Abstract
Targeting of proteins to appropriate sub-cellular compartments is a crucial process in all living cells. Secretory and membrane proteins usually contain an N-terminal signal peptide, which is recognised by the signal recognition particle (SRP) when nascent polypeptide chains emerge from the ribosome. The SRP-ribosome nascent chain complex is then targeted through its GTP-dependent interaction with SRP-receptor to the protein-conducting channel on endoplasmic reticulum membrane in eukaryotes or plasma membrane in bacteria. A universally conserved component of SRP1, 2, SRP54 or its bacterial homolog, fifty-four homolog (Ffh), binds the signal peptides which have a highly divergent sequence divisible into a positively charged n-region, an h-region commonly containing 8-20 hydrophobic residues and a polar c-region 3-5. No structure has been reported that exemplified SRP54 binding of any signal sequence. We have produced a fusion protein between Sulfolobus solfataricus SRP54 and a signal peptide connected via a flexible linker. This fusion protein oligomerises in solution, through interaction between the SRP54 and signal peptide moieties belonging to different chains, and it is functional, able to bind SRP RNA and SRP-receptor FtsY. Here we present the crystal structure at 3.5 Å resolution of an SRP54-signal peptide complex in the dimer, which reveals how a signal sequence is recognised by SRP54.
Mammalian SRP consists of 7SL RNA and six proteins (SRP54, SRP19, SRP68, SRP72, SRP14 and SRP9)6 whereas the simplest bacterial SRP consists of only 4.5S RNA and Ffh2,7,8. SRP54 (Ffh) comprises three domains: the N-terminal four helix bundle (N domain) and a Ras-like GTPase domain (G domain), which together form the NG domain9, and the C-terminal methionine-rich M domain which associates with SRP RNA and the signal sequence10,11. The SRP-ribosome nascent chain (RNC) complex is docked to the protein-conducting channel via a GTP-dependent interaction between the NG domains of SRP54 and a membrane-associated SRP-receptor, FtsY in bacteria or the SRα-SRβ dimer in eukaryotes1. The crystal structure of the M domain of Thermus aquaticus Ffh revealed a groove lined by flexible hydrophobic residues, which was bound by the ‘finger loop’ of a neighbouring M domain and to which the signal sequence was proposed to bind10. It has been suggested that the flexible hydrophobic residues contribute to the structural plasticity necessary for SRP to bind signal sequences of different lengths and amino acid sequence10. Here we present a crystal structure of SRP54 with a signal sequence bound.
We cloned the SRP54 gene from Sulfolobus solfataricus into pET15b, together with a synthetic gene encoding the signal anchor sequence of the yeast dipeptidyl aminopeptidase B (DPAP-B)12 (Fig. S1). Linker sequences of various lengths were inserted between the SRP54 and the signal sequence (Table S1). The SRP54-signal sequence fusion protein, designated SRP54* hereafter, was highly expressed in E. coli. Analytical gel filtration showed that, with a linker of 11-39 residues SRP54* formed a mixture of dimers and higher order oligomers (Fig. S2). With a linker of six residues it formed only high molecular weight oligomers or aggregates. No monomers were ever observed in solution even with the longest linker, which suggests that as the signal sequence moiety at the C-terminus of SRP54* emerges from the ribosome it is immediately captured on the ribosome by the SRP54 moiety of a previously released molecule of SRP54*. In support of this view, SRP54 (or Ffh) was found to bind the ribosome through its N domain both in the presence and absence of SRP RNA and can be cross-linked to the ribosomal protein L23 near the nascent chain exit channel13,14. This observation is consistent with the cryoEM structure of the ribosome-SRP complex15,16. Thus it is highly plausible for the emerging DPAP-B signal sequence to be captured in its physiological orientation on the M domain by another SRP54* molecule bound to the ribosome through its N domain. When Leu-455 in the hydrophobic core of the signal peptide was mutated to Arg, this mutant was eluted in the void volume, suggesting that the signal sequence no longer binds to the M domain and causes aggregation. SRP54* forms a complex with helix 8 of SRP RNA (Fig. S3), and binds the NG domain of FtsY only in the presence of Mg2+ and GMPPNP, showing that the important functions of SRP54 are unaffected by the linker1 (Fig. S4).
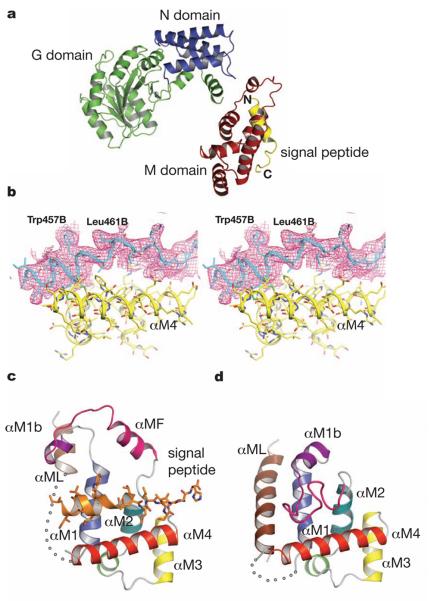
SRP54* with an 11mer linker crystallised in space group P41212 with one monomer per asymmetric unit. The structure was solved by the MAD method using a methylmercury derivative of a single cysteine mutant (N177C) and refined to an Rfree of 32.3% at 3.5Å resolution (Table S2 and Fig. 1a and b). From an unbiased electron density map calculated by combining the Hg MAD phases with molecular replacement phases from a homologous NG domain (PDB:1J8M), followed by solvent flattening (Fig. S5), the orientation and register of the signal peptide relative to the M domain have been established unambiguously. In SRP54* the signal peptide binds to a groove formed between helix αM4 and helices αM1 and αM2 oriented perpendicular to αM4 (Fig. 1c). From these three helices 11 conserved residues (Val332, Ile336, Leu339, Ile374, Trp377, Leu417, Trp420, Met424, Leu427, Leu428, Val431) form a continuous hydrophobic surface over the groove (Fig. 2 and S1b). The indole rings of Trp377 and Trp420 cluster together creating a bulge in the groove surface (Fig. 2b). Eight residues (Ile450-Trp457) in the h-region of the signal peptide (Fig. S1) are in α-helical conformation, but the helix extends to Leu461 with a bend at Gly458 that complements the Trp bulge in the groove floor. The signal peptide helix interacts extensively with hydrophobic residues on the binding groove as summarized in Fig. 2d. The polar c-region forms a more irregular structure near the N-terminus of αM4 containing three glutamates, while the n-region is poorly ordered. By matching the h-region with the hydrophobic part of αM4 and the c-region with the hydrophilic, the orientation of the signal peptide relative to the M domain shown by SRP54* is most likely biological, which will dictate the orientation of the M domain relative to the exit channel in the ribosome.
Figure 1. Structural of SRP54 in complex with a signal sequence.
a, Overall structure of Sulfolobus solfataricus SRP54-signal peptide fusion protein. b, The sigmaA-weighted and B-factor (−50A2) sharpened 2Fo - Fc map is shown over the signal peptide. The map is contoured at about 1 sigma. The atomic model is shown as bonds overlaid by ribbons along the backbone. c, The M domain of SRP54* with a bound signal peptide (residues Gly449B-His468B). Helices are colour-coded as in Fig. S1 and the poorly ordered region (residues 308-325) is represented by a dotted curve. d, The M domain of free SRP54 (PDB: 1QZX)19. The disordered region (residues 320-326) is shown as a dotted curve.
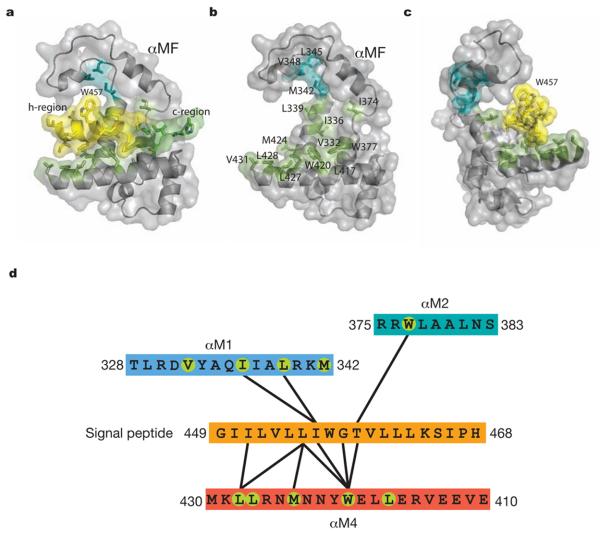
Figure 2. Interaction of the M domain with a signal peptide.
a, Surface representation with the h- and c-regions of the signal sequence coloured yellow and blue, respectively. b, Same view as in a but the signal peptide is omitted to reveal the signal peptide binding pocket. Met342, Leu345 and Val348 (cyan) may be involved in coupling the M domain to the NG domain via the linker. c, The bulky Trp457 side chain of the signal peptide limits its rotational freedom in the binding pocket. d, Contact diagram between the signal peptide and the binding groove with a 5.5Å cut-off. Green circles indicate surface residues of the binding groove as highlighted in Fig. S1b.
The M domain binds signal peptides with considerable sequence variations in the h-region, including a synthetic peptide with poly-Leu for its h-region17. Clues to the structural basis are embedded in the SRP54* structure. Firstly, the signal sequence helix and αM4 interact by the most commonly occurring 4-4 “ridges-into-grooves” helix packing, with a crossing angle of −40° in the Chothia notation18. Because this crossing angle results in the extensive interaction between the h-region of the signal peptide and αM4 of the M domain, we expect the 4-4 helix packing with αM4 to be conserved in signal peptide recognition. The closest approach is 7.1Å (calculated for Cα atoms) between Leu455 and Met424, which belong respectively to the Ile451-Leu455-Thr459 ridge in the signal peptide helix and Trp420-Met424-Leu428 ridge in αM4. The ridge on αM4 contains two conserved residues that in homologous cases are frequently Leu and Met (Fig. S1b). Secondly, in SRP54 the signal peptide is bent at Gly458 to accommodate two adjacent Trp residues (Trp377, Trp420) in the groove. In the absence of signal peptide (PDB:1QZX) Trp420 adopts a different rotamer19 so these indole rings are not clustered together, suggesting that the signal peptide binding can involve an induced fit mechanism to maximise the hydrophobic interactions with particular signal sequences. However, unlike the signal peptide the backbone structure in αM2-αM4 is consistently rigid among homologous structures (PDB: 2FFH; 1QZX)10,19. Thirdly, additional hydrophobic residues lining the groove are available for interaction with different or longer signal peptides. The minimal length of the h-region is eight residues3-5 and this roughly corresponds to nine residues of the DPAP-B signal peptide forming extensive contacts. Lastly, the bulky side chain of Trp457 in the DPAP-B signal peptide faces away from the groove and restricts the rotational freedom of the signal peptide in its binding groove (Fig. 2c). The majority of SRP-dependent signal peptides verified experimentally20 (Fig. S6) contain one or two consecutive aromatic amino acids (Phe-Tyr in SfmC and Phe-Phe in FocC), which may similarly restrict the orientation of the signal peptide. The exclusion of the n- and c-regions from the hydrophobic groove likewise restricts the longitudinal position of the signal peptide with respect to the groove.
In the absence of signal sequence (PDB: 1QZX and 1QZW)19 the finger loop drapes across the binding groove and inserts some of its six hydrophobic residues (Leu355, Ile357, Met358, Leu359, Leu367, and Ile369) in the signal sequence binding site (Fig. 1d). In SRP54* the finger loop region forms a lid above the bound signal peptide in a conformation roughly resembling the finger loop of Thermus aquaticus Ffh (PDB: 2FFH)10: residues 362-371 form a helix (αMF), linked to αM1b via a loop in an extended conformation (Fig. 1c). Our structure and the cryoEM model of the signal peptide-bound M domain (EMD1264, PDB:2J37) are in good agreement except in the finger loop/helix15. In the EM model this sequence was interpreted as a loop but the density appears more consistent with our observation that it partly forms a helix (αMF) (Fig. S7). However, the cryoEM map suggests αMF closes on the signal sequence to provide additional stabilizing interactions in the SRP-RNC complex; this movement is restricted in SRP54* by lattice contacts of the αM1b-αMF loop (Fig. S8).
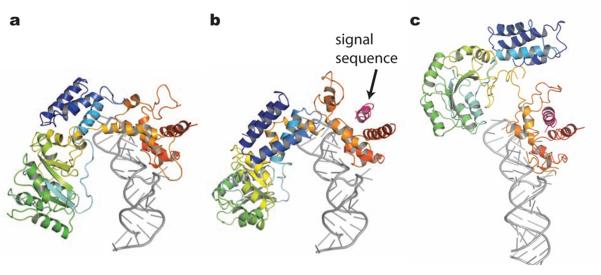
The binding of SRP to the RNC complex causes a large movement of the NG domain with respect to the M domain (Fig. 3), such that the SRP54 M domain interacts with ribosomal RNA helices 24, 59 and 50 and protein L24, and the SRP54 NG domain interacts through the N domain with the ribosomal protein L23 and L2915,16. The structure of SRP54* shows that the binding of signal sequence to the M domain alone is insufficient to induce this large domain re-arrangement. The linker helix (αML) between the NG and M domains is disordered in SRP54* (Fig. 1c), but in the signal peptide-free SRP54 (PDB: 1QZX, 1QZW)19 αML and αM1 partially block the signal peptide binding site (Fig. 1c and 1d). It has been suggested that the orientation of the linker helix relative to the NG and M domains markedly changes upon ribosome binding. Consequently, the linker helix could couple the signal peptide binding and ribosome binding. It has been proposed that the SRP RNA has a role in signal recognition via electrostatic interactions between the backbone of the RNA and the positively charged residues in the n-region21,22. When the M domain of SRP54* is overlaid onto the M domain of bacterial or mammalian SRP-RNA complex19,21,23, the n-region of the signal peptide is too far from SRP-RNA to make direct contact (Fig. S9). The cryoEM models of the E. coli and mammalian SRP-RNC complexes (PDB: 2J28 and 2J37) consistently indicate no interaction between the signal sequence and SRP RNA15, thus making the involvement of SRP-RNA in signal sequence recognition unlikely. The positively charged residues in the n-region may be conserved for other reasons, for example, for its interaction with the phospholipid head group during protein translocation24.
Figure 3. Structural change of SRP54 upon signal sequence binding.
a, Sulfolobus solfataricus SRP54 bound to RNA (PDB: 1QZW)19. b, model of the SRP54*-RNA complex. The M domain of SRP54* was overlaid onto that of the SRP54-RNA complex (PDB: 1QZW)19. c, E. coli signal recognition particle bound to the ribosome (PDB: 2J28)15.
In conclusion, SRP54* has provided a first glimpse of signal peptide binding to SRP. However additional structures with different signal sequences in the future will be required to fill out the repertoire of accommodations, as well as assessing in a statistically meaningful way the relative importance of each contact position for the recognition.
Summary Method
A fusion protein (SRP54*) consisting of Sulfolobus solfataricus SRP54 and yeast dipeptidyl aminopeptidase B signal sequence was produced in E. coli. Crystal structure of SRP54* was determined by the MAD method using a methylmercury derivative of a mutant (N177C).
Methods
Plasmid for protein expression
The sequence encoding Sulfolobus solfataricus SRP54 residues 2–432 was amplified by PCR from genomic DNA and cloned into the NcoI-XhoI site of pET15b. Four overlapping oligonucleotides encoding the signal anchor sequence from S. cerevisiae dipeptidyl-aminopeptidase B (KLIRVGIILVLLIWGTVLLLKSIPHH) and a penta-histidine tag, were cloned into the BamHI site in such a way that a BamHI site is retained only on the 5′ end. This signal sequence was used for a cryo-EM study of the SRP-ribosome-nascent chain complex12. A pair of oligonucleotides encoding various linker sequences was cloned between the XhoI and BamHI sites (Table S1). The 11 residue linker sequence was (ARSGSGSGSGS). A single cysteine mutant N177C was generated by a PCR based mutagenesis.
Protein expression and purification
Rosetta (DE3) pLysS cells (Novagen) harbouring pET15-SRP54* or pET15-SRP54*(N177C) were grown in 2xTY media with 50μg/ml Ampicillin and 25μg/ml Chloramphenicol, and protein expression was induced at an OD600 of 0.7 with 1 mM IPTG for 3 hr at 25°C. Harvested cells were suspended in 20 mM Hepes pH 7.4, 1 M NaCl, 10 % glycerol and EDTA-free protease inhibitor cocktail (Roche) and lysed by sonication. The clarified lysate was applied to a Ni-NTA agarose (QIAGEN) column equilibrated in 20 mM Hepes pH 7.4, 500 mM NaCl, 20 mM imidazole and eluted with a linear gradient to 20 mM Hepes pH 7.4, 500 mM NaCl, 320 mM imidazole. The peak fractions were applied to a hydroxyapatite (Bio-Rad) column equilibrated in 20 mM Tris-HCl pH 7.4, 200 mM NaCl and eluted with a linear gradient of 0-12 % ammonium sulfate in the same buffer. The pooled fractions were dialysed against 20 mM Tris-HCl pH 7.4, 200 mM NaCl and applied to a heparin-Sepharose column (Amersham Biosciences) equilibrated with the same buffer. The protein was eluted with a linear gradient of 200 mM-1 M NaCl in 20 mM Tris-HCl pH 7.4. Peak fractions were pooled and dialysed against 20 mM Tris HCl pH 7.4, 100 mM NaCl. At this salt concentration, dimers remained soluble whereas all higher oligomers precipitated.
Crystallization
SRP54* dimer crystals were grown by hanging drop vapour diffusion at 295 K, by mixing equal volumes of protein (18 mg.ml−1) and reservoir solution containing 5-7 % PEG 4000, 100 mM Bis-Tris pH 5.5, 100 mM NaCl, 2 % Polypropylene glycol P400, 5-50 mM Mg(OAc)2. Single crystals were obtained by streak seeding from existing SRP54* crystals. Purified SRP54*(N177C) was pre-reacted with 0.5 mM methylmercury nitrate and crystallised under the same condition. The native and derivative crystals grew in the space group P41212 and appeared within 5-7 days. The crystals were equilibrated with 25 % PEG 4000, 15 % Glycerol, 100 mM Bis-Tris pH 5.5, 100 mM NaCl and flash frozen in liquid nitrogen.
Structure determination
The native dataset and three-wavelength MAD datasets (peak, inflection and remote) of the methylmercury derivative were collected at 100 K on beamline ID14-1 and ID14-4 at the European Synchrotron Radiation Facility in Grenoble, France. The data were processed with MOSFLM/SCALA/TRUNCATE25,26. The single methylmercury site was determined in AutoSHARP27 and refined in SHARP27 with phase calculation for the resolution range of 46-4.19 Å. The overall FOM was 0.354 (0.416 acentric, 0.152 centric), increased to 0.399 for centric after solvent flipping in SOLOMON28. MAD phases before Solomon were combined with native amplitudes to 3.5Å resolution to calculate the initial map. This showed unambiguous density for the NG domain, into which residues 2-307 of the wild type structure (PDB: 1QZX)19 were placed readily with only minor adjustments of the coordinates. Additional α-helical density for the M domain was observed and αM1-αM1b (residues 327–352) and αM2–αM4 (residues 374–431) were built by several rounds of manual model rebuilding in COOT29 and refinement (see below) in CNS30. The signal sequence (residues 449-468) was built into an additional rod-like density, running roughly anti-parallel to αM4, while the linker connecting the M domain and the signal sequence (residues 432-448) was disordered. Refinement led to a 2Fo - Fc map showing convincing densities for residues 353-373, formerly termed the “finger loop”10, but now showing residues 362-371 as an α-helix which we renamed the finger helix (αMF). This interpretation was validated in two ways. Firstly, an unbiased map calculated by combining the Hg MAD phases with molecular replacement phases (Fig. S5a), where the search model was a homologous NG domain (PDB: 1J8M), showed clearly all elements in our model including the signal peptide and finger helix. Secondly, a simulated annealing omit map, omitting both the signal peptide and the finger loop sequence, confirmed their polypeptide path (Fig. S5b). The signal peptide and finger loop were rebuilt into the connected densities in the omit map before refinement of the whole structure. The structure was refined against the MAD phases in CNS30 with simulated annealing and restrained individual B-factors to a crystallographic R factor of 30.2% (Rfree=32.3%) for resolution 58-3.5 Å, with good stereochemistry (Table S2) and good Ramachandran statistics (83.3% in preferred regions; 15.5% in allowed regions and 1.2% in disallowed regions). Due to the modest resolution, harmonic restraints (10-20 kcal /mol Å2) were imposed on all atom positions throughout refinement, together with main chain hydrogen bond restraints, which resulted in better geometry and R-factors. All figures are drawn with Pymol31.
Mass Spectrometry
Mass spectrometry data were acquired using a modified QTOF2 instrument (Waters) configured for nanoflow ESI in positive ion mode essentially as described previously32. Spectra under native conditions were collected in 500 mM ammonium acetate.
Supplementary Material
Acknowledgements
We thank members of the Nagai group for assisting with data collection and Oliver Daumke for help with the GTP binding and GTPase assays. We thank Jennifer Kavran for helpful discussion. This paper is dedicated to the memory of César Milstein who discovered the signal peptide. This project was supported by the Medical Research Council of the UK. Claudia Janda was supported by a HFSP grant.
Footnotes
Atomic coordinates and structure factors for SRP54* have been deposited in the PDB data bank under accession number 3KL4.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 3.Emanuelsson O, Brunack S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protoc. 2007;4:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 4.Gierasch LM. Signal sequences. Biochemistry. 1989;28:923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 7.Luirink J, et al. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature. 1992;359:741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- 8.Poritz MA, et al. An E.coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 9.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 10.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 11.Zopf D, Bernstein HD, Johnson AE, Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 13.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstain B. Distinct modes of signal recognition interaction with the ribosome. Science. 2002;297:1345–1238. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 15.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 16.Schaffitzel C, et al. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 17.Kendall DA, Bock SC, Kaiser ET. Idealization of the hydrophobic segment of the alkaline phosphatase signal peptide. Nature. 1986;321:706–708. doi: 10.1038/321706a0. [DOI] [PubMed] [Google Scholar]
- 18.Chothia C, Levitt M, Richardson D. Structure of proteins: Packing of α-helices and plated sheets. Proc. Natl. Acad. Sci. USA. 1997;74:4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 2005;187:2983–2991. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 22.Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nature Struct. Mol. Biol. 2004;11:1049–1053. doi: 10.1038/nsmb853. [DOI] [PubMed] [Google Scholar]
- 23.Kuglstatter A, Oubridge C, Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nature Struc. Biol. 2000;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 24.van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapport TA. X-ray structure of a protein-conducting Channel. Nature. 2003;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 25.Leslie AGW. The integration of macromolecular diffraction data. Acta Crystallogr. 2006;D 62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 26.Evans PR. Scaling and assessment of data quality. Acta Crystallogr. 2006;D 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 27.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 28.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. 1996;D 52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The PyMOL Molecular Graphics System. 2002. http://www.pymol.org.
- 32.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.