Abstract
There is growing evidence that strains of Mycobacterium tuberculosis differ in pathogenicity and transmissibility, but little is understood about the contributory factors. We have previously shown that increased expression of the principal sigma factor, SigA, mediates the capacity of M. tuberculosis strain 210 to grow more rapidly in human monocytes, compared with other strains. Strain 210 is part of the widespread W-Beijing family of M. tuberculosis strains and includes clinical isolate TB294. To identify genes that respond to changes in SigA levels and that might enhance intracellular growth, we examined RNA and protein expression patterns in TB294-pSigA, a recombinant strain of TB294 that overexpresses sigA from a multicopy plasmid. Lysates from broth-grown cultures of TB294-pSigA contained high levels of Eis, a protein known to modulate host–pathogen interactions. DNA microarray analysis indicated that the eis gene, Rv2416c, was expressed at levels in TB294-pSigA 40-fold higher than in the vector control strain TB294-pCV, during growth in the human monocyte cell line MonoMac6. Other genes with elevated expression in TB294-pSigA showed much smaller changes from TB294-pCV, and the majority of genes with expression differences between the two strains had reduced expression in TB294-pSigA, including an unexpected number of genes associated with the DNA-damage response. Real-time PCR analyses confirmed that eis was expressed at very high levels in TB294-pSigA in monocytes as well as in broth culture, and further revealed that, like sigA, eis was also more highly expressed in wild-type TB294 than in the laboratory strain H37Rv, during growth in monocytes. These findings suggested an association between increased SigA levels and eis activation, and results of chromatin immunoprecipitation confirmed that SigA binds the eis promoter in live TB294 cells. Deletion of eis reduced growth of TB294 in monocytes, and complementation of eis reversed this effect. We conclude that SigA regulates eis, that there is a direct correlation between upregulation of SigA and high expression levels of eis, and that eis contributes to the enhanced capacity of a clinical isolate of M. tuberculosis strain 210 to grow in monocytes.
INTRODUCTION
Molecular epidemiological studies have shown that a small percentage of Mycobacterium tuberculosis strains cause a large proportion of cases (Barnes et al., 1997; Bishai et al., 1998; Small et al., 1994), implying that some strains spread more effectively than others. M. tuberculosis strain 210 is widely distributed in the south-west and south-central USA (Barnes et al., 1997; Weis et al., 2002; Yang et al., 1998), and isolates of this strain grow more rapidly in human macrophages than other strains (Theus et al., 2005; Zhang et al., 1999), suggesting an enhanced capacity to avoid host defences. The ability of M. tuberculosis to grow in human mononuclear phagocytes is a central feature of its pathogenic potential.
Strain 210 is a member of the widespread W-Beijing family of strains, and growing evidence indicates that the altered expression patterns of some genes contribute to the success of these strains (Reed et al., 2007; Sinsimer et al., 2008; Tsenova et al., 2005; Wu et al., 2004). We have previously shown that increased expression of the principal sigma factor SigA mediates the capacity of isolates of M. tuberculosis strain 210 to grow more rapidly in human monocytes and in mice (Wu et al., 2004). SigA is believed to be the principal sigma factor, based on sequence homology to σ70 of Escherichia coli and evidence that the gene is essential in mycobacteria (Gomez et al., 1998; Hu & Coates, 1999). Although SigA is predicted to control housekeeping functions, our previous study (Wu et al., 2004) and work by others (Collins et al., 1995; Steyn et al., 2002) suggest that SigA also regulates genes involved in host–pathogen interactions. We wanted to identify genes regulated by SigA that enhance the ability of M. tuberculosis strain 210 to grow in host cells.
An earlier report suggested that the eis gene (Rv2416c) of M. tuberculosis is a member of the SigA regulon, given the similarity of the putative −10 and −35 regions of the eis promoter to SigA-like promoters (Roberts et al., 2004). The eis gene was originally identified by the Friedman laboratory based on the ability of the M. tuberculosis gene to enhance survival of the non-pathogenic species Mycobacterium smegmatis in macrophages, hence the name ‘eis’ for ‘enhanced intracellular survival’ (Wei et al., 2000). Subsequent studies from the same laboratory indicate that the Eis protein is present in cytosolic and cell envelope fractions of M. tuberculosis, and that it is released into culture supernatants and into the cytoplasm of infected macrophages (Dahl et al., 2001; Samuel et al., 2007). In one study, sera from 40 % of pulmonary tuberculosis patients were positive for anti-Eis antibody, indicating that the protein is antigenic (Dahl et al., 2001).
Bioinformatic analyses suggest that Eis is an acetyltransferase (Samuel et al., 2007), and additional evidence indicates that it modulates the host immune response (Lella & Sharma, 2007; Samuel et al., 2007), but the precise function of Eis is unclear. An eis deletion mutant induces higher levels of TNF-α and lower levels of IL-10 in monocytes than those induced by control strains (Samuel et al., 2007), suggesting that Eis induces secretion of IL-10, which is known to inhibit TNF-α production. However, the eis mutant shows no growth defect in a macrophage cell line or in mice (Samuel et al., 2007). Eis reduces TNF-α and IL-4 production by mitogen-stimulated human T-cells (Lella & Sharma, 2007), indicating that Eis affects T-cell function.
In this study, we used TB294, which is a clinical isolate of M. tuberculosis strain 210, and a recombinant strain of TB294 that overexpresses sigA, to identify SigA-regulated genes that might affect host–pathogen interactions. Our findings confirm that eis is a member of the SigA regulon and support a role for Eis in intracellular growth of M. tuberculosis.
METHODS
Bacterial strains and culture conditions.
TB294 (Table 1) is a member of M. tuberculosis strain 210 and was isolated from a patient with tuberculosis (Zhang et al., 1999). Transformants of TB294 bearing a sense sigA (TB294-pSigA) and vector control plasmid (TB294-pCV) were generated, as previously described (Wu et al., 2004). H37Rv was obtained from the Sanger Center. Mycobacteria were grown with shaking at 37 °C in 5 % CO2 in Middlebrook 7H9 broth (Difco) containing 10 % oleic acid–albumin–glucose–catalase (OADC; Difco) and either 0.2 % (v/v) glycerol or 0.05 % Tween 80. Transformant strains were cultured with 20 μg kanamycin sulphate ml−1. The E. coli NovaBlue strain (Novagen) was used for cloning, and was grown in Luria–Bertani broth or on Luria–Bertani agar (Difco).
Table 1.
M. tuberculosis strains and oligonucleotides used in this study
*TB294-pSigA and TB294-pCV are equivalent, respectively, to the ‘TB294 sense transformant’ and ‘TB294 vector control’ described in our previous publication (Wu et al., 2004).
†Restriction sites used in cloning are underlined.
Infection of human mononuclear phagocytes.
The human monocytic cell line MonoMac6 (Ziegler-Heitbrock et al., 1988) was cultured in RPMI medium containing 10 % heat-inactivated fetal bovine serum, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 9 μg bovine insulin ml−1. MonoMac6 cells were infected with single-cell suspensions of M. tuberculosis in RPMI containing 10 % heat-inactivated human serum at an m.o.i. of 1 : 4, as previously described (Wu et al., 2004; Zhang et al., 1999). At different times after infection, the supernatant was aspirated, monocytes were lysed with distilled water for 10 min and incubated with 0.09 % SDS for 10 min, and then 20 % BSA was added. Bacterial suspensions in cell lysates were ultrasonically dispersed, serially diluted, and plated in triplicate at 37 °C on 7H10 agar (Difco) supplemented with 10 % OADC and 0.5 % (v/v) glycerol. c.f.u. were counted after 2–3 weeks.
RNA isolation.
For mycobacteria cultured in broth, RNA was extracted as described previously (Wu et al., 2004). To isolate RNA from intracellular mycobacteria, bacteria were harvested from MonoMac6 cells, as described by Wu et al. (2004), and then RNA was extracted using the method described for broth-grown cultures, except that following extraction, the bacterial RNA was further purified with the MICROBEnrich kit (Ambion).
Relative quantification of mRNA by real-time PCR.
A mixture of 37 genome-directed primers designed to prime all ORFs in the M. tuberculosis genome was used for reverse transcription (Talaat et al., 2000). Reaction mixtures (20 μl) included 500 ng RNA denatured for 5 min at 65 °C, 0.5 mM of each dNTP, 1.0 μg of a mixture of 37 genome-directed primers, 10 U RNase inhibitor (Promega), 2 μl 10× buffer RT (Qiagen), and 4 U Omniscript reverse transcriptase (Qiagen). Reactions were performed at 37 °C for 60 min, followed by heating to 93 °C for 5 min to inactivate the reverse transcriptase.
Primers and probes for real-time PCR were designed with primer express software (Applied Biosystems), and probes were labelled with 5′-fluorescein phosphoramidite and 3′-tetramethyl-6-carboxyrhodamine (TAMRA). The sequences of the primers and probes are shown in Table 1. PCR conditions were identical for all reactions, which were performed in triplicate. Assays were performed using the ABI Prism 7700 (Applied Biosystems) and were normalized for 16S rRNA, as described previously (Wu et al., 2004).
DNA microarray analysis.
The M. tuberculosis DNA microarray consisted of 4295 70-mer oligonucleotides representing the 3924 predicted ORFs of the H37Rv strain (http://www.sanger.ac.uk) and 371 probes designed to detect sequences in the CDC1551 strain (http://www.tigr.org). The arrays were prepared by spotting oligonucleotides (Tuberculosis Genome Set version 1.0, Operon Biotechnologies) onto poly-l-lysine-coated glass microscope slides, as described previously (Pang et al., 2007). Total RNA from two independent experiments was prepared from M. tuberculosis harvested from infected MonoMac6 cells, as described above. cDNA synthesis, labelling, hybridization, scanning, image acquisition, normalization, filtering and fold-change calculations were performed as described previously (Pang et al., 2007), and for each experiment, a dye flip was performed. A one-class t test was run using the MEV software (Saeed et al., 2003), with P values based on t distribution and an overall alpha set to 0.05. Expression changes were considered significant if there was at least a twofold or higher change in expression between TB294-pSigA and TB294-pCV in at least three of the four array experiments, and the P value was ≤0.05.
Construction of eis deletion mutant and complemented strain.
The eis gene was deleted using the pNIL/pGOAL vector system, which yields unmarked deletions (Parish & Stoker, 2000). Genomic DNA was prepared from TB294 for use as the PCR template using the Wizard Genomic DNA Purification kit (Promega). Synthetic oligonucleotide primer sets (EisL and EisR, Table 1), complementary to regions that flank the eis gene, were designed to amplify fragments of ∼1.1 kb on either side of eis. The PCR products were sequenced and cloned into the BamHI and HindIII sites of p2NIL, yielding a construct with the two flanking fragments of eis. The lacZ–sacB cassette from pGOAL17 (Parish & Stoker, 2000) was inserted into the PacI site of the vector containing the eis flanking fragments to produce a suicide delivery vector for transformation. TB294 was electroporated with the final construct, and double-crossover recombinants were obtained by a two-step selection method, as described by Parish & Stoker (2000). Deletion of eis was confirmed by PCR.
To complement the eis deletion, a 1.3 kb DNA fragment containing the eis gene and 143 bp of upstream sequence was amplified by PCR using the EisC primer set (Table 1). The PCR product was cloned into the HpaI–KpnI site of the integrating kanamycin-resistant plasmid pMV361 (Stover et al., 1991). The resulting plasmid was electroporated into the eis deletion mutant, followed by plating and selection for kanamycin-resistant colonies.
Protein analysis.
To isolate protein from broth cultures of M. tuberculosis, protein was extracted using the B-PER kit (Pierce) according to the manufacturer's instructions, except for the additional step of disrupting the mycobacteria with a FastPrep FP120 shaker and lysing matrix B. The amount of protein in each sample was quantified by the bicinchoninic acid method (Pierce). Using 30 μg protein extract in each sample, 10 % SDS-PAGE was performed under reducing conditions (Laemmli, 1970). Coomassie brilliant blue R-250 staining and destaining solutions (Bio-Rad) were used to detect protein on the gel. A band of approximately 42 kDa was excised and sent to Amprox for analysis by MS.
Chromatin immunoprecipitation (ChIP).
To study DNA–protein interactions in live mycobacteria, we used procedures based on our recently published method (Rodrigue et al., 2007). Briefly, a 50 ml culture of M. tuberculosis was grown to OD600 0.5 in 7H9 medium, incubated with formaldehyde to cross-link DNA–protein complexes, and then the formaldehyde was neutralized with glycine. Sonication was used to disrupt the cells and to shear the genomic DNA into 250–800 bp fragments. From the sonicate, 100 μl was saved as the input DNA control. The remaining sonicate was used for immunoprecipitation with paramagnetic M-450 beads (Dynal Biotech) that had been pretreated by coating with sheep anti-mouse IgG, and then incubating with antibodies to SigA or GroEL2 (control antibody). Anti-GroEL2 antibody was kindly provided by Dr John Belisle, Colorado State University. For the anti-SigA antibody, we used mAb 2G10 (Neoclone Biotechnology), which is a monoclonal antibody to the σ70 subunit of E. coli RNA polymerase (Strickland et al., 1988), and which recognizes SigA of M. smegmatis (Predich et al., 1995) and M. tuberculosis (Wu et al., 2004). Following incubation for at least 3 h at 4 °C, the immunoprecipitated samples were washed and the bound material was eluted, as described previously (Rodrigue et al., 2007). The supernatant was then collected and treated at 37 °C for 2 h with proteinase K to remove complexed protein. The DNA was purified by phenol/chloroform extraction, precipitated with 2-propanol, and washed with 70 % ethanol. Immunoprecipitated DNA was resuspended in 50 μl Tris-EDTA buffer. The sample was analysed by PCR, using the EisP set of primers (Table 1), which amplifies a 143 bp region of the eis promoter.
RESULTS
We have previously shown that the capacity of clinical M. tuberculosis strains to grow in human macrophages correlates with their intracellular expression levels of sigA (Wu et al., 2004). Of particular interest, we found that strains of the widespread W-Beijing family, including strain 210, express higher levels of sigA and have enhanced growth in human monocytes, compared with other strains. Moreover, transformants of M. tuberculosis strain 210 that overexpress sigA from a multicopy plasmid show a further increase in intracellular growth, compared with a vector control strain (Wu et al., 2004). Based on these findings, we hypothesized that upregulation of SigA in these strains increases expression of SigA-dependent virulence genes that influence intracellular growth. To identify such genes, we took a two-pronged approach by examining expression patterns in the SigA-overexpressing strain (TB294-pSigA) at both the protein and RNA levels. As described below, both methods of analysis revealed that the eis gene (Rv2416c) was highly expressed in TB294-pSigA.
Upregulation of sigA increases expression of Eis
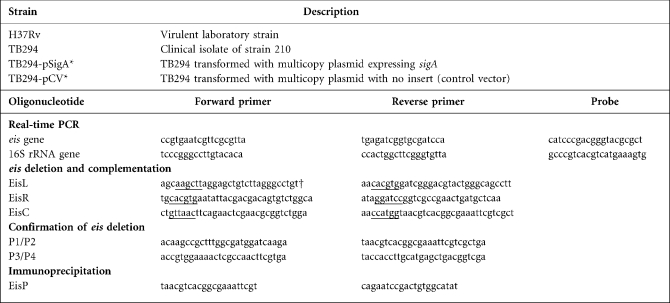
To identify proteins that were upregulated in the presence of high levels of SigA, we prepared lysates from broth-grown cultures of strain TB294-pSigA and the parental strain that had been transformed with the control vector (strain TB294-pCV). Using SDS-PAGE, a prominent band of approximately 42 kDa was identified in TB294-pSigA, but not in TB294-pCV (Fig. 1a). This band was excised from the gel, digested with trypsin and subjected to MS. The pattern of peptides obtained indicated that the band was Eis (Fig. 1b).
Fig. 1.
Production of Eis protein by TB294-pSigA. (a) Analysis of bacterial cell lysates. Recombinant strains TB294-pCV and TB294-pSigA were cultured in broth to OD600 0.6–0.8, and cell lysate proteins were extracted and separated by SDS-PAGE. The arrow indicates a protein of approximately 42 kDa from TB294-pSigA that was excised and analysed by MS. Lane M, molecular size markers, shown in kDa. (b) Peptides identified by MS. The amino acid sequence of the eis gene was obtained from Tuberculist (http://genolist.pasteur.fr/TubercuList/). Peptides identified by MS analysis of the 42 kDa protein band are underlined.
Expression analysis of recombinant strains growing in MonoMac6 cells
We had previously observed that strains transformed with pSigA, including TB294-pSigA, have increased growth in the human monocytic cell line MonoMac6, compared with vector control strains (Wu et al., 2004). To identify SigA-regulated genes that were more highly expressed in TB294-pSigA than in TB294-pCV during intracellular growth, we infected MonoMac6 cells and used DNA microarray analysis to compare gene expression at 6 days post-infection (Supplementary Table S1). In two independent experiments, eis was the most highly expressed gene in TB294-pSigA compared with TB294-pCV, with a mean fold difference of 40.38±19.18 (se). Rv2415c, which lies downstream of eis, showed the next highest difference in expression, with a mean fold difference of 8.7±2.25 (se). The other 19 genes with significantly higher expression in TB294-pSigA had mean fold changes of ≤4.3 compared with expression levels in the control strain. The majority of these genes are predicted to have biosynthetic and/or metabolic functions, including moaC3, moaX/gphA, ppsA, ppsC, guaA, accA3, nuoK and metC (Supplementary Tables S1 and S2). Rv3193c, Rv3194c, and lpqB, which encode potential secreted or membrane/surface proteins, were also expressed at higher levels in TB294-pSigA.
Forty-seven of the 77 genes showing significant differences in expression between the two strains, including eis, were in gene clusters (Supplementary Table S2). However, the eis/Rv2415c cluster was one of only five clusters that were expressed at higher levels in TB294-pSigA (Supplementary Table S2). Most of the differentially expressed genes had lower expression in TB294-pSigA (Supplementary Tables S1 and S2), and interestingly, as described in the Discussion, the majority of these genes are regulated by RecA-NDp promoters (Supplementary Table S2), a type of promoter associated with RecA-independent activation in response to DNA damage (Gamulin et al., 2004).
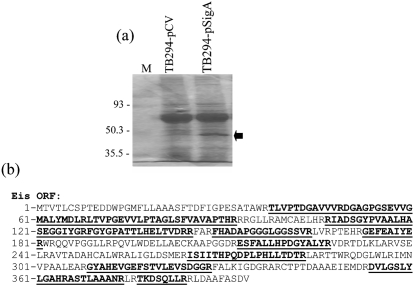
To confirm that eis was expressed at higher levels in TB294-pSigA, we performed three additional experiments, in which TB294-pSigA and TB294-pCV were grown in MonoMac6 cells for 6 days or in 7H9 medium, and then eis mRNA was quantified by real-time PCR. In 7H9 medium, eis expression was over 200-fold greater in TB294-pSigA than in TB294-pCV (Fig. 2a), consistent with the high levels of Eis protein detected in lysates of broth-grown TB294-pSigA (Fig. 1a). In MonoMac6 cells, the eis expression level was more than 100-fold higher in TB294-pSigA than in TB294-pCV (Fig. 2a), which supported the results of DNA microarray analyses. Overall, these results suggested that increasing sigA expression results in enhanced eis expression, during growth in monocytes and in broth.
Fig. 2.
Expression patterns of eis in recombinant and wild-type TB294. M. tuberculosis strains were cultured in MonoMac6 cells for 6 days, or in broth to OD600 0.6–0.8. RNA was extracted and eis expression was examined by real-time PCR. The ratio of eis mRNA to 16S rRNA was calculated for each sample and is shown as eis mRNA levels. Because amounts of eis mRNA were low in comparison to 16S rRNA, ratios were multiplied by 1000 to give values ≥1. Mean values and standard errors for three experiments are shown. (a) Comparison of eis expression in TB294-pSigA and TB294-pCV. White bars, TB294-pCV; black bars, TB294-pSigA. (b) Comparison of eis expression in wild-type TB294 and H37Rv. White bars, H37Rv; black bars, TB294.
eis is expressed at higher levels in wild-type TB294 than in H37Rv during intracellular growth
We have previously found that, in broth culture, wild-type TB294 and H37Rv have similar growth rates and similar expression levels of sigA. However, during intracellular growth, sigA expression is five- to 10-fold higher in TB294, compared with H37Rv, and TB294 grows more rapidly than H37Rv in human macrophages (Wu et al., 2004). To determine whether eis expression levels reflected sigA expression patterns in these strains, we cultured wild-type strain TB294 and H37Rv in broth and in MonoMac6 cells, and measured eis mRNA by real-time PCR (Fig. 2b). eis expression levels were similar in the broth cultures, but in MonoMac6 cells, eis expression decreased by approximately 10-fold in H37Rv from the levels detected in broth. In contrast, eis expression did not decrease in TB294 and even increased slightly, so that the overall levels in TB294 were 11-fold higher than in H37Rv during intracellular growth. These results further supported close correlations between sigA and eis expression levels, and between high expression levels of these genes and enhanced intracellular growth.
SigA binds to the eis promoter in live M. tuberculosis
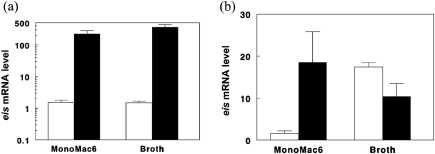
The results described above demonstrated that M. tuberculosis strains that overexpress SigA also produce elevated levels of eis mRNA and Eis protein. An earlier report indicated that the −35 and −10 regions of the eis promoter resemble the consensus sequence for SigA-like promoters (Roberts et al., 2004). To determine whether SigA directly regulates eis, we performed ChIP on broth-grown TB294, using a monoclonal anti-SigA antibody to recover SigA–DNA complexes (Fig. 3a). Using primers that flanked the predicted −35 and −10 regions of the eis promoter region (Fig. 3b), we performed PCR on the recovered promoter pool to determine whether this region bound SigA. A PCR product was obtained after immunoprecipitation with anti-SigA antibody (Fig. 3a, lane 1), but not after immunoprecipitation with a control antibody (lane 2). PCR product was also obtained with the positive controls of input DNA (lane 3) and M. tuberculosis genomic DNA (lane 5), but not with the negative control (lane 4). These results indicate that SigA binds the eis promoter, and suggest that the high expression levels of eis in TB294-pSigA and in wild-type TB294 during intracellular growth (Fig. 2) result directly from the increased expression of SigA that occurs in these strains during infection (Wu et al., 2004).
Fig. 3.
SigA binds to the eis promoter in live M. tuberculosis TB294. (a) ChIP analysis. TB294 was cultured in 7H9 broth to OD600 0.8 and then ChIP was performed, using anti-SigA antibody (lane 1), or anti-GroEL2 antibody as a negative control (lane 2). PCR was performed on the immunoprecipitated samples using primers designed to amplify a 143 bp region of the eis promoter (arrow). Positive controls included PCR amplification of input DNA (lane 3) and chromosomal DNA (lane 5). The negative PCR control was without added DNA (lane 4). (b) Region of the eis promoter amplified in ChIP analysis. Sequences of the primers used to amplify the 143 bp region are underlined. The predicted −10 and −35 regions (in bold type) of the SigA-like promoter and the transcriptional start point (bold capital A) were identified by Roberts et al. (2004).
An eis deletion mutant has reduced growth in MonoMac6 cells
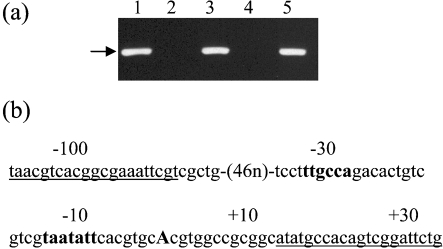
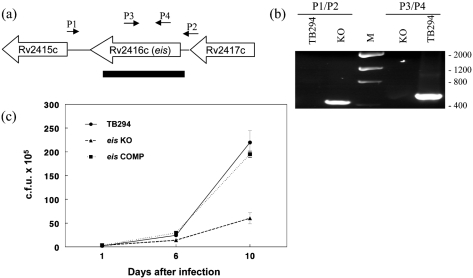
To determine whether eis is associated with the increased intracellular growth of isolate TB294, we generated an eis deletion mutant of this strain, using the p2NIL/pGOAL vector system (Parish & Stoker, 2000). We confirmed the presence of the deletion in the knockout strain TB294-eisKO by PCR (Fig. 4a, b). Next, we generated the complemented strain TB294-eisCOMP by introducing a copy of the eis gene under the control of its own promoter, using an integrating plasmid. Reintroduction of eis was confirmed by PCR (data not shown).
Fig. 4.
Effect of disruption of the eis gene on growth of TB294 in MonoMac6 cells. (a) Schematic diagram showing location of the eis deletion. The eis gene (Rv2416c) and flanking genes are indicated. Black rectangle, region deleted from TB294-eisKO. P1–P4, primers used to confirm the deletion by PCR. (b) Confirmation of eis deletion in TB294-eisKO. PCR was performed using genomic DNA from wild-type TB294 (TB294) or TB294-eisKO (KO), and primer sets P1/P2 or P3/P4, as indicated. For location of primers see (a). Reactions were run on an agarose gel and stained with ethidium bromide. Primers P1 and P2 yielded an amplicon from TB294-eisKO consistent in size with the predicted 442 bp fragment spanning the deleted region. The 1.7 kb region from wild-type TB294 was not amplified under the conditions used. Primers P3 and P4, which are located inside the deleted region, generated the expected amplicon of approximately 560 bp from wild-type TB294. No product was generated from TB294-eisKO with primers P3 and P4. Lane M, DNA low-mass ladder, sizes (in bp) are indicated to the right of the panel. (c) Comparison of intracellular growth of TB294, TB294-eisKO and TB294-eisCOMP. MonoMac6 cells were infected with the three strains at an m.o.i. of 1 : 4, and c.f.u. were measured at 1, 6 and 10 days after infection. Mean±sd for three independent experiments is shown.
To evaluate the role of eis during intracellular growth of TB294, we cultured the wild-type strain, TB294-eisKO and TB294-eisCOMP in broth and in MonoMac6 cells. All three strains grew at nearly identical rates in broth (data not shown), indicating that eis is not required for growth in vitro. However, in MonoMac6 cells, TB294-eisKO had reduced growth at 10 days post-infection, compared with the parental strain (Fig. 4c). Normal growth was restored in TB294-eisCOMP (Fig. 4c), indicating that the absence of eis in the deletion mutant was responsible for the growth defect.
DISCUSSION
We have previously shown that SigA influences a central pathogenic property of M. tuberculosis, namely, the ability to multiply in human monocytes (Wu et al., 2004). Our study suggests that this effect of SigA is mediated in part through the eis gene. We found a strong correlation between upregulation of sigA and high expression levels of eis in transformants and wild-type strains, and showed that SigA binds the eis promoter in live M. tuberculosis. Deletion of eis reduced the capacity of M. tuberculosis to replicate in monocytes, and this defect was restored by complementation with eis. These findings indicate that SigA regulates expression of eis, which contributes to the pathogenicity of M. tuberculosis.
Understanding the mechanisms by which increasing levels of Eis affect intracellular growth requires further investigation, but the differences in expression levels may have clinical significance. In this study, we found that, during growth in monocytes, eis is expressed at higher levels in TB294 than in H37Rv, and this may contribute to the greater intracellular growth of this isolate (Wu et al., 2004). It has also been reported that a clinical M. tuberculosis isolate upregulates expression of eis in activated human macrophages, whereas H37Rv does not (Cappelli et al., 2001). In contrast to our findings that deletion of eis results in reduced growth of TB294 in monocytes, another study found that an eis deletion mutant of H37Rv has no growth defect in the macrophage-like cell line U937 (Samuel et al., 2007). However, as expression of eis is already reduced in H37Rv, compared with TB294, during infection (Fig. 2b), it may be that further reducing its expression has little effect on host-cell interactions. Recent studies suggest that the effects of a particular gene on the host response can depend upon the background strain (Sinsimer et al., 2008). In addition, U937 cells are relatively undifferentiated monocytoid cells, whereas the MonoMac6 cells used in the current study are more differentiated and closer in phenotype to primary human monocytes. These distinctions may result in different responses to the effects of Eis.
A role for eis in the survival of mycobacteria in monocytes/macrophages was first recognized by Friedman and colleagues, who used an H37Rv plasmid library to identify genes that could enhance survival of M. smegmatis in U937 cells (Wei et al., 2000). M. smegmatis clones transformed with a plasmid containing the eis gene showed approximately two- to fivefold greater survival than other clones. On the basis of Southern blot analysis, the same study concluded that M. smegmatis did not contain a native eis gene, and suggested that eis may be present only in pathogenic mycobacteria. However, recently available genomic sequence data (www.tigr.org) indicate that the M. smegmatis genome contains a homologue (gene MSMEG_3513) encoding a protein with 58 % amino acid identity to Eis. In addition to the M. smegmatis gene, a search against the NCBI non-redundant protein sequence database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), using the blastp program (Altschul et al., 1997), revealed that the environmental mycobacteria Mycobacterium gilvum and Mycobacterium vanbaalenii also contain genes encoding proteins with >50 % amino acid identity to Eis. It is possible that Eis of M. tuberculosis has functions lacked by the homologous genes. Eis is a member of the GNAT superfamily of acetyltransferases (Samuel et al., 2007), and although there are some conserved motifs within these enzymes, they also have extensive sequence variation and a broad array of roles (Vetting et al., 2005). Comparative analyses of the mycobacterial orthologues may give insights into the regions of the M. tuberculosis gene that are important in host–pathogen interactions, and shed light on the function of M. tuberculosis Eis. Alternatively, it is possible that the mycobacterial enzymes have similar functions, but that the native gene of M. smegmatis is only expressed at low levels during intracellular growth, and that, in the original study (Wei et al., 2000), this deficiency was overcome by overexpression of eis from the plasmid clone.
Because Eis contributes to intracellular mycobacterial growth and eis expression varies in different strains, it is important to identify the factors that regulate eis expression. Our analyses indicate a direct correlation between upregulation of SigA and activation of eis. In addition, we showed that SigA binds the eis promoter, consistent with an analysis of the eis promoter region that found that the −10 and −35 regions were similar to those of the E. coli σ70 consensus sequence (Roberts et al., 2004). However, it is not clear that the eis promoter is itself especially sensitive to SigA-RNA polymerase levels such that the upregulation would result directly from an increase in the availability of the holoenzyme for promoter binding. Changes in sigma factor levels affect the competition between the sigma factors for core RNA polymerase, and therefore can markedly affect gene activation, particularly at weak promoters, but these effects may be mediated through other transcriptional regulators (Bernardo et al., 2006; Farewell et al., 1998; Grigorova et al., 2006; King et al., 2004; Shah & Wolf, 2004; Typas et al., 2007). It may be that a transcriptional activator of eis is the direct beneficiary of increased amounts of SigA-RNA polymerase and that upregulation of the activator aids in recruitment or stabilization of the holoenzyme at the eis promoter. For example, evidence indicates that SigA regulates expression of some virulence genes through interaction with the transcription factor WhiB3 (Steyn et al., 2002). Mutational analyses of the eis promoter show that sequences upstream of the −35 region are required for optimal expression of a reporter gene in mycobacteria (Roberts et al., 2004), suggesting that as yet unidentified transcriptional activators contribute to eis expression.
In addition to eis and the adjacent gene Rv2415c, 19 other genes were expressed at higher levels in TB294-pSigA than in TB294-pCV during growth in monocytes (Supplementary Table S1). Given that SigA is the principal sigma factor, we had anticipated that a larger number of genes would show increased expression. However, as discussed above, not all target genes will be equally affected by changes in levels of a sigma factor. Moreover, sigA is already expressed at higher levels in TB294-pCV than in H37Rv during infection (Wu et al., 2004), so the sensitivity of some target genes to further increases in SigA levels may be diminished. We did, however, identify at least two other genes besides eis that may contribute to the enhanced capacity of TB294-pSigA to grow in monocytes. Rv0986 is associated with host–pathogen interactions (Be et al., 2008; Jain et al., 2006; Rosas-Magallanes et al., 2007; Talaat et al., 2004) and was upregulated by approximately 2.5-fold, and nuoK (Rv3155), which encodes a subunit of a NADH dehydrogenase involved in inhibition of apoptosis (Velmurugan et al., 2007), was also upregulated (Supplementary Table S1).
Another intriguing discovery was that of the 56 genes with lower expression in TB294-pSigA, 37 are predicted to be regulated by a RecA-NDp promoter, or are clustered with another gene regulated by this type of promoter (Supplementary Table S2). These genes include the DNA repair genes recA, recX, radA, ogt and alkA. The RecA non-dependent promoter (RecA-NDp) motif was identified upstream of genes that are activated in response to DNA damage, independently of RecA and LexA (Gamulin et al., 2004). Most DNA repair genes in M. tuberculosis that are induced by DNA damage are regulated independently of RecA (Rand et al., 2003), and promoter sequences containing the RecA-NDp motif have been shown to be required for the RecA-independent induction of recA itself and Rv2719c (Brooks et al., 2006; Davis et al., 2002; Gopaul et al., 2003), both of which are expressed at lower levels in TB294-pSigA.
Rv3614c–Rv3620c was another notable group of genes expressed at a lower level in TB294-pSigA (Supplementary Table S2). These genes are components of the ESX-1 secretion system, which is primarily associated with mycobacterial virulence (Fortune et al., 2005; MacGurn et al., 2005; Raghavan et al., 2008), although in M. smegmatis, the ESX-1 system is also involved in DNA conjugation (Coros et al., 2008; Flint et al., 2004). The basis for the downregulation of the RecA-NDp and ESX-1 genes is unknown, but we speculate that these genes are regulated by an alternative sigma factor that is at a competitive disadvantage in the presence of increased levels of SigA, or has reduced expression in TB294-pSigA. Promoters for both sigG and sigH contain the RecA-NDp motif (Gamulin et al., 2004), and additional evidence links sigG to the DNA-damage response (Lee et al., 2008). However, sigH actually showed a slight increase in expression (1.5-fold) in TB294-pSigA (data not shown), and sigG (Rv0182c) expression was below detectable levels in our array analysis, although its neighbouring genes (Rv0181c, Rv0184 and Rv0185) did have lower expression levels in TB294-pSigA (Supplementary Table S2). The association between upregulation of SigA and reduced expression of genes involved in the major DNA damage response therefore remains to be determined.
A complete understanding of the factors regulating expression of eis, and of sigA itself, during intracellular growth of the W-Beijing isolate TB294 also awaits further study, but our results provide further evidence of a significant role for both genes in host–pathogen interactions.
Acknowledgments
We thank Dr John Belisle for provision of the anti-GroEL2 antibody, under NIH contract N01-AI40091, and we thank Professor Tanya Parish, Barts and the London Queen Mary's School of Medicine and Dentistry, for the pNIL/pGOAL vectors. Funding for the study was provided by NIH grant AI44935 (to P. F. B.), the Margaret E. Byers Cain Chair for Tuberculosis Research, the Cain Foundation for Infectious Disease Research, and the Center for Pulmonary and Infectious Disease Control.
Abbreviations
ChIP, chromatin immunoprecipitation
Footnotes
Two supplementary tables, listing the results of DNA microarray analyses of recombinant strains grown for 6 days in human monocyte cells and features of selected genes showing differential expression in TB294-pSigA compared with TB294-pCV, with associated references, are available with the online version of this paper.
The microarray data used in this study have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE13780 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=pbsltuqwkukqexu&acc=GSE13780.
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, P. F., Yang, Z., Preston-Martin, S., Pogoda, J. M., Jones, B. E., Otaya, M., Eisenach, K. D., Knowles, L., Harvey, S. & Cave, M. D. (1997). Patterns of tuberculosis transmission in central Los Angeles. JAMA 278, 1159–1163. [PubMed] [Google Scholar]
- Be, N. A., Lamichhane, G., Grosset, J., Tyagi, S., Cheng, Q. J., Kim, K. S., Bishai, W. R. & Jain, S. K. (2008). Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J Infect Dis 198, 1520–1528. [DOI] [PubMed] [Google Scholar]
- Bernardo, L. M., Johansson, L. U., Solera, D., Skärfstad, E. & Shingler, V. (2006). The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of σ54-dependent transcription. Mol Microbiol 60, 749–764. [DOI] [PubMed] [Google Scholar]
- Bishai, W. R., Graham, N. M., Harrington, S., Pope, D. S., Hooper, N., Astemborski, J., Sheely, L., Vlahov, D., Glass, G. E. & Chaisson, R. E. (1998). Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA 280, 1679–1684. [DOI] [PubMed] [Google Scholar]
- Brooks, P. C., Dawson, L. F., Rand, L. & Davis, E. O. (2006). The mycobacterium-specific gene Rv2719c is DNA damage inducible independently of RecA. J Bacteriol 188, 6034–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli, G., Volpe, P., Sanduzzi, A., Sacchi, A., Colizzi, V. & Mariani, F. (2001). Human macrophage gamma interferon decreases gene expression but not replication of Mycobacterium tuberculosis: analysis of the host–pathogen reciprocal influence on transcription in a comparison of strains H37Rv and CMT97. Infect Immun 69, 7262–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, D. M., Kawakami, R. P., deLisle, G. W., Pascopella, L., Bloom, B. R. & Jacobs, W. R., Jr (1995). Mutation of the principal sigma factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 92, 8036–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros, A., Callahan, B., Battaglioli, E. & Derbyshire, K. M. (2008). The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol 69, 794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, J. L., Wei, J., Moulder, J. W., Laal, S. & Friedman, R. L. (2001). Subcellular localization of the intracellular survival-enhancing Eis protein of Mycobacterium tuberculosis. Infect Immun 69, 4295–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, E. O., Springer, B., Gopaul, K. K., Papavinasasundaram, K. G., Sander, P. & Böttger, E. C. (2002). DNA damage induction of recA in Mycobacterium tuberculosis independently of RecA and LexA. Mol Microbiol 46, 791–800. [DOI] [PubMed] [Google Scholar]
- Farewell, A., Kvint, K. & Nyström, T. (1998). Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol 29, 1039–1051. [DOI] [PubMed] [Google Scholar]
- Flint, J. L., Kowalski, J. C., Karnati, P. K. & Derbyshire, K. M. (2004). The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci U S A 101, 12598–12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune, S. M., Jaeger, A., Sarracino, D. A., Chase, M. R., Sassetti, C. M., Sherman, D. R., Bloom, B. R. & Rubin, E. J. (2005). Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A 102, 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamulin, V., Cetkovic, H. & Ahel, I. (2004). Identification of a promoter motif regulating the major DNA damage response mechanism of Mycobacterium tuberculosis. FEMS Microbiol Lett 238, 57–63. [DOI] [PubMed] [Google Scholar]
- Gomez, M., Doukhan, L., Nair, G. & Smith, I. (1998). SigA is an essential gene in Mycobacterium smegmatis. Mol Microbiol 29, 617–628. [DOI] [PubMed] [Google Scholar]
- Gopaul, K. K., Brooks, P. C., Prost, J. F. & Davis, E. O. (2003). Characterization of the two Mycobacterium tuberculosis recA promoters. J Bacteriol 185, 6005–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorova, I. L., Phleger, N. J., Mutalik, V. K. & Gross, C. A. (2006). Insights into transcriptional regulation and σ competition from an equilibrium model of RNA polymerase binding to DNA. Proc Natl Acad Sci U S A 103, 5332–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. & Coates, A. R. (1999). Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, S. K., Paul-Satyaseela, M., Lamichhane, G., Kim, K. S. & Bishai, W. R. (2006). Mycobacterium tuberculosis invasion and traversal across an in vitro human blood–brain barrier as a pathogenic mechanism for central nervous system tuberculosis. J Infect Dis 193, 1287–1295. [DOI] [PubMed] [Google Scholar]
- King, T., Ishihama, A., Kori, A. & Ferenci, T. (2004). A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol 186, 5614–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., Geiman, D. E. & Bishai, W. R. (2008). Role of stress response sigma factor SigG in Mycobacterium tuberculosis. J Bacteriol 190, 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lella, R. K. & Sharma, C. (2007). Eis (enhanced intracellular survival) protein of Mycobacterium tuberculosis disturbs the cross regulation of T-cells. J Biol Chem 282, 18671–18675. [DOI] [PubMed] [Google Scholar]
- MacGurn, J. A., Raghavan, S., Stanley, S. A. & Cox, J. S. (2005). A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol 57, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Pang, X., Vu, P., Byrd, T. F., Ghanny, S., Soteropoulos, P., Mukamolova, G. V., Wu, S., Samten, B. & Howard, S. T. (2007). Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153, 1229–1242. [DOI] [PubMed] [Google Scholar]
- Parish, T. & Stoker, N. G. (2000). Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146, 1969–1975. [DOI] [PubMed] [Google Scholar]
- Predich, M., Doukhan, L., Nair, G. & Smith, I. (1995). Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol Microbiol 15, 355–366. [DOI] [PubMed] [Google Scholar]
- Raghavan, S., Manzanillo, P., Chan, K., Dovey, C. & Cox, J. S. (2008). Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454, 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, L., Hinds, J., Springer, B., Sander, P., Buxton, R. S. & Davis, E. O. (2003). The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol 50, 1031–1042. [DOI] [PubMed] [Google Scholar]
- Reed, M. B., Gagneux, S., Deriemer, K., Small, P. M. & Barry, C. E., III (2007). The W/Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and is constitutively upregulated for the DosR dormancy regulon. J Bacteriol 189, 2583–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, E. A., Clark, A., McBeth, S. & Friedman, R. L. (2004). Molecular characterization of the eis promoter of Mycobacterium tuberculosis. J Bacteriol 186, 5410–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue, S., Brodeur, J., Jacques, P. E., Gervais, A. L., Brzezinski, R. & Gaudreau, L. (2007). Identification of mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J Bacteriol 189, 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Magallanes, V., Stadthagen-Gomez, G., Rauzier, J., Barreiro, L. B., Tailleux, L., Boudou, F., Griffin, R., Nigou, J., Jackson, M. & other authors (2007). Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect Immun 75, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., Braisted, J., Klapa, M., Currier, T. & other authors (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Samuel, L. P., Song, C. H., Wei, J., Roberts, E. A., Dahl, J. L., Barry, C. E., III, Jo, E. K. & Friedman, R. L. (2007). Expression, production and release of the Eis protein by Mycobacterium tuberculosis during infection of macrophages and its effect on cytokine secretion. Microbiology 153, 529–540. [DOI] [PubMed] [Google Scholar]
- Shah, I. M. & Wolf, R. E. J. (2004). Novel protein–protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase α subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J Mol Biol 343, 513–532. [DOI] [PubMed] [Google Scholar]
- Sinsimer, D., Huet, G., Manca, C., Tsenova, L., Koo, M. S., Kurepina, N., Kana, B., Mathema, B., Marras, S. A. & other authors (2008). The phenolic glycolipid of Mycobacterium tuberculosis differentially modulates the early host cytokine response but does not in itself confer hypervirulence. Infect Immun 76, 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, P. M., Hopewell, P. C., Singh, S. P., Paz, A., Parsonnet, J., Ruston, D. C., Schecter, G. F., Daley, C. L. & Schoolnik, G. K. (1994). The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med 330, 1703–1709. [DOI] [PubMed] [Google Scholar]
- Steyn, A. J., Collins, D. M., Hondalus, M. K., Jacobs, W. R., Jr, Kawakami, R. P. & Bloom, B. R. (2002). Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc Natl Acad Sci U S A 99, 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., Bansal, G. P., Young, J. F., Lee, M. H. & other authors (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. [DOI] [PubMed] [Google Scholar]
- Strickland, M. S., Thompson, N. E. & Burgess, R. R. (1988). Structure and function of the σ-70 subunit of Escherichia coli RNA polymerase. Monoclonal antibodies: localization of epitopes by peptide mapping and effects on transcription. Biochemistry 27, 5755–5762. [DOI] [PubMed] [Google Scholar]
- Talaat, A. M., Hunter, P. & Johnston, S. A. (2000). Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat Biotechnol 18, 679–682. [DOI] [PubMed] [Google Scholar]
- Talaat, A. M., Lyons, R., Howard, S. T. & Johnston, S. A. (2004). The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A 101, 4602–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theus, S. A., Cave, M. D. & Eisenach, K. D. (2005). Intracellular macrophage growth rates and cytokine profiles of Mycobacterium tuberculosis strains with different transmission dynamics. J Infect Dis 191, 453–460. [DOI] [PubMed] [Google Scholar]
- Tsenova, L., Ellison, E., Harbacheuski, R., Moreira, A. L., Kurepina, N., Reed, M. B., Mathema, B., Barry, C. E., III & Kaplan, G. (2005). Virulence of selected Mycobacterium tuberculosis clinical isolates in the rabbit model of meningitis is dependent on phenolic glycolipid produced by the bacilli. J Infect Dis 192, 98–106. [DOI] [PubMed] [Google Scholar]
- Typas, A., Barembruch, C., Possling, A. & Hengge, R. (2007). Stationary phase reorganisation of the Escherichia coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J 26, 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan, K., Chen, B., Miller, J. L., Azogue, S., Gurses, S., Hsu, T., Glickman, M., Jacobs, W. R., Jr, Porcelli, S. A. & Briken, V. (2007). Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog 3, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting, M. W., de Carvalho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L. & Blanchard, J. S. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 433, 212–226. [DOI] [PubMed] [Google Scholar]
- Wei, J., Dahl, J. L., Moulder, J. W., Roberts, E. A., O'Gaora, P., Young, D. B. & Friedman, R. L. (2000). Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol 182, 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, S. E., Pogoda, J. M., Yang, Z., Cave, M. D., Wallace, C., Kelley, M. & Barnes, P. F. (2002). Transmission dynamics of tuberculosis in Tarrant county, Texas. Am J Respir Crit Care Med 166, 36–42. [DOI] [PubMed] [Google Scholar]
- Wu, S., Howard, S. T., Lakey, D. L., Kipnis, A., Samten, B., Safi, H., Gruppo, V., Wizel, B., Shams, H. & other authors (2004). The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis strains in vivo. Mol Microbiol 51, 1551–1562. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Barnes, P. F., Chaves, F., Eisenach, K. D., Weis, S. E., Bates, J. H. & Cave, M. D. (1998). Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol 36, 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Gong, J., Yang, Z., Samten, B., Cave, M. D. & Barnes, P. F. (1999). Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J Infect Dis 179, 1213–1217. [DOI] [PubMed] [Google Scholar]
- Ziegler-Heitbrock, H. W., Thiel, E., Futterer, A., Herzog, V., Wirtz, A. & Riethmuller, G. (1988). Establishment of a human cell line (Mono Mac 6) with characteristics of mature monocytes. Int J Cancer 41, 456–461. [DOI] [PubMed] [Google Scholar]