ubiquitin and proteasome system, and Ufd3, a ubiquitin-binding cofactor of Cdc48. We observed that these partners are required for the Ubp3–Bre5-dependent and starvation-induced selective degradation of yeast mature ribosomes, also called ribophagy. By contrast, proteasome-dependent degradation does not participate in this process. Our data favour the idea that these factors cooperate to recognize and deubiquitinate specific substrates of ribophagy before their vacuolar degradation.
Introduction
Covalent conjugation of ubiquitin to target proteins and also the reversal of this process, deubiquitination, represent highly dynamic post-translational regulations that have a key role in virtually all cellular functions. Ubiquitin-specific processing proteases (UBPs) that release ubiquitin from conjugated proteins represent the most widespread deubiquitinating enzymes across species, with 16 UBPs encoded by the Saccharomyces cerevisiae genome but none being essential for cell viability.
The yeast Ubp3 forms a symmetric heterotetrameric complex with its essential positive regulator Bre5 (Li et al, 2005, 2007). The Ubp3–brefeldin A sensitivity 5 (Bre5) complex is involved in various cellular functions, such as transcription elongation (Kvint et al, 2008), DNA repair by non-homologous end joining (Bilsland et al, 2007) and protein kinase C-mediated signalling (Wang et al, 2008). We reported previously that this deubiquitinating enzyme exerts its activity on specific targets. Among those targets are Sec23 and Sec27, components of the coatomer protein complex (COP) II and COPI complexes involved in transport between the endoplasmic reticulum and the Golgi apparatus. Ubp3 specifically cleaves off the first conjugated ubiquitin from Sec23 and Sec27, thus protecting these proteins from proteasome-mediated degradation (Cohen et al, 2003a, 2003b). Beside these ubiquitin- and proteasome-dependent pathways, the Ubp3–Bre5 complex has been implicated in autophagy, another major degradative pathway involved in cell survival on starvation. It regulates not only the cytoplasm-to-vacuole targeting pathway through its action on Atg19 (Baxter et al, 2005), but also the starvation-induced degradation of mature ribosomes, called ribophagy (Kraft et al, 2008).
In this study, we show that Ubp3–Bre5 interacts with both Cdc48—a chaperone-like protein that has a key role in the proteasomal escort pathway—and Ufd3, one of the ubiquitin-binding adaptors of Cdc48. These newly described Ubp3 partners are required for efficient ribophagy but are dispensable for non-selective degradation of cytoplasmic proteins by the autophagic process. However, inhibiting the degradative function of the 26S proteasome does not prevent ribophagy. We thus propose that the ability of Cdc48 to associate with Ufd3 and Ubp3–Bre5 might allow Cdc48 to function as a molecular platform at which ubiquitinated substrates of ribophagy are recognized and deubiquitinated before vacuolar degradation.
Results And Discussion
Identification of Ubp3–Bre5 partners
To identify components of the ubiquitin proteasome system (UPS) that cooperate with Ubp3–Bre5, we searched for partners of this complex. For this purpose, cell extracts from genomically protein A (PrA)-tagged BRE5 or UBP3 strains were prepared by cryolysis (Alber et al, 2007). PrA-tagged proteins were isolated on immunoglobulin G-conjugated magnetic beads and bound proteins were analysed by tandem nano liquid chromatography mass spectrometry (supplementary Fig S1A and supplementary Table SI online). This approach allowed the identification of about 400 partners for the Bre5–Ubp3 complex, with more than 60% of the targets interacting with both proteins. Importantly, previously described substrates of the Ubp3–Bre5 complex, such as COPI and COPII components, were identified. The main surprise from this Ubp3–Bre5 interactome is its similarity with the recently published eIF3 interactome (Sha et al, 2009), in particular the representation of factors involved in ribosomal biogenesis and translation that constitute 48% of identified proteins (supplementary Fig S1B and supplementary Table SI online). Finally, Ubp3–Bre5 associates with various proteins of the UPS, including subunits of the 26S proteasome and the AAA-ATPase Cdc48.
Ubp3–Bre5 complex interacts with Cdc48
The chaperone-like Cdc48 is an essential component of the UPS. It has a major role in the proteasomal escort pathway through its ability to bind directly and indirectly to ubiquitinated proteins via specific substrate-recruiting cofactors and its interaction with other key factors of the UPS (Jentsch & Rumpf, 2007). Cdc48 also controls the balance between ubiquitination and degradation as well as deubiquitination and stabilization through its ability to interact with both the E4 multiubiquitination enzyme Ufd2 and the deubiquitinating enzyme Otu1 (Rumpf & Jentsch, 2006).
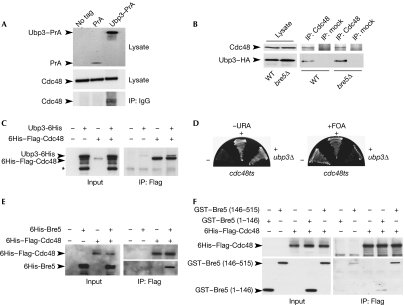
Cdc48 was pulled out by using both Ubp3–PrA and Bre5–PrA with 5 and 4 unique peptides identified covering 8.2% and 6.8% of sequence, respectively, but was not detected in control experiments (supplementary Fig S1A online; supplementary Table S1 online). Immunoprecipitation assays in cells expressing genomically tagged Ubp3–haemagglutinin (HA) show that Ubp3–HA co-immunoprecipitated with Cdc48 even in cells deleted for BRE5, indicating that Cdc48 interacts with Ubp3 in a Bre5-independent manner (Fig 1B). This result was confirmed in vitro by the direct interaction between recombinant 6His–Flag-Cdc48 and Ubp3-6His (Fig 1C). In addition, the combination of UBP3 deletion with a CDC48 thermosensitive allele induced a synthetic lethal growth phenotype at 23°C (Fig 1D), indicating a genetic link between Cdc48 and Ubp3.
Figure 1.
Ubp3–Bre5 deubiquitination complex interacts physically and genetically with Cdc48. (A) Lysates from untagged cells or cells expressing protein A (PrA) or Ubp3–PrA were affinity purified on immunoglobulin G-coupled magnetic beads. Lysates or bound proteins were analysed by western blotting using Cdc48 antibodies (lower panels) or only secondary antibodies (upper panel). (B) Lysates from wild-type (WT) or bre5Δ cells expressing Ubp3–HA were immunoprecipitated (IP) using anti-Cdc48 or mock antibodies and analysed by western blotting with the indicated antibodies. Anti-Flag–agarose beads were used to immunoprecipitate purified recombinant 6His–Flag-Cdc48 (1 μg) in the absence or presence of recombinant (C) Ubp3–6His (3 μg), (E) 6His–Bre5 (1.2 μg) or (F) GST–Bre5 mutant proteins (1 μg). Anti-Flag–agarose beads were used as the control. Approximately, 3% of input or 50% of bound proteins were analysed by western blotting using anti-6His (C,E (upper panel), F (upper panel)), anti-Bre5 (E, lower panel), or anti-GST (F, lower panel). (*) indicates the major degradation product of Ubp3–6His. (D) The cdc48-6 thermosensitive mutant cells were transformed (+) or not (−) with p426-UBP3 (URA3) plasmid, before the deletion of the genomic copy of UBP3 gene (ubp3Δ). Transformants were streaked subsequently on 5-fluoroorotic acid (5-FOA) medium. Bre5, brefeldin A sensitivity 5; Cdc48, cell division cycle 48; GST, glutathione-Stransferase; HA, haemagglutinin; IP, immunoprecipitated; PrA, protein A; Ubp3, ubiquitin-specific processing protease 3; WT, wild type.
Consistent with mass spectrometry analysis, recombinant 6His–Flag-Cdc48 also binds directly to 6His-Bre5 or glutathione-S-transferase (GST)–Bre5 (Fig 1E and data not shown). In addition, immunoprecipitation assays in cells expressing genomically tagged Bre5–green fluorescent protein (GFP) show that the Cdc48–Bre5 interaction occurs even in cells deleted for UBP3, indicating that Cdc48 interacts with Bre5 in a Ubp3-independent manner (supplementary Fig S2 online). To investigate further whether Bre5, Ubp3 and Cdc48 can form a ‘menage-à-trois' rather than mutually exclusive complexes, the interaction of recombinant 6His–Flag-Cdc48 with either the amino-terminal nuclear transport factor 2-like domain (GST–Bre5 (1–146)) responsible for Ubp3 binding, or the carboxy-terminal domain of Bre5 (GST–Bre5 (146–515)) was analysed by immunoprecipitation using Flag antibodies. As shown in Fig 1F, Cdc48 associated clearly with the C-terminal domain of Bre5, indicating that Ubp3–Bre5 and Cdc48 form a tripartite complex with Ubp3 and Cdc48 interacting with two distinct regions of Bre5.
Together, these data indicate that the Ubp3–Bre5 deubiquitination complex interacts with Cdc48, as shown previously for Otu1.
Ubp3 interacts with Ufd3, a cofactor of Cdc48
Cdc48 interacts directly with the ubiquitin-binding protein Ufd3, which recruits Cdc48 to ubiquitinated substrates, but also competes with Ufd2 on the Cdc48-binding site, thus reinforcing deubiquitination promoted by Cdc48-bound Otu1 (Ghislain et al, 1996; Mullally et al, 2006; Rumpf & Jentsch, 2006).
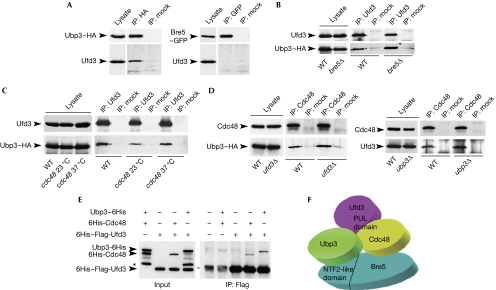
Epistatic miniarray profiles indicate an alleviating interaction between Ubp3–Bre5 and Ufd3, suggesting a potential interaction between these proteins (Collins et al, 2007). We observed a clear co-immunoprecipitation of Ufd3 with Ubp3–HA but not with Bre5–GFP (Fig 2A). Moreover, Ubp3–HA co-precipitated with Ufd3 in both wild-type and bre5Δ cells (Fig 2B), thus indicating that Ubp3 and Ufd3 interact in vivo in a Bre5-independent manner. To determine whether interaction between Ufd3 and Ubp3 requires functional Cdc48, immunoprecipitation assays were performed in the cdc48-6 thermosensitive mutant at 23°C and 37°C. As shown in Fig 2C, Ubp3–HA co-precipitated with Ufd3 at both temperatures, whereas expression of mutant Cdc48 was not affected by shifting the temperature to 37°C (data not shown). Functional integrity of Cdc48 is therefore dispensable for interaction between Ubp3 and Ufd3 in vivo. Finally, co-immunoprecipitation of Cdc48 with Ubp3 or Ufd3 was unaffected by the deletion of UFD3 or UBP3, respectively (Fig 2D), suggesting that these interactions occur directly and independently. Indeed, recombinant 6His–Flag-Ufd3 was able to bind directly not only to 6His-Cdc48 as described previously (Mullally et al, 2006), but also to Ubp3-6His (Fig 2E). A similar binding to Ufd3 was observed when both Cdc48 and Ubp3 were added together (data not shown), indicating that interactions between Ufd3 with Cdc48 and Ubp3 are independent. In agreement with immunoprecipitation assays (Fig 2A), no binding was detected in vitro between recombinant Ufd3 and Bre5 (data not shown).
Figure 2.
Ubp3 interacts with Ufd3. (A–D) Lysates from wild-type and mutant cells expressing Ubp3–HA or Bre5–GFP were immunoprecipitated using the indicated antibodies. Immunoprecipitates were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and analysed by western blotting with the indicated antibodies. (E) Anti-Flag–agarose beads were used to immunoprecipitate recombinant 6His–Flag-Ufd3 (1 μg) in the presence or absence of 6His-Cdc48 (1 μg) or Ubp3-6His (1 μg). Anti-Flag–agarose beads were used as a control. Approximately, 3% of input or 50% of bound proteins were analysed by western blotting using 6His antibodies. (*) indicates the major degradation product of Ubp3-6His and (°) is a non-specific band associated with the agarose beads. (F) Model illustrating the interaction between Ubp3–Bre5 and Cdc48 complexes. It should be noted that the PUL domain of Ufd3 that mediates its association with Cdc48 is not sufficient for binding to Ubp3. Bre5, brefeldin A sensitivity 5; Cdc48, cell division cycle 48; GFP, green fluorescent protein; HA, haemagglutinin; NTF2, nuclear transport factor 2; Ubp3, ubiquitin-specific processing protease 3; Ufd3, ubiqutin fusion degradation 3; WT, wild type.
Together these results show that Ubp3, Ufd3 and Cdc48 form non-mutually exclusive pair-off interactions and support the idea that these proteins could constitute an at least transient tripartite complex and even tetrapartite with Bre5 (Fig 2F).
Cdc48 and Ufd3 are required for ribophagy
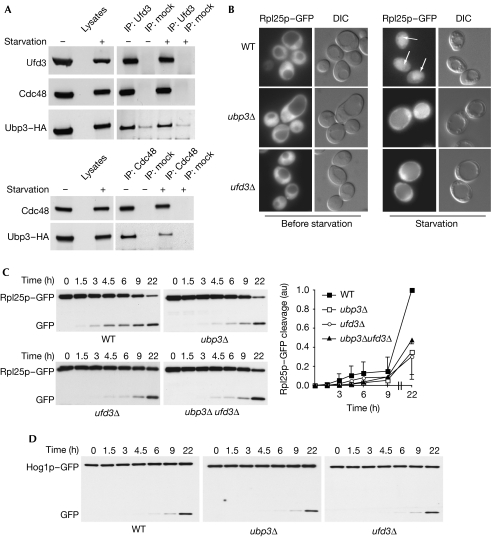
Recently, it was shown that the deubiquitinating activity of the Ubp3–Bre5 complex is involved in the control of 60S ribosomal particle degradation on starvation. This selective autophagic pathway induces a relocalization of ribosomal particles from the cytoplasm to the vacuole, in which they are degraded by vacuolar proteases. Ubiquitination of ribosomal proteins or ribosome-associated proteins is increased in ubp3Δ cells, suggesting that ribophagy could be regulated directly by ubiquitination (Kraft et al, 2008). To examine whether Cdc48 and Ufd3 are involved in this autophagic process, we first confirmed that the Ubp3–Cdc48–Ufd3 complex was preserved on nitrogen starvation (Fig 3A). Ribophagy efficiency was then analysed in cells deleted for UBP3 or UFD3, or mutated for CDC48. A GFP-tagged version of the ribosomal protein Rpl25 (Rpl25–GFP) was expressed in wild-type and mutant cells and GFP localization was analysed by epifluorescence microscopy. Rpl25–GFP is incorporated entirely in ribosomes (Gadal et al, 2001; Kraft et al, 2008) and therefore was located throughout the cytoplasm in both wild-type and mutant cells before starvation, but the fluorescent signal was detected in the vacuole of starved wild-type cells (Fig 3B). This relocalization corresponds to the transport of Rpl25–GFP to the vacuole, resulting in the degradation of the Rpl25 moiety and vacuolar accumulation of GFP. Both autophagy and functional vacuole are required for this process as degradation of Rpl25–GFP did not occur in pep4Δ or atg7Δ cells (Kraft et al, 2008; data not shown). In agreement with Kraft et al (2008), Rpl25–GFP remained located in the cytoplasm of starved ubp3Δ cells, indicating inhibition of ribophagy (Fig 3B). Strikingly, deletion of UFD3 also led to an inhibition of ribophagy with all mutant cells accumulating Rpl25–GFP in the cytoplasm (Fig 3B). The cleavage of GFP can also be followed by western blotting of total extracts prepared from starved cells by using GFP antibodies. Consistent with the fluorescence-based assay, a significant GFP cleavage can be observed at 22 h of starvation. As shown in Fig 3C, deletion of UBP3 or UFD3 led to a delayed GFP cleavage on starvation and deletion of both genes did not show a significant additive effect. In addition, this double knock-out mutant did not display an increased sensitivity to the TOR inhibitor rapamycin as compared with single deletion mutants (supplementary Fig S3 online), indicating that Ubp3 and Ufd3 probably function in the same pathway. By contrast, degradation of the 40S ribosomal subunit was not affected on deletion of UBP3 or UFD3 (Kraft et al, 2008; data not shown). Although deletion of UFD3 has been reported to decrease ubiquitin levels (Johnson et al, 1995), the effects of UFD3 deletion on ribophagy were not suppressed by the overexpression of a 6His-tagged version of ubiquitin (supplementary Fig S4 online), indicating that Ufd3 has a more direct function in this autophagic pathway.
Figure 3.
Ubp3–Bre5 and Ufd3 control ribophagy. (A) Lysates from wild-type cells expressing Ubp3–HA were cultured in rich medium or were subjected to 6 h of nitrogen starvation before immunoprecipitation using Ufd3 (upper panel) or Cdc48 (lower panel) antibodies. Immunoprecipitates were resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and analysed by western blotting with the indicated antibodies. (B) Cells expressing Rpl25–GFP were grown to mid-log phase in rich medium (before starvation), starved in medium containing 0.17% yeast nitrogen base without amino acids for 24 h (starvation) and examined by both fluorescence microscopy (Rpl25–GFP) and interferential contrast (DIC). Vacuolar accumulation of the GFP signal is indicated by arrows. Cells expressing (C) Rpl25–GFP or (D) Hog1–GFP were starved for the indicated period of time. Degradation of GFP-tagged proteins was then analysed by western blotting in whole-cell extracts using GFP antibodies. To avoid loading or experimental variations, the ratio between cleaved GFP and full-length protein was quantified at the times indicated in three independent experiments as described previously (Kraft et al, 2008). Quantification of the experiment presented in this figure is also shown as supplementary Fig S8 online. Bre5, brefeldin A sensitivity 5; Cdc48, cell division cycle 48; GFP, green fluorescent protein; HA, haemagglutinin; Ubp3, ubiquitin-specific processing protease 3; Ufd3, ubiqutin fusion degradation 3; WT, wild type.
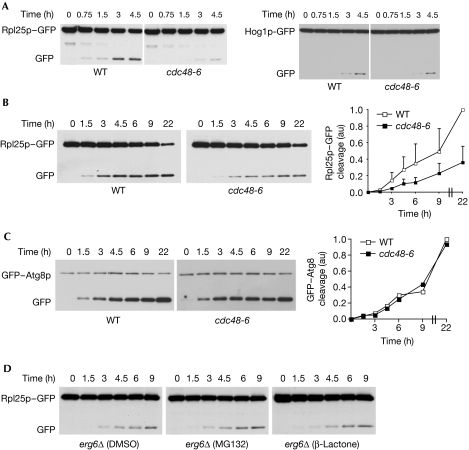
The defect in ribophagy of the 60S subunit could also be observed in a thermosensitive mutant of CDC48 both at the restrictive and the permissive temperature (Fig 4A,B; supplementary Fig S5 online). Ubp3 has been described to be involved specifically in ribophagy and not in general autophagy (Kraft et al, 2008). To analyse whether Ubp3 partners behave similarly, nitrogen starvation-induced cleavage of Hog1–GFP, a cytoplasmic control protein, was followed in wild-type and mutant cells. As shown in Fig 3D and 4A, neither deletion of UBP3 or UFD3 nor mutation of CDC48 were able to significantly affect the degradation of Hog1–GFP by non-selective autophagy. In addition, general autophagy analysed by the starvation-induced degradation of GFP–Atg8 was not inhibited in cdc48-6 mutant cells (Fig 4C) or on deletion of UBP3 or UFD3 (supplementary Fig S6 online). Together these data clearly show that selective and efficient ribophagy not only requires the Ubp3–Bre5 complex but also its binding partners Ufd3 and Cdc48. Although we tested different interaction mutants of Cdc48, Bre5 and Ufd3, the pair-wise interactions between the different components of the complex Bre5–Ubp3–Cdc48–Ufd3 so far prevent the design of useful mutants for demonstrating fully that these proteins act together and not separately in the ribophagic function. The similar defects observed in ubp3Δ, ufd3Δ and ubp3Δufd3Δ mutant cells (Fig 3C; supplementary Fig S3 online) suggest strongly that Ubp3–Bre5 and Ufd3 function as a complex in the ribophagic pathway. However, further experiments are required to validate the functionality of the newly identified complex.
Figure 4.
Cdc48 but not the proteasome controls ribophagy. Wild-type (WT) or cdc48-6 cells expressing Rpl25–GFP, Hog1–GFP or GFP–Atg8 were starved and shifted to (A) 37°C or starved at (B,C) 25°C for the indicated period of time. Degradation of GFP-tagged proteins was then analysed by western blotting in whole-cell extracts using GFP antibodies and quantified as in Fig 3. (D) erg6Δ cells expressing Rpl25–GFP were starved at 25°C in the absence or presence of MG132 (50 μM) or clasto-lactacystin β-lactone (20 μM) for the indicated period of time. Degradation of GFP-tagged proteins was then analysed as described previously. The increase of total polyubiquitinated proteins on treatment with proteasome inhibitors was confirmed by using ubiquitin antibodies (supplementary Fig S7 online). Cdc48, cell division cycle 48; GFP, green fluorescent protein; WT, wild type.
Mutations of Cdc48 strongly affect polyubiquitination and subsequent proteasomal degradation. To determine whether ribophagy was mediated by the catalytic activity of the proteasome, starvation-induced degradation of Rpl25–GFP was analysed in wild-type cells on treatment with various proteasome inhibitors. As shown in Fig 4D, neither MG132, an inhibitor of the chymotrypsin-like activity of the 20S proteasome, nor clasto-lactacystin β-lactone, an inhibitor of both trypsin-like and chymotrypsin-like activity of the 20S proteasome, led to an inhibition of ribophagy, whereas treatment with these proteasome inhibitors led to the accumulation of polyubiquitinated proteins (supplementary Fig S7 online), indicating that the proteolytic activity of the proteasome is probably not involved in this process.
The deubiquitinating complex Ubp3–Bre5 and its newly described binding partners Ufd3 and Cdc48 are required for an efficient ribophagy, most probably through the regulation of a yet to be identified ubiquitinated substrate. Although an Ubp3-independent role of Cdc48 in ribophagy cannot be formally excluded, the insensitivity of this process towards proteasome inhibitors makes a Cdc48-mediated proteasomal escort pathway unlikely. Cdc48 has been proposed to act as a segregase that liberates ubiquitinated proteins from non-modified partners (Braun et al, 2002). Cdc48 together with Ufd3 might thus function to recognize and isolate a ubiquitinated target crucial for ribophagy and facilitate its deubiquitination by Ubp3–Bre5. In addition, the ability of Ubp3–Bre5 to associate with ribosomes (and/or translasosome) probably strengthens the efficiency of this process. We propose that efficient ribophagy requires the Ubp3–Bre5–Ufd3–Cdc48-dependent elimination of specific ubiquitinated target(s) by deubiquitination before vacuolar degradation.
Methods
Yeast strains and media. The S. cerevisiae strains and plasmids used in this study are listed in supplementary Table S2 online. Yeast cultures were grown either in rich medium (yeast peptone dextrose; YPD) or in synthetic medium containing 0.67% yeast nitrogen base with ammonium sulphate, 2% dextrose, and supplemented with appropriate nutrients. Cells were starved in medium containing 0.17% yeast nitrogen base without amino acids, 2% dextrose (Kraft et al, 2008) in the absence or presence of MG132 (50 μM) or clasto-lactacystin β-lactone (20 μM).
Preparation of yeast total extracts. Yeast cells grown in YPD or synthetic medium were collected during the exponential growth phase (A600 of 1.5 or 0.8, respectively). Total protein extracts were prepared by the NaOH–trichloroacetic acid lysis method (Cohen et al, 2003a). Rabbit polyclonal antibodies to Sec23p, Cdc48 and Ufd3 were provided by B. Lesch and R. Schekman, T. Sommer and S. Jentsch, respectively. Chicken polyclonal antibodies to Bre5 were raised against recombinant His-Bre5.
Co-immunoprecipitation experiments. Yeast cells were grown up to an A600=1.5. Cells were collected and frozen rapidly in liquid nitrogen before cryolysis (Alber et al, 2007). Grindate was resuspended in ice-cold immunoprecipitation (IP) buffer (50 mM HEPES (pH 7.5), 75 mM NaCl, 1 mM dithiothreitol, 0.1% Triton-X 100, 5% glycerol, protease inhibitors cocktail), homogenized and cleared by centrifugation. Alternatively, cells were lysed at 4°C with glass beads in IP buffer. Lysates were cleared by centrifugation and incubated with indicated antibodies together with protein G-coupled Dynabeads (Invitrogen) for 2 h at 4°C. Beads were washed with IP buffer and bound proteins were eluted by heating samples at 60°C for 10 min in Laemmli sample buffer before western blot analysis using appropriate antibodies and chemiluminescence protein immunoblotting reagents (Pierce).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank S. Jentsch and T. Sommer for providing us with reagents; E. Hurt and M. Peter for Rpl25–GFP- and Hog1–GFP-expressing vectors, respectively; and K. Wilkinson for Cdc48 and Ufd3 expression plasmids. We also thank L. Pintard and members of C. Dargemont's lab for helpful discussions. This study was funded by grants from the Ministère de la Recherche, the Association for Research against Cancer and the Fondation pour la Recherche Médicale. M.B. is supported by the Centre National pour la Recherche Scientifique.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alber F et al. (2007) Determining the architectures of macromolecular assemblies. Nature 450: 683–694 [DOI] [PubMed] [Google Scholar]
- Baxter BK, Abeliovich H, Zhang X, Stirling AG, Burlingame AL, Goldfarb DS (2005) Atg19p ubiquitination and the cytoplasm to vacuole trafficking pathway in yeast. J Biol Chem 280: 39067–39076 [DOI] [PubMed] [Google Scholar]
- Bilsland E, Hult M, Bell SD, Sunnerhagen P, Downs JA (2007) The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair (Amst) 6: 1471–1484 [DOI] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J 21: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C (2003a) Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5: 661–667 [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Dargemont C (2003b) Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J Biol Chem 278: 51989–51992 [DOI] [PubMed] [Google Scholar]
- Collins SR et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, Tollervey D, Hurt E (2001) A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J 20: 3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J 15: 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S (2007) Cdc48 (p97): a ‘molecular gearbox' in the ubiquitin pathway? Trends Biochem Sci 32: 6–11 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2008) Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell 30: 498–506 [DOI] [PubMed] [Google Scholar]
- Li K, Zhao K, Ossareh-Nazari B, Da G, Dargemont C, Marmorstein R (2005) Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor. J Biol Chem 280: 29176–29185 [DOI] [PubMed] [Google Scholar]
- Li K, Ossareh-Nazari B, Liu X, Dargemont C, Marmorstein R (2007) Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J Mol Biol 372: 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally JE, Chernova T, Wilkinson KD (2006) Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain. Mol Cell Biol 26: 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21: 261–269 [DOI] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA (2009) The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell 36: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu M, Ayalew M, Ruff JA (2008) Down-regulation of Pkc1-mediated signaling by the deubiquitinating enzyme Ubp3. J Biol Chem 283: 1954–1961 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.