ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes
Initiation of transcription by RNA polymerase II is regulated by activators that recruit coactivator complexes to gene promoters. Here, Krebs and colleagues show that the ATAC and Mediator coactivator complexes form a stable meta structure and regulate the transcription of particular non-coding RNA genes. Their work suggests that the formation of stable meta-complexes may facilitate the combined actions of coactivator complexes on specific target genes.
Keywords: GCN5, PCAF, SAGA, LUZP1, snRNA biogenesis
Abstract
The Ada-Two-A-containing (ATAC) histone acetyltransferase and Mediator coactivator complexes regulate independent and distinct steps during transcription initiation and elongation. Here, we report the identification of a new stable molecular assembly formed between the ATAC and Mediator complexes in mouse embryonic stem cells. Moreover, we identify leucine zipper motif-containing protein 1 as a subunit of this meta-coactivator complex (MECO). Finally, we demonstrate that the MECO regulates a subset of RNA polymerase II-transcribed non-coding RNA genes. Our findings establish that transcription coactivator complexes can form stable subcomplexes to facilitate their combined actions on specific target genes.
Introduction
Gene expression is a tightly regulated process. Initiation of transcription by RNA polymerase II (Pol II) is believed to be the outcome of a sequence of events beginning with the binding of specific activators to their cognate binding sites. This initial step triggers the recruitment of coactivator complexes and general transcription factors at promoters to allow the loading of Pol II into the preinitiation complex (PIC) to achieve transcription initiation. In this process, coactivators have crucial roles through their enzymatic and non-enzymatic functions. For example, they allow chromatin modification and opening, as well as enhancement of the formation of initiation complexes (Thomas & Chiang, 2006).
The histone acetyltransferases (HATs) general control of amino-acid synthesis 5 (GCN5 or KAT2A) and p300/CBP-associated factor (PCAF or KAT2B) are encoded by paralogue genes and share 70% sequence identity. GCN5 and PCAF are mutually exclusive subunits of two functionally distinct, but related, multisubunit coactivator complexes: Spt–Ada–Gcn5 acetyltransferase (SAGA) and Ada-Two-A-containing (ATAC; Wang et al, 2008; Nagy et al, 2010). These complexes have been shown to differentially regulate both locus-specific gene expression and global chromatin structure through their enzymatic activities (HAT activity and histone deubiquitination).
The Mediator complex (MED) is another multisubunit key transcription coactivator complex. Even though the complete mechanism by which this complex controls transcription is not fully understood, the MED is believed mainly to be required to mediate molecular bridges among activators, coactivators and general transcription factors to enhance or repress the formation of PIC leading to transcription initiation (Malik & Roeder, 2005). The existence of at least two functional forms of the MED complex has been reported. In the presence of the MED kinase module, the MED (known as TRAP) represses transcription. By contrast, when the kinase module is lost and the subunit MED26 is instead incorporated to the complex, the MED (known as PC2) activates transcription (Malik & Roeder, 2005; Paoletti et al, 2006).
To gain more insight into the molecular mechanisms of ATAC function, we carried out a comprehensive analysis of the ATAC interactome. We showed that in certain cell types, including mouse embryonic stem cells (mESCs), the ATAC and MED complexes form a highly stable meta-coactivator complex (MECO) and that the MECO associates and regulates transcription of a subset of Pol II-transcribed non-coding RNA (ncRNA) genes.
Results And Discussion
ATAC interacts stably with the active Mediator complex
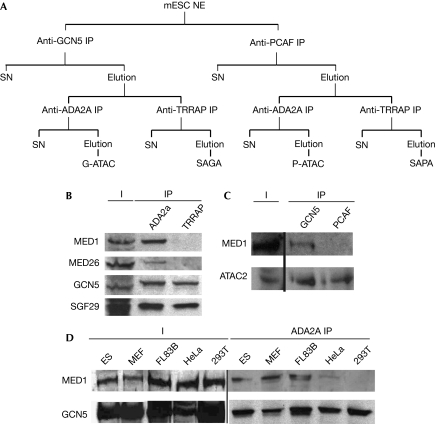
To identify partners of the GCN5- (G-) and PCAF- (P-) containing complexes in mESC, we used a tandem immunoprecipitation (IP) strategy. First, complexes containing either one or the other HAT paralogues were separated. Then the ATAC and SAGA complexes were separated successively by immunoprecipitations using antibodies specific to each type of complex (Fig 1A). At this stage, the bead–antibody-bound complexes were washed with 500 mM KCl containing IP buffer. The four preparations were analysed by mass spectrometry (MS) to identify potential interacting partners of the complexes in mESCs. In the four complex preparations, all the known core subunits of the SAGA and ATAC complexes were identified with high confidence (Table 1; supplementary Fig S1A online). Unexpectedly, in the G-ATAC preparation, 19 subunits of the MED were also identified (Table 1). By contrast, no MED subunits were identified in the different SAGA immunoprecipitations or in the P-ATAC preparation (Table 1; supplementary Fig S1A online; data not shown). This observation indicates a new specific interaction between G-ATAC and the MED complexes in mESCs. Remarkably, whereas the MED subunit MED26 was identified with good confidence, MED kinases CDK8 or CDK11 were not detectable in our preparation. These data, together with the fact that we also identified subunits of Pol II (Table 1), indicate strongly that the active PC2-Pol II form of the MED is the one that associates preferentially with the GCN5-containing ATAC.
Figure 1.
Ada-Two-A-containing and the Mediator complex interact stably in a cell-specific manner. (A) Schematic representation of the ATAC and SAGA tandem immunoprecipitation (IP). First, GCN5- (G-) or PCAF- (P-) containing complexes were isolated by the corresponding IPs followed by peptide competition elution. From these pools ATAC (α-ADA2a IP) or SAGA (α-TRRAP IP) complexes were isolated, respectively. (B) Equivalent amounts of purified ATAC (ADA2a IP) and SAGA (TRRAP IP) complexes (normalized for their GCN5 content) were analysed by western blot against ATAC (GCN5, SGF29) and MED subunits (MED1, MED26). Input (I) and immunoprecipitated fractions are shown. (C) Equivalent amounts of GCN5- and PCAF-containing complexes (purified by either an α-GCN5 IP or an α-PCAF IP, respectively, and normalized for their ATAC2 content) were analysed by western blot with the indicated antibodies. (D) Cell extracts were prepared from the indicated mouse (ES, MEF and FL83B) and human (HeLa and 293T) cell lines. From these, ATAC complexes were prepared by a single α-ADA2a IP and the complexes analysed by western blot with the indicated antibodies. ATAC, Ada-Two-A-containing; ES, embryonic stem; GCN5, general control of amino-acid synthesis 5; mESC, mouse embryonic stem cell; MED, mediator complex; MEF, mouse embryonic fibroblast; NE, nuclear extract; PCAF, p300/CBP-associated factor; SAGA, Spt–Ada–Gcn5 acetyltransferase; SAPA, Spt–Ada-PCAF-acetyltransferase; SGF29, SAGA-associated factor 29; SN, supernatant.
Table 1. Results of mass spectrometry analysis of GCN5- and PCAF-ATAC showing the percentage of coverage and the number of unique peptides found for each subunit.
| Uniprot ID | GCN5-ATAC | PCAF-ATAC | ||
|---|---|---|---|---|
| Coverage (%) | Unique peptides | Coverage (%) | Unique peptides | |
| ATAC core | ||||
| YETS2 | 67.4 | 64 | 47.3 | 41 |
| ZZZ3 | 56.5 | 36 | 24.3 | 17 |
| SGF29 | 53.6 | 13 | 45.7 | 8 |
| MBIP1 | 46.3 | 14 | 36.7 | 11 |
| WDR5 | 44 | 10 | 40.7 | 8 |
| TAD2L | 42.9 | 15 | 28.4 | 9 |
| TAD3L | 36.1 | 11 | 16.4 | 5 |
| CSR2B | 27.9 | 20 | 14.8 | 11 |
| GCNL2 | 23.4 | 19 | — | — |
| PCAF | — | — | 21.2 | 14 |
| NC2B | 4 | 1 | 4 | 1 |
| Mediator | ||||
| MED27 | 31.5 | 5 | — | — |
| MED15 | 21.6 | 12 | — | — |
| MED28 | 21.3 | 2 | — | — |
| MED20 | 20.8 | 4 | — | — |
| MED17 | 19.1 | 9 | — | — |
| MED31 | 18.3 | 2 | — | — |
| MED4 | 15.6 | 2 | — | — |
| MED14 | 15 | 13 | — | — |
| MED21 | 13.9 | 1 | — | — |
| MED26 | 12.9 | 5 | — | — |
| MED8 | 12.3 | 2 | — | — |
| MED18 | 8.7 | 2 | — | — |
| MED6 | 8.1 | 2 | — | — |
| MED24 | 7.1 | 5 | — | — |
| MED12 | 6.1 | 8 | — | — |
| MED16 | 6 | 4 | — | — |
| MED23 | 5.7 | 6 | — | — |
| MED1 | 3.3 | 4 | — | — |
| MED13 | 1.2 | 2 | — | — |
| RNA Pol II | ||||
| RPB1 | 4.8 | 7 | — | — |
| RPB2 | 2.9 | 3 | — | — |
| ATAC, Ada-Two-A-containing; GCN5, general control of amino-acid synthesis 5; PCAF, p300/CBP-associated factor; RPB, RNA polymerase B subunit. | ||||
To validate further the G-ATAC–MED interaction and the lack of interaction between the SAGA and MED complexes, we separated equivalent amounts (normalized to their GCN5 content) of ATAC (ADA2a IP) and SAGA (TRRAP IP) complexes by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and tested for the presence of MED subunits in these immunopurified complexes by western blot. In agreement with the MS data, MED1 was detected in ATAC, but not in SAGA immunoprecipitations (Fig 1B). The association between G-ATAC and MED was also confirmed using different antibodies specific to either ATAC (α-ATAC2) or MED (α-MED1). These immunoprecipitations further confirmed the results obtained with the α-ADA2a IP (supplementary Fig S1B,C online). Moreover, the G-ATAC–MED interaction was not mediated by DNA because when the nuclear extracts were treated with either DNAse I or ethidium bromide we obtained the same co-immunoprecitation results as before (supplementary Fig S2 online). When the presence of MED subunits was investigated in the GCN5- and PCAF-containing complexes by loading equivalent amounts of purified G-ATAC and P-ATAC (normalized for ATAC2; Fig 1C), the MED1 subunit was detected only in the GCN5-containing complexes. This result confirms our MS data and indicates, for the first time, a functional difference between G-ATAC and P-ATAC. Moreover, this indicates that GCN5 probably has a structural role in the formation of the interaction(s) between ATAC and the MED complexes. However, as GCN5 is shared between SAGA and ATAC, further ATAC-specific subunits and new MECO subunits could also be involved in the interaction (see below).
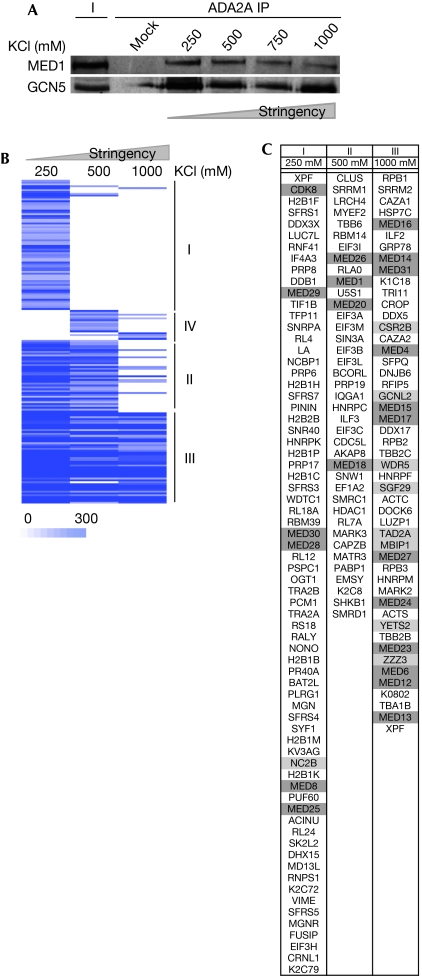
Next, we analysed the G-ATAC–MED stability by increasingly concentrated salt washes of the bead–antibody-bound complexes. This experiment indicated that G-ATAC–MED interaction is extremely stable as the α-ADA2a IP co-purified MED1 up to 1 M KCl washes (Fig 2A). This result indicates that, in contrast to the previously reported low-stability interaction between hSAGA and MED (Liu et al, 2008), the G-ATAC–MED complex seems to be a very stable MECO structure formed by the assembly of ATAC and MED in vivo.
Figure 2.
Identification of candidates for constituting the Ada-Two-A-containing–Mediator molecular bridge. (A) ATAC complexes were immunoprecipitated with an α-ADA2a antibody and the bead–antibody-bound complexes were washed with the indicated salt-containing immunoprecipitation (IP) buffer (from 250 to 1000 mM) three times before elution. The purified complexes were then analysed by western blot with the indicated antibodies. (B) ADA2a-containing complexes were prepared as in Fig 1A except that the bead–antibody-bound complexes were washed with 250, 500 and 1000 mM KCl-containing buffers. Proteins in these three ATAC complex preparations were analysed by mass spectrometry. The heat-map represents the clustering analysis of the proteins identified in the ATAC purifications under different salt stringencies. Proteins were grouped according to their enrichment score variation (from 0 to 300 MASCOT unit) in the different conditions. Three groups representing gradual stability in the complex were isolated: I lost at 500 mM; II lost at 1000 mM; III stable in all conditions tested; VI only present at high stringencies. (C) Table presenting the Uniprot ID for the proteins for cluster categories I–III. Light grey denotes ATAC subunits and medium grey MED subunits. ATAC, Ada-Two-A-containing; GCN5, general control of amino-acid synthesis 5; MED, mediator complex.
Altogether, the above data show that in mESCs, G-ATAC, but not SAGA nor P-ATAC, specifically co-purifies with a fraction of the PC2 MED26-containing MED.
The ATAC–MED interaction is not restricted to mESC
To analyse whether this newly identified ATAC–MED complex is formed specifically in mESCs because of their particular pluripotent state, or whether this complex can be formed in other cell types, we repeated the ATAC immunopurification in different mouse cell lines representing different stages of differentiation, and in human cell lines from which ATAC was originally purified (HeLa and HEK293Tl; Wang et al, 2008; Nagy et al, 2010). Interestingly, we observe that this interaction can be found in cell lines other than mESC, as MED1 also co-purifies with GCN5 in fibroblastic lineages (MEF, FL83B). In agreement with previous reports, this interaction could not be observed in HEK293T cells and was very weak in HeLa cells. These data show that the association of the ATAC and the MED is not restricted to pluripotent cells, and that its abundance varies in the different cell lines tested, indicating that the formation of this MECO complex is dependent on the cellular context. This result also indicates that a bridging factor might exist between the two complexes in some cell types, but not in others.
Candidates to form the bridge between the two complexes
To isolate candidate bridging factors between ATAC and MED, we analysed by MS different ATAC preparations in which the bead–antibody-bound ATAC complexes were washed with increasing salt concentrations. As the G-ATAC–MED complex is stable up to 1 M KCl washes (Fig 2A), we prepared G-ATAC complexes at three selected washing stringencies (250, 500 and 1000 mM KCl) and analysed by MS the associated proteins in each condition. Next, all the identified proteins in each individual list were clustered according to their enrichment scores, to group proteins behaving similarly in the three different conditions (Fig 2B). Four distinct groups of proteins could be isolated. Group III contained proteins observed at all stringencies, which were also unaffected by the 1 M KCl wash. Groups I–II and IV represented groups of proteins that disappear (groups I–II) or appear (group IV) with the increasing stringency of washes used. Remarkably, we observed that almost all ATAC subunits are highly stable and are in group III (Fig 2C), except the 10 kDa NC2β subunit. Furthermore, most MED subunits were found in group III, meaning that the interaction between ATAC and MED is maintained at all stringencies. Consequently, group III is also likely to contain factors that are potentially involved in the molecular bridge that could have a role in the strong association of G-ATAC with MED. By excluding proteins that are also found in control immunoprecipitations (α-GST) and in other non-ATAC preparations from group III, we could isolate a restricted subset of candidates (supplementary Table S2 online), including the leucine zipper motif-containing protein 1 (LUZP1; Fig 2C; Sun et al, 1996). As LUZP1 presented the highest enrichment score among the candidates (supplementary Table S2 online), we further characterized the function of this protein in the formation of MECO.
LUZP1 is part of the bridge between ATAC and Mediator
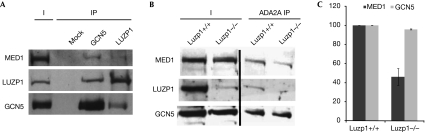
To verify whether LUZP1 is a bona fide subunit of the MECO, we compared the ATAC and MED contents of α-GCN5 and α-LUZP1 immunoprecipitations (Fig 3A). We detected LUZP1 in the GCN5 IP and more importantly, we could detect both GCN5 and MED1 in the α-LUZP1 IP, showing that LUZP1 is associated with the newly identified ATAC–MED meta-complex.
Figure 3.
Leucine zipper motif-containing protein 1 contributes to the molecular bridge between Ada-Two-A-containing and Mediator complexes. (A) Proteins were immunoprecipitated with antibodies raised against GCN5 or LUZP1 from embryonic stem cell extracts and immunoprecipitated complexes were analysed by western blot using antibodies against ATAC (GCN5) and MED (MED1) subunits as well as LUZP1. Input (I) and immunoprecipitation (IP) fractions are shown. (B) Whole-cell extracts were prepared from either Luzp1 (+/+) or Luzp1 (−/−) cells. From these extracts ATAC complexes were immunoprecipitated with α-ADA2a antibody. Complexes were analysed by western blot with the indicated antibodies. (C) The quantity of MED1 and GCN5 in Luzp1 (−/−) cells was determined relative to the quantity detected in Luzp1 (+/+) cells using the Quantity One software. Mean and range in the value over two biological replicates were calculated. ATAC, Ada-Two-A-containing; GCN5, general control of amino-acid synthesis 5; LUZP1, leucine zipper motif-containing protein 1; MED, mediator complex.
To determine whether LUZP1 has a role in the establishment of the interaction between ATAC and MED, we purified the ATAC complex from mESCs derived from Luzp1 knockout mice (Lee et al, 2001) and determined the quantity of the MED remaining associated with ATAC (Fig 3B). In Luzp1−/− mESCs, we observed a marked (∼50%) decrease in the interaction of MED with ATAC (Fig 3C). These data indicate that LUZP1 facilitates the formation of the molecular bridge between ATAC and MED, but also that further factors or post-translational modifications could be required for this interaction.
MECO is required for the activity set of ncRNA genes
Two lines of evidence indicate that the newly identified MECO would enhance transcription. First, the subunit composition of the ATAC-associated MED corresponds to the active form of the MED (MED26-containing, CDK-free, Pol II-associated). Second, ATAC is a HAT complex, known to be involved in transcription stimulation of certain stress-regulated genes (Nagy et al, 2010). To gain more insight into the in vivo function of the ATAC–MED complex, we undertook chromatin immunoprecipitation against LUZP1 and GCN5 followed by high-throughput sequencing (ChIP-seq) to determine the common loci to which the LUZP1-containing ATAC–MED complex binds. After bioinformatics analysis, 46 LUZP1 binding sites were identified with high confidence, which most probably represented only the most significant LUZP1 binding events. This low number might be because of the dynamic behaviour of LUZP1 in vivo limiting its crosslinking to chromatin. When comparing these LUZP1 binding sites with those of the genome-wide GCN5 density map obtained after ChIP-seq using mESCs, we observed that most LUZP1 sites lack GCN5, indicating that LUZP1 could also be detected on the genome in an ATAC–MED-independent manner (group I, Fig 4A). When the genome-wide LUZP1 binding sites were compared with those bound by GCN5 and Pol II (available from mESCs for Pol II; Mikkelsen et al, 2007), we identified a number of sites that were bound by all three factors (group II, Fig 4A). In agreement with the three ChIP-seq data sets, our ChIP-quantitative PCR (CRIP-qPCR) validation indicated that these sites were bound by LUZP1, GCN5, Pol II and MED1 (Fig 4C). Surprisingly, we observed that all the identified, bound genes belong to the family of genes expressing unspliced ncRNAs (Fig 4B), most of them being small nuclear RNA (snRNA) genes. To investigate how general is the recruitment of MECO subunits on snRNA promoters, we systematically analysed GCN5, Pol II and LUZP1 levels at all mouse snRNA promoters. We observed that the relevant GCN5 levels are detected systematically at active Pol II-transcribed snRNA promoters, indicating that the regulation of the expression of snRNAs by MECO would be a widespread mechanism (supplementary Fig S3 online).
Figure 4.
The Ada-Two-A-containing–Mediator meta-complex is bound to transcriptionally active non-coding RNA loci together with leucine zipper motif-containing protein 1 in mouse embryonic stem cells. (A) Two genome-wide ChIP-seq experiments were carried out in mESC using both LUZP1 and GCN5 polyclonal antibodies. The tag density over a 2 kb region around the identified LUZP1 binding sites was then compared with those of GCN5 or Pol II. The plot is delimited by bars representing cut-off (15 reads/kb as a cut-off value) used in further subdivision of the data. (I) Loci bound only by LUZP1; (II) loci bound by LUZP1, GCN5 and Pol II. (B) Analysis of the functional category of the transcripts neighbouring the group II loci (±500 bp). (C) Validation of the ChIP-seq data by ChIP-qPCR quantification of LUZP1–MECO binding on control loci (white bars) and loci from category I (LUZP1 only, black bars) and II (LUZP1–MECO, grey bars) using the indicated antibodies. (D) Quantification of non-coding gene expression changes on MECO subunit knockdowns. Measurements of the efficiencies of siRNA knockdowns against MECO subunits are presented in supplementary Fig S4 online. The relative levels of expression of MECO target genes after knockdown of GCN5 (light grey bars), ATAC2 (medium grey bars) and MED1 (dark grey bars) by siRNA relative to scramble siRNA (white bars) were quantified by reverse transcription–qPCR. Mean and standard deviation over three biological replicates were calculated. P-values were calculated using an unpaired t-test for triplicates (*P<0.05; **P<0.01). ATAC, Ada-Two-A-containing; IP, immunoprecipitation; ChIP-qPCR, chromatin immunoprecipitation-quantitative PCR; ChIP-seq, chromatin immunoprecipitation-coupled high-throughput sequencing; chr, chromosome; GCN5, general control of amino-acid synthesis 5; LUZP1, leucine zipper motif-containing protein 1; MECO, meta-coactivator complex; MED, mediator complex; mESC, mouse embryonic stem cell; ncRNA, non-coding RNA; siRNA, small interfering RNA; snRNA, small nuclear RNA.
To characterize the function of MECO at the identified ncRNA promoters, we first tested their expression in Luzp1−/− ES cell lines. Under these conditions no marked decrease in the tested ncRNA gene expression levels was observed (supplementary Fig S5 online). We hypothesize that the partial effect of LUZP1 knockout on MECO formation is not sufficient to reduce the expression of these genes. Next, we carried out a series of knockdowns against catalytic (GCN5) and structural subunits (ATAC2 and MED1) of ATAC and MED complexes and measured the changes in the expression levels of the four MECO target genes by quantitative PCR after reverse transcription of RNA (RT–qPCR). The downregulation of the three MECO subunits was around 60% (supplementary Fig S4 online). Under these conditions we observed a marked decrease (between 20–60%) in the relative expression levels of the target genes (snRNA and long ncRNA; Fig 4D). This result indicates that MECO is a main regulator of the tested ncRNAs.
Interestingly, transcriptional mechanisms of these snRNAs are known to be different from those of messenger RNAs (Jawdekar & Henry, 2008). Although the precise mechanistic action of this bridged ATAC–MED MECO and its integration into the snRNA biosynthesis mechanisms need to be studied further on the newly identified genomic loci, our results clearly indicate a new role for the identified MECO complex in the expression of these ncRNA transcripts.
We have shown the existence of a new MECO formed in cells by the stable association of the G-ATAC and the active PC2 form of the MED complexes. The integrity of this complex depends partly on the presence of LUZP1 in MECO and this complex is required for the active transcription of a particular class of non-coding transcripts. Our findings support the new idea that coactivator networks are functionally interconnected and, under particular conditions, can be modulated in stable meta structures to potentiate their actions at particular sites on the genome. Moreover, it indicates that ncRNA genes have different needs for chromatin-modifying coactivator assemblies than have the broadly studied protein-coding genes.
Methods
Protein immunopurification and MS analysis. Detailed IP procedure can be found in supplementary methods online. For MS analysis, mESC nuclear extracts were subjected to tandem IP followed by peptide competition elution. Retrieved purified complexes were concentrated and loaded on acrylamide gels. Gel lanes were cut into slices and each was washed and trypsinized. Proteolytic peptides were then collected and analysed by nanoflow liquid chromatography–tandem mass spectrometry.
ChIP-seq. ChIP was carried out as described previously (Nagy et al, 2010). For ChIP-seq analysis, 300 μg of chromatin (DNA) was incubated with GST (mock), LUZP1 and GCN5 antibodies. Retrieved purified DNA was sequenced using Illumina Genome Analyzer II following the manufacturer's recommendations.
Peptides and antibodies. Previously described α-GCN5 (2676), α-PCAF (2760), α-ADA2a (2AD2A1), α-TRRAP (2TRA1B3), α-ATAC2 (2734) and α-GST (TFII1D10; Nagy et al, 2010); α-mLUZP1 (Sun et al, 1996); and commercial MED1 (sc-5334) and MED26 (sc-48776) were used for IP, western blotting and ChIP experiments, respectively.
Bioinformatics analysis. Detailed bioinformatics procedures can be found in the supplementary methods online, and see supplementary Table S1 online for quality control. Briefly, LUZP1 enrichment clusters were detected by using model-based analysis of ChIP-seq (MACS; Zhang et al, 2008). After repeat masking, peaks were ranked and a cut-off was determined according to ChIP-qPCR validation results. Loci were annotated using a genomic position annotation tool (Krebs et al, 2008). Average read densities were collected in GCN5 and Pol II (GSE12241; Mikkelsen et al, 2007) raw data sets and compared with a mock ChIP data set.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Note added in proof. The raw and processed ChIP-seq datasets have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE21717.
Supplementary Material
Acknowledgments
We are grateful to M. Orpinell and D. Devys for critical reading of the paper, M. Meinsterernst for antibodies, R. Poot and M. Ballarino for advice, A. Dierich for help in mESC culture, the IGBMC high-throughput sequencing platform, G. Duval, P. Eberlin, IGBMC purchase service for their help, and K. Bezstarosti for technical support. A.R.K. is the recipient of a fellowship from INSERM-Région Alsace and the Association pour la Recherche sur le Cancer. This work was funded by grants from Agence Nationale de la Recherche (GenomATAC; ANR-09-BLAN-0266) and the European Union (EUTRACC, LSHG-CT-2007-037445).
Footnotes
The authors declare that they have no conflict of interest.
References
- Jawdekar GW, Henry RW (2008) Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta 1779: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs A, Frontini M, Tora L (2008) GPAT: retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics 9: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Chang AC, Sun DS, Hsu CY, Chang NC (2001) Restricted expression of LUZP in neural lineage cells: a study in embryonic stem cells. J Biomed Sci 8: 504–511 [DOI] [PubMed] [Google Scholar]
- Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E (2008) STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol 28: 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2005) Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L (2010) The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell Mol Life Sci 67: 611–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci USA 103: 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DS, Chang AC, Jenkins NA, Gilbert DJ, Copeland NG, Chang NC (1996) Identification, molecular characterization, and chromosomal localization of the cDNA encoding a novel leucine zipper motif-containing protein. Genomics 36: 54–62 [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41: 105–178 [DOI] [PubMed] [Google Scholar]
- Wang YL, Faiola F, Xu M, Pan S, Martinez E (2008) Human ATAC is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem 283: 33808–33815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.