This Keystone Symposium brought together a broad group of experts to examine the current advances in stem-cell biology, with a particular focus on induced pluripotent stem cells, and to discuss both their potential and limitations in therapeutic applications.
Abstract
The Keystone Symposium on Stem Cell Differentiation and Dedifferentiation attracted nearly 500 participants affiliated with academia, the biotechnology industry, scientific publishing and clinical practice. This broad group examined current advances in stem-cell biology, with a particular focus on induced pluripotent stem cells, and discussed both their potential and limitations in therapeutic applications.
The Keystone Symposium on Stem Cell Differentiation and Dedifferentiation held in February this year was co-organized by Fiona Watt (CRUK, Cambridge Research Institute, UK) and Shinya Yamanaka (Center for iPS Cell Research and Application, Kyoto U., and Gladstone Institute of Cardiovascular Disease, UCSF). The five-day meeting kicked off with a presentation from James Thomson (Morgridge Institute, Wisconsin), a pioneer in human embryonic stem cell (ESC) derivation. Thomson began with what was to rapidly emerge as a recurring theme in the meeting—that human stem cells generated by induced pluripotency show varied phenotypes that can be moderated by recloning, but probably reflect the different cellular origins of the somatic cells from which they arose.
The good, bad and unpredictable
Induced pluripotent stem cells (iPSCs) were elaborated originally by Kazutoshi Takahashi (Center for iPS Cell Research and Application, Kyoto U.) and Yamanaka (Takahashi & Yamanaka, 2006) in a breath-taking experiment published in 2006. The pair showed that the forced expression of four factors (OCT4, SOX2, c-MYC and KLF4) could reprogramme differentiated fibroblasts to an ESC-like state, albeit at low frequency and after substantial time lag (Fig 1). These iPSCs could be used to generate embryonic chimaeras, but not adult mice in vivo, and failed to establish germ-line transmission. Technical refinements followed the publication of their work in quick succession, as the possibilities of using this approach for cell transplantation therapy, modelling human disease and establishing novel drug screening approaches were realized across the globe. Although these developments successfully addressed many early concerns, including the development of vector-free approaches (Yu et al, 2009; Okita et al, 2010; Stadtfeld et al, 2008), one major hurdle still exercises the iPSC community: although iPSCs are capable of extensive differentiation in vitro, they generate tumours on transplantation in vivo (Miura et al, 2009) at an alarming rate.
…one major hurdle still exercises the iPSC community: although iPSCs are capable of extensive differentiation in vitro, they generate tumours on transplantation in vivo…
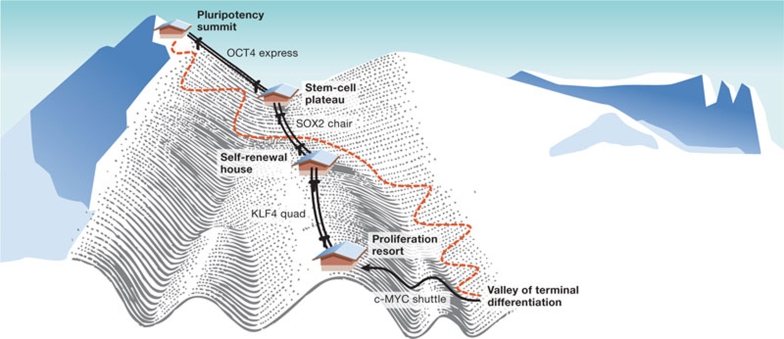
Figure 1.
A skier's guide to negotiating Waddington's slopes. Self-renewing, tissue-specific stem cells might get back to the ‘pluripotency summit' by a single ride on Schöler's ‘OCT4 express' (Kim et al, 2009). If, however, you follow the slopes too far and a thrilling run takes you all the way to the ‘valley of terminal differentiation', you will definitely need the ‘four factor pass' to get back to the top. (Always read the label: reprogramming might not be complete and redifferentiation is subject to status; limitations apply and tumours might arise; the order of lifts on the map is approximate and might change according to conditions.) Compared with Waddington's bleak landscape, today's map provides a lot more information, but is still far from complete; other routes to pluripotency might exist, but skiers explore them at their own risk.
At the Keystone meeting Yamanaka presented a careful evaluation of this problem, comparing around 20 independent iPSC and ESC lines in primary and secondary neurosphere differentiation assays. He showed that the frequency and extent of tumour growth after transplantation correlated with the proportion of differentiation-resistant Nanog-expressing cells—marked with green fluorescent protein (GFP)—detected in secondary neurosphere cultures. About half of all tested iPSC lines scored more than 20% Nanog–GFP in these secondary colonies, although even among these ‘unsafe' clones, it was possible to subclone lines in which Nanog expression was extinguished successfully, in addition to lines in which Nanog expression was not silenced properly. As these phenotypically distinct subclones, originating from the same reprogrammed cell, were extremely difficult to distinguish on most other grounds, Yamanaka cautioned that heterogeneity among iPSCs will probably be an important issue in moving the application forward for direct clinical benefit.
In this context, Hideyuki Okano (Keio U. School of Medicine, Tokyo) demonstrated that transplantation of pre-evaluated ‘safe' mouse iPSC-derived neural precursors into a mouse model of spinal cord injury resulted in graft-derived neurogenesis and myelination, various non-cell-autonomous trophic effects and a long-lasting recovery of locomotive function, but no tumour formation. ‘Unsafe' iPSCs, however, induced a partial recovery but eventual deterioration that coincided with a burgeoning tumour mass. Yamanaka and Okano also provided data showing that iPSC heterogeneity was more pronounced when the target cells used were derived from adult rather than embryonic tissues—an indication that the developmental history of the differentiated cell might be important for the overall success of epigenetic reprogramming.
Rudolf Jaenisch (Whitehead Institute for Biomedical Research, Boston) elaborated on this theme, suggesting that although most differentiated cells might be susceptible to iPSCs, full reprogramming probably required numerous cellular divisions, and that increasing division rate (for example, by inhibiting p53/p21) accelerates reprogramming. By providing Nanog to the iPSC factor cocktail, he suggested, reprogramming occurs successfully with fewer cell divisions (Hanna et al, 2009).
Christoph Bock (Broad Institute, Harvard) showed emerging data from a tripartite collaboration with the Kevin Eggan (Harvard U.) and Alexander Meissner (Harvard U.) labs that aims to profile and compare systematically the epigenomes of some 30 pluripotent cell lines, including 10 from iPSC sources. An ‘epigenetic score card' that is designed to evaluate the performance of lines according to their gene expression, DNA methylation and differentiation capacities is under development. These metrics could help to establish which characteristics of iPSCs are shared with ESCs and which are unique to reprogrammed cells, and also whether these features offer predictive clues about the suitability of certain cells for safe cell replacement therapy.
…the developmental history of the differentiated cell might be important for the overall success of epigenetic reprogramming
Achieving reprogramming through the use of small molecular drugs—rather than by conventional reprogramming factors—is a focus of Eggan's work. In a series of ‘chemical reprogramming' experiments, Eggan and colleagues showed that chromatin modifiers such as VPA and TSA (which both inhibit histone deacetylase activity) could potentiate reprogramming by OCT4, c-MYC and KLF4 in the absence of SOX2. Eggan presented an elegant screening assay to identify drugs that would allow reprogramming by these three factors without VPA. Of the 800 compounds tested, 3 scored in this ‘replacement of SOX2' assay, 2 of which were established TGFβ pathway inhibitors. An analogous approach is being set up to systematically identify candidate ‘replacers' for each of the established reprogramming factors, as well as to reveal crucial signalling networks that underlie sequential steps in pluripotent reprogramming.
Stem cells and new disease models
iPSC approaches provide an opportunity to derive patient-specific stem cells that can be used for cell replacement therapies, as well as forming the basis for new models of human disease. Chad Cowan (Harvard Stem Cell Institute) described his successful attempts to produce and characterize disease-specific iPSC lines from adult pancreatic tissue, as well as strategies to efficiently convert pluripotent cells into renewable sources of human fat cell precursors using the regulated expression of crucial regulators and modified culture conditions.
The meeting was also reminded of the remarkable progress that has been made during a decade of research to optimize the generation of lineage-specific and functionally competent cell types from a variety of pluripotent sources, including epiblast stem cells, embryonic germ cells, ESCs and iPSCs. Austin Smith (The Wellcome Trust Centre for Stem Cell Research, Cambridge, UK) and Azim Surani (The Gurdon Institute, Cambridge, UK) carefully defined the ontogeny, transcriptional networks and signalling cascades that distinguish these cell types, as well as experiments to explain their functional inter-relatedness. Rapid progress in this area, pioneered largely by the Smith and Surani laboratories, has begun to uncover the precise molecular details of a delicate balance required to keep stem cells in an undifferentiated state, and moreover, the epigenetic barriers that arise during specification that not only discourage lineage interconversion (Bao et al, 2009) but also ultimately give differentiation its direction.
…chromatin modifiers such as VPA and TSA […] could potentiate reprogramming by OCT4, c-MYC and KLF4 in the absence of SOX2
A hotly debated topic was the potential of iPSCs as new and vastly improved models for disease and toxicity testing. The sheer number of attendees from biotech companies suggests that industry sees tremendous scope in this arena. Clearly, the substantial and sustained investment that is required seems to be outweighed by the prospect of turning post-mitotic differentiated cells from diseased tissues into a potentially inexhaustible source of iPSCs to model the disease process, and to develop drugs to treat them. Contributions in this area were numerous but largely preliminary. Rita Perlingeiro (U. Minnesota) showed elegant experiments in which the manipulation of PAX3 and PAX7 levels was used to efficiently generate ESC-derived skeletal muscle that improved contractile function after transplantation, and indicated that this approach was being extended using iPSCs. There was general concern that although iPSCs might be useful for modelling diseases caused by rare alleles with relatively strong effects, such as cystic fibrosis, it might not be as useful for examining common alleles with modest effects. Jaenisch questioned whether there was evidence that a disease-relevant phenotype was necessarily expected with all iPSCs. In cases such as Parkinson disease, in which significant changes in dopaminergic function evolve over several years in patients, iPSCs might be unable to offer an appropriate model. Intriguingly, Jaenisch showed evidence that X-chromosome inactivation, a developmentally regulated epigenetic event that is induced as ESCs differentiate, is influenced by oxidative stress. Inhibition of oxidative stress by, for example, culturing blastocyts in low (5%) oxygen delayed Xist expression substantially, while culturing in high (20%) oxygen prompted Xist expression by most cells. Experiments of this kind—using oxidative stress to artificially ‘age' the system—could, Jaenisch argued, offer important advantages for using iPSCs to model certain diseases.
Planned pathways of descent
Micha Drukker from Irv Weissman's laboratory (Standford U.) with collaborator Yoav Soen (Weizmann Institute) presented interesting data from a study aimed at the identification of the earliest lineage precursors that emerge during early differentiation of human ESCs. Drukker and Soen screened more than 500 commercially available antibodies to cell-surface antigens and monitored the responses of the cells to BMP4- or MEF-conditioned media. He then sorted populations that showed a bimodal marker distribution. An analysis of gene expression in these sorted fractions identified clustered features that were reminiscent of visceral endoderm, mesendoderm, allantois or trophectoderm. The results of this Herculean experiment suggest that human ESCs can be readily and rapidly programmed towards four types of stem cell that are thought to be anatomically and functionally distinct in the developing embryo. These experiments promise that the prospective isolation of human embryonic precursors from ESCs, as well as potentially from iPSCs, might not be too far off.
A hotly debated topic was the potential of iPSCs as new and vastly improved models for disease and toxicity testing
A short talk by Lawrence Stanton (Genome Institute, Singapore) on the properties of SOX factors produced a memorable reminder of the complexity of pluripotency and differentiation networks—as well as perhaps the most quoted phrase of the meeting. Stanton showed genome-wide comparisons of SOX2 (important for pluripotency) and SOX17 (important for endoderm differentiation) factors that individually bind equivalently to a canonical (CTTTGTT) as well as a compressed motif in vitro. However, in partnership with OCT4, these factors show a marked preference for the canonical (SOX2) or the compressed (SOX17) motif. Remarkably, the substitution of a single amino-acid residue in the DNA-binding domain of SOX17 protein conferred SOX2-like binding properties and converted SOX17 into a pluripotency-inducing factor. These data elegantly illustrated an emerging theme in the meeting: how highly related families of DNA binding factors, such as the SOX group, hand over the control of gene regulation during development to ensure continuity and stage-specific changes in gene expression. In other words: “To change your fate you may need to change your SOX.” After five days of discussion, of climbing up and down the slopes in Colorado, ski-lifts and passes, it was a well-placed reminder that it was indeed time to go back home and change your socks.
References
- Bao S et al. (2009) Nature 461: 1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J et al. (2009) Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim et al. (2009) Nature 461: 649–65319718018 [Google Scholar]
- Miura K et al. (2009) Nat Biotechnol 27: 743–745 [DOI] [PubMed] [Google Scholar]
- Okita K et al. (2010) Nat Protoc 5: 418–428 [DOI] [PubMed] [Google Scholar]
- Stadtfeld et al. (2008) Science 322: 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Yu J et al. (2009) Science 324: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]