Summary
DICER is a central regulator of microRNA maturation. However little is known about mechanisms regulating its expression in development or disease. While profiling miRNA expression in differentiating melanocytes, two populations were observed: some upregulated at the pre-miRNA stage, and others upregulated as “mature” miRNAs (with stable pre-miRNA levels). Conversion of pre-miRNAs to fully processed miRNAs appeared to be dependent upon stimulation of DICER expression—an event found to occur via direct transcriptional targeting of DICER by the melanocyte master transcriptional regulator MITF. MITF binds and activates a conserved regulatory element upstream of DICER’s transcriptional start site upon melanocyte differentiation. Targeted KO of DICER is lethal to melanocytes, at least partly via DICER-dependent processing of the pre-miRNA-17~92 cluster thus targeting BIM, a known pro-apoptotic regulator of melanocyte survival. These observations highlight a central mechanism underlying miRNA regulation which could exist for other cell types during development.
Introduction
MicroRNAs (miRNAs) are short noncoding RNAs that inhibit mRNA translation or stability (Carthew and Sontheimer, 2009; Du and Zamore, 2007; Filipowicz et al., 2008; Maroney et al., 2006; Neilson and Sharp, 2008; Schier and Giraldez, 2006). Numerous fundamental processes, including development, differentiation and tumorigenesis, are regulated by miRNAs (Calin and Croce, 2006; Chang and Mendell, 2007; Ma and Weinberg, 2008; Medina and Slack, 2008; Schickel et al., 2008).
Functional mature miRNAs arise through several post-transcriptional processing steps which include cleavage by Drosha/DGCR8 to pre-miRNA, export, and digestion by the RNase III endonuclease DICER, which also mediates loading onto RISC complexes (Bushati and Cohen, 2007; Filipowicz et al., 2008; Kim et al., 2009; Schickel et al., 2008; Winter et al., 2009; Zeng and Cullen, 2006). Even though a group of miRNAs called mirtrons can bypass Drosha by instead using the splicing machinery (Berezikov et al., 2007; Ruby et al., 2007), there is no evidence of DICER-independent miRNAs, suggesting that DICER is indispensable for miRNA biogenesis.
DICER activity is regulated by co-factors such as TRBP (TAR RNA-binding protein), PACT (PKR activator) and KH-type splicing regulatory protein (KSRP) (Chendrimada et al., 2005; Haase et al., 2005; Jaskiewicz and Filipowicz, 2008; Lee et al., 2006; Trabucchi et al., 2009), as well as by an ATP mediated control of its helicase/ATPase domain (Asada et al., 2008; Bernstein et al., 2001; Liu et al., 2003; Zhang et al., 2002). DICER regulation has been explored in development where it may be differentially expressed and localized within the cell (Barbato et al., 2007). Studies show post-transcriptional regulation of miRNA at the DICER level during development, via either differential expression or activity of DICER (Obernosterer et al., 2006; Wulczyn et al., 2007). DICER cleavage activity and mature-miRNA expression in mammals are restricted to certain tissues and cell types, suggesting tissue-specific regulation of its activity (Obernosterer et al., 2006). Transcriptional regulation of DICER expression remains incompletely understood. Mammalian genomes encode one DICER gene with two alternative 5′ UTRs (the coding regions are similar) (Jaskiewicz and Filipowicz, 2008).
Microphthalmia-associated transcription factor (MITF) is a tissue-restricted master regulator of melanocytes, and is essential for their proliferation, survival, and differentiation (Levy et al., 2006). Melanocyte differentiation/pigmentation is stimulated by Melanocyte Stimulating Hormone (MSH). MSH binds the Melanocortin Receptor 1 (MC1R) to stimulate cAMP production, which in turn activates CREB/ATF1 to transcribe MITF, which transcriptionally targets numerous genes associated with melanocyte differentiation (Levy et al., 2006). The sun-tanning pigment response is associated with keratinocyte DNA damage, followed by p53-mediated induction of MSH expression and secretion (Cui et al., 2007). Human redheads typically harbor variant forms of MC1R that cannot respond to MSH, thus likely explaining their poor or absent tanning response. Of note, topical administration of the adenylate cyclase agonist forskolin (forsk) was shown to “rescue” this pathway in a mouse mc1r “redhead” model, inducing strong melanin synthesis, skin darkening, and UV photo-protection (D’Orazio et al., 2006). Forsk can thus serve as a means to induce melanocytic differentiation, even in cells harboring nonresponsive MC1R variants.
MITF also plays a vital role in lineage survival, and MITF mutation in numerous species (including humans) results in major loss of melanocyte viability. Correspondingly MITF appears to be an amplified oncogene in a fraction of human melanomas (Garraway et al., 2005). The profound resistance of melanomas to numerous triggers of apoptosis (including cytotoxic chemotherapy) is incompletely understood, but may involve lineage specific survival mechanisms. For example BCL-2, a direct transcriptional target of MITF, is required for melanocyte survival, based upon loss of the melanocyte lineage in Bcl-2 null mice (Kamada et al., 1995; Veis et al., 1993; Yamamura et al., 1996), thus linking lineage-specific transcriptional control of pigmentation to survival (McGill et al., 2002). The BCL-2 family regulates apoptosis propensity in many contexts (Youle and Strasser, 2008). Pro-survival (anti-apoptotic) members of the family include BCL-2 while pro-apoptotic members include BIM (Cartlidge et al., 2008; Ewings et al., 2007; O’Connor et al., 1998). Whereas Bcl-2 KO mice display premature de-pigmentation due to melanocyte apoptosis (Nishimura et al., 2005; Veis et al., 1993), additional deletion of both BIM alleles prevents this defect, restoring pigmentation (Bouillet et al., 2001). This finding places BIM as a critical player in the regulation of melanocyte apoptosis and possibly also of melanomagenesis.
Expression and pro-apoptotic activity of BIM are regulated by several different signaling systems, including the ERK, p38 and JNK MAP kinase pathways, via transcriptional and post-transcriptional mechanisms (Cai et al., 2006; Cartlidge et al., 2008; Ewings et al., 2007; Ley et al., 2005; O’Connor et al., 1998). miRNAs also take part in post-transcriptional regulation of BIM. Multiple studies have shown that the miRNA-17~92 cluster directly targets BIM, and that downregulation of BIM expression is tumorigenic and affects normal development of many tissues (Fontana et al., 2008; Koralov et al., 2008; Mendell, 2008; Ventura et al., 2008; Xiao et al., 2008).
Here we observe that DICER expression is strongly induced during melanocyte differentiation via direct transcriptional regulation by MITF. Through miRNA profiling we identify one class of miRNAs upregulated at the pre-miRNA level and one class upregulated as mature-miRNAs (with unchanged pre-miRNA). The latter class appears to reflect DICER induction during lineage differentiation. We also demonstrate that DICER KO is lethal to melanocytes both in-vivo and in-vitro, and may involve post-transcriptional processing of the miR-17~92 cluster targeting BIM.
Results
miRNA expression profiles in melanocytes
To assess expression changes in miRNAs during melanocyte differentiation, we utilized human primary melanocytes, generated from discarded foreskin tissue. Since the donor’s genetic background is unknown and therefore the responsiveness of MC1R to MSH may be variable, we stimulated the cells with forsk, which directly activates adenyl cyclase, thus mimicking a key step in the pigmentation/differentiation pathway. Human primary melanocytes from 7 different donors were stimulated with forsk for 16hrs. Induction of differentiation was confirmed by documenting upregulation of MITF and known MITF transcriptional target genes by forsk (Bertolotto et al., 1998; Price et al., 1998) (data not shown and see below). Once validated, the treated and vehicle-control treated cultures were subjected to miRNA expression profiling. The distribution of expression values for each miRNA in the differentiated versus resting cell groups was compared using several statistical tests (Figure 1 and Table S1). We further investigated the levels of both primary and mature miRNAs upon treatment with forsk.
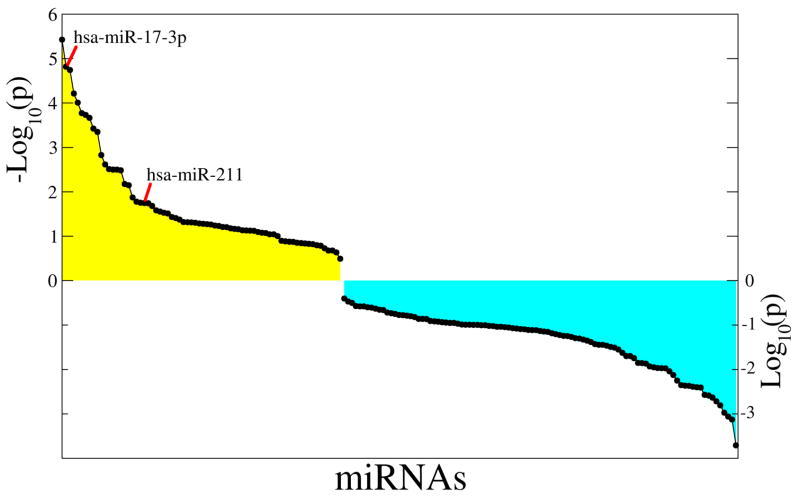
Figure 1. MicroRNA expression profile upon melanocyte stimulation by forskolin.
Primary human melanocytes were serum depleted for 12 hours, followed by treatment with 20uM forsk (or DMSO control) for 16 hours. Total RNA was harvested, and miRNA expression array analysis was performed. Fold-changes between paired control and activated samples were ranked, and p-values for the rank distribution of fold-changes corresponding to given miRNAs were computed using the hypergeometric test. The figure shows the ranked log-transformed p-values of up-regulated (yellow) and down-regulated miRNAs (cyan). A full list of differentially regulated miRNAs can be found in Table S1.
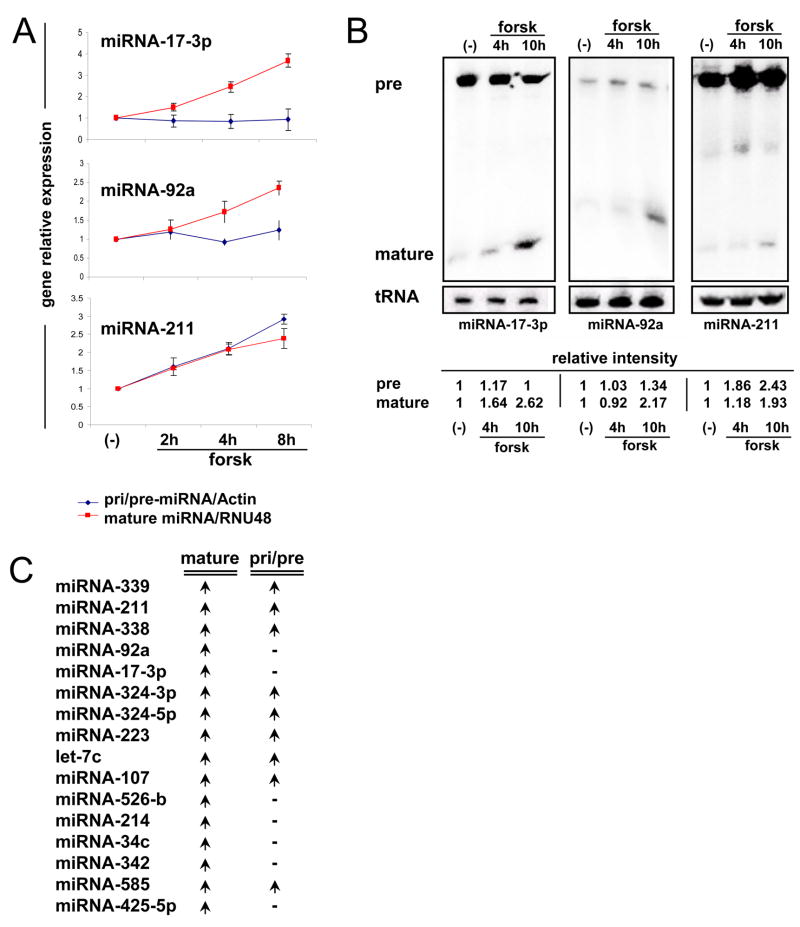
As shown in Figure 2, we utilized qRT-PCR and Northern blot analyses to examine both pre-miRNA and mature miRNA levels. One notable set of miRNAs was miR-17-3p and miR-92a, both of which belong to the same miRNA cluster. Mature and pre levels of all six miR-17~92 cluster members were analyzed. Four of the six showed induction upon melanocyte differentiation and the largest changes were observed for miR-17-3p and miR-92 (Figure S1). We also measured miR-211 levels, as it is an intronic miRNA hosted by the known MITF target gene TRPM1 (Miller et al., 2004) and, therefore, is likely to be transcriptionally regulated by MITF. Increases in mature miR-17-3p, miR-92a and miR-211 were seen at 4 or 14h of stimulation; however, among the precursor forms, only pre-miR-211 levels were increased, while no significant change was observed in pre-miR-17-3p or pre-miR-92a (Figure 2A and B). Upregulation of mature miRNAs, without an accompanying increase in the pre-miRNA level, suggested post-transcriptional regulation of these miRNAs. To assess if such post-transcriptional modulation was unique to miR-17 in melanocytes, we similarly examined precursor levels for 16 miRNAs that were differentially expressed at the 5% significance level of both hypergeometric test and t-test. We observed that 7 of the 16 miRNAs (~43%) were similarly upregulated post-transcriptionally (but not at the pre-miRNA level), suggesting that this mode of regulation may be relatively common (Figure 2C, and S2 and S3D). There are several potential mechanisms that could explain this, including altered DICER level, DROSHA complex activity, and pre-miRNA export or stability. We therefore examined the expression of DICER and DROSHA during melanocyte differentiation.
Figure 2. miRNA17~92 cluster is post-transcriptionally regulated upon melanocyte stimulation.
(A) Primary human melanocytes were stimulated with forsk for the indicated times and then total RNA was harvested for qRT-PCR analysis, mature-miRNA expression (Red) and pre-miRNA gene expression (Blue). Mature-miRNAs were normalized to RNU48 and pre-miRNAs to Actin and are presented relative to non-stimulated cells. Error bars indicate mean ±SD of 5 independent experiments from different primary melanocyte donors.
(B) Total RNA from primary melanocytes treated as in (A) subjected to Northern blot analysis. Band intensity was normalized to tRNA and presented as fold-induction.
(C) Similar qRT-PCR analysis was performed on 16 different miRNAs. The table presents the indicated mature-miRNA and pre-miRNA expression status upon melanocyte stimulation. An arrow indicates upregulation and a line represents no change in the gene expression. Detailed pre-miRNA expression patterns upon melanocyte stimulation can be found in Figure S2.
DICER protein and RNA levels are upregulated during melanocyte differentiation
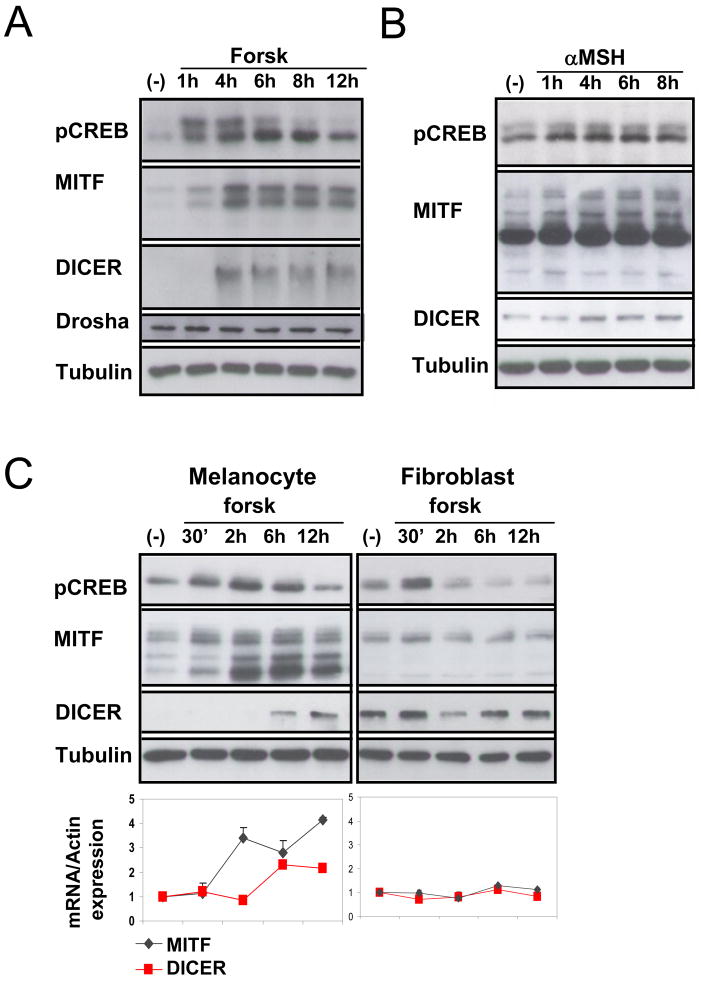
Upon differentiation of melanocytes with either αMSH or forsk, phosphorylation of CREB is observed, followed by upregulation of MITF expression (Price et al., 1998) (Figure 3A and B). We observed a striking increase in DICER protein level in stimulated human primary melanocytes, which occurred shortly after upregulation of MITF. Levels of DROSHA protein did not change upon melanocyte stimulation (Figure 3A). In addition to elevated protein levels, DICER mRNA levels also increased upon melanocyte differentiation (Figure 3C and Figure S3A), consistent with the possibility that DICER may be transcriptionally upregulated during melanocyte differentiation.
Figure 3. Lineage specific DICER upregulation upon melanocyte stimulation.
(A and B) Primary human melanocytes were stimulated with forsk or αMSH for the indicated times and harvested for immunoblot analysis using monoclonal antibodies. Tubulin was used as loading control.
(C) Primary human melanocytes or primary human fibroblasts were stimulated with forsk for the indicated times and harvested for immunoblot and qRT-PCR analysis. Tubulin was the loading control. mRNA expression results were normalized to Actin and are relative to non stimulated (−) values. The data show fold of stimulation and represent mean ±SD of 3 experiments performed in triplicate.
To test whether DICER upregulation by forsk/cAMP signaling is a ubiquitous phenomenon, we compared regulation of DICER expression by forsk in both human primary fibroblasts and melanocytes generated from 5 different donors. Both cell types were stimulated with forsk followed by total RNA and protein analyses. No increase in DICER protein or RNA level was observed in the treated fibroblasts (Figure 3C right panel) despite upregulation of pCREB. Thus DICER upregulation seen in melanocytes upon forsk treatment does not occur in other cell lineages and may therefore utilize lineage specific functional intermediate(s).
DICER upregulation is MITF-dependent
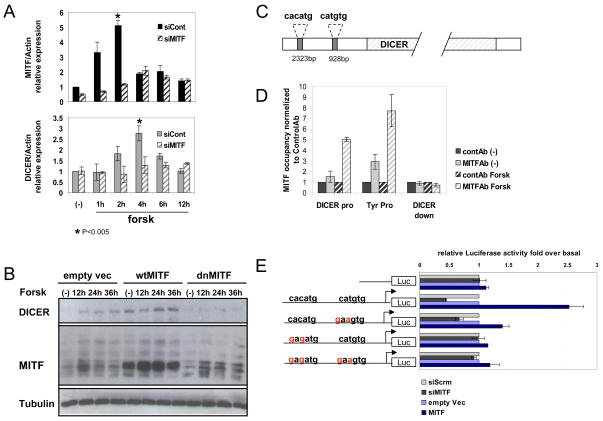
The possibility that DICER regulation in response to cAMP signaling is lineage specific led us to explore the possibility that MITF may directly upregulate DICER. Human primary melanocytes were transfected with siRNA targeting MITF (or control siRNA) prior to forsk treatment (Figure 4A). Differentiation of primary melanocytes led to 5- and 3-fold increases in MITF and DICER mRNA levels respectively. In contrast, introduction of siMITF blunted upregulation of MITF levels and resulted in virtually no induction of DICER mRNA. Moreover, upregulation of miRNAs 17 and 92 upon forsk stimulation was seen to be MITF dependent (Figure S3B). As a complementary approach, we also utilized adenoviruses encoding either nuclear-targeted GFP (control), wild-type (wt) MITF, or dominant-negative (dn) MITF, followed by forsk treatment. These adenoviral vectors have previously been shown to efficiently modulate MITF activity (McGill et al., 2002). In differentiating melanocytes, dnMITF significantly blocked DICER upregulation upon forsk stimulation. In addition, melanocytes that were infected with wt MITF showed significantly higher levels of endogenous DICER, even without stimulation, relative to controls, whereas dnMITF did not appear to affect basal DICER levels (Figure 4B). These data suggest that MITF resides upstream in the transcriptional regulation of DICER expression during differentiation of primary human melanocytes.
Figure 4. DICER is an MITF target gene.
(A) Primary human melanocytes were transfected with siMITF or scrambled siRNA. At 48 hrs post-transfection, cells were serum-depleted for 12 hours, followed by forsk stimulation. Total RNA from stimulated melanocytes was used for qRT-PCR analysis. Graphs present MITF (upper) or DICER (lower) mRNA levels upon stimulation, in either siMITF-transfected cells (dash bars) or cells transfected with control siRNA (full bars). Results are normalized to Actin and are relative to non stimulated (−) values. The data show fold of induction relative to siCont and represent mean ±SD of 5 replicates.
(B) Primary human melanocytes were infected with either MITF wild-type (wt), MITF dominant-negative (dn) or empty vector adenovirus (MOI:200). 72 hours post infection, cells were serum-depleted for 12 hours, stimulated with forsk for the indicated time, and harvested for immunoblot analysis. Tubulin was used as loading control.
(C) Diagram of the human DICER gene promoter region showing two putative MITF binding sites (filled gray boxes) at positions 2323 and 928 nucleotides upstream of the transcription start site.
(D) Binding of MITF to the endogenous DICER promoter region in forsk activated and resting (−) primary human melanocytes. Chromatin immunoprecipitations were performed using log-phase primary human melanocytes. Protein:chromatin-crosslinked complexes were immunoprecipitated with either MITF antibody or control antibody. PCR primers spanning either DICER promoter (“pro”) or downstream (“down”) regions were employed. Tyrosinase promoter (“Tyr pro”) primers were used as a positive control. The data show promoter occupancy relative to control Ab and represent mean ±SD of 3 independent experiments.
(E) UACC62 melanoma cells were transiently transfected with DICER promoter reporter plasmids, in addition to siMITF, control siRNA, over expression vector for MITF, or empty vector. The same set of experiments was performed on DICER promoter wt, mutated E-boxes or empty reporter plasmids as indicated. After 48h, firefly luciferase was measured and normalized to renilla luciferase (transfection control). The data show fold of stimulation and represent mean ±SD of three experiments performed in triplicate.
MITF regulation of the DICER promoter
Examining the DICER promoter in both human and mouse revealed two conserved E-box elements, the MITF consensus binding sequences, within ~2kb upstream of the DICER transcription start site (Figure 4C). Chromatin immunoprecipitation generated from activated and resting primary melanocytes revealed forsk-induced occupancy of the DICER promoter by MITF upon stimulation. The same anti-MITF antibody showed no binding to a downstream region of the DICER gene, while binding was detected upstream of the known MITF target gene for tyrosinase (positive control) (Yasumoto et al., 1994) (Figure 4D). In addition, a luciferase reporter driven by the DICER promoter region exhibited MITF responsiveness which was lost upon mutation of either of the two E-box elements, although a stronger affect was observed when the more upstream E-box was mutated. Moreover activity of the same reporter was suppressed upon introduction of siMITF, also in an E-box dependent fashion (Figure 4E). These results indicate that MITF occupies and may directly regulate the endogenous DICER promoter.
MITF and DICER expression correlate in vivo
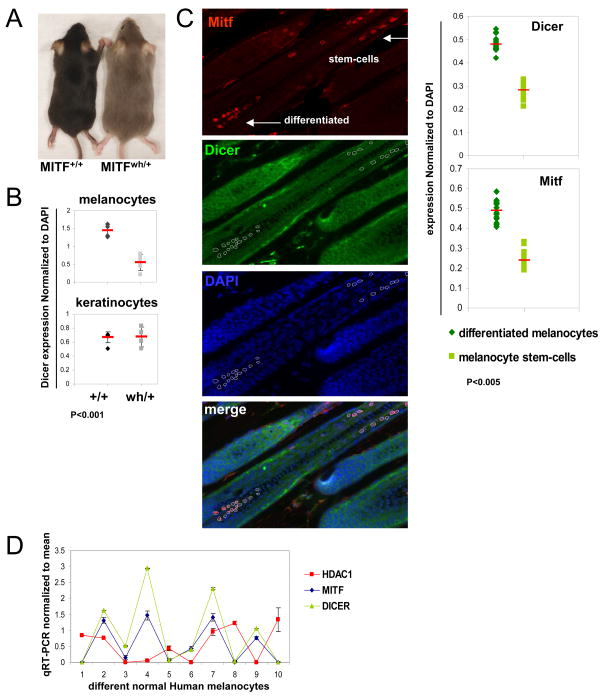
If MITF were transcriptionally regulating DICER, then DICER levels might parallel MITF in melanocytes in vivo. To test this, we examined skin from mutant mice (mitfwh/+) heterozygous for a dominant negative allele of Mitf, with the resulting Mitf net activity <50% of normal (Steingrimsson et al., 1994). Fur on these mice (on a C57BL/6 background) appears homogeneously grayish in color (Figure 5A). The coat color is due to a distinct pigment/color on each hair, and is not due to a mixture of black and white hairs (DEF, K. Wakamatsu, S. Ito, unpublished data). Moreover the color pattern is stable and not associated with progressive whitening (or progressive melanocyte cell death, data not shown). We carried out 3-color immunofluorescence staining for Mitf, Dicer, and DAPI (multiple independent skin sections per mouse), and quantified the staining intensity by normalizing the Dicer signal to DAPI from the same cells (Figure 5B and Figure S4A). In mitfwh/+ heterozygotes we observed a significant decrease in Dicer within melanocytes (identified as Mitf positive) as compared to Dicer expression in melanocytes of Mitf wt (mitf+/+) littermates, normalized to DAPI in the same cells. As a control, the identical analysis was carried out in adjacent keratinocytes (identified by their location within hair follicles and Mitf negativity). In keratinocytes no significant difference in Dicer expression was observed between mitfwh/+ and mitf+/+ genetic backgrounds (Fig 5B and Figure S4A).
Figure 5. MITF and DICER expression levels correlate in-vivo.
(A) Dorsal view of MITF (mitfwh/+) heterozygote mice compared to wt MITF (mitf+/+) mice, indicating altered pigmentation (which does not change upon aging).
(B) DICER expression intensity in hair follicle melanocytes and keratinocytes was measured by immunofluorescence staining of the indicated mouse skin sections (see also Figure S4). DICER intensity expression level was normalized to DAPI staining and plotted.
(C) Three-color co-immunofluorescence staining of C57BL/6 skin sections indicates expression of DICER (green, cytoplasmic), MITF (red, nuclear) and DAPI (blue, nuclear) (400× magnification). MITF-stained differentiated melanocytes and melanocyte stem cells, both appear red (arrows). The merged image shows co-localization of DICER and MITF in melanocytes (“merge”). DICER and MITF staining intensities in melanocyte stem cells and differentiated melanocytes were measured (circled areas), normalized to DAPI staining, and plotted. (D) MITF and DICER mRNA levels in primary human melanocytes from 10 different donors. HDAC1 was used as a control gene, expression levels were relative to Actin. Expression values for each gene were normalized to a mean of zero and a standard deviation of one.
To further study the expression of Dicer and Mitf in vivo, we examined melanocyte stem cells and differentiated melanocytes in C57BL/6 hair follicles. These melanocyte sub-populations were studied because Mitf and its melanogenic target genes are upregulated in the differentiated vs. stem-cell populations (Osawa et al., 2005), and because these populations can be discriminated by their distinct locations within the follicles (bulge vs. bulb regions) (Nishimura et al., 2005). Similar to the 3-color quantitative immunofluorescence technique used in Figure 5B, increased levels of both Mitf and Dicer were seen in differentiated melanocytes as compared to melanocyte stem cells (Figure 5C), consistent with the possibility that Mitf could regulate Dicer expression within the melanocyte lineage in vivo.
We also checked the correlation of MITF and DICER mRNA levels by performing qRT-PCR for MITF and DICER on a series of human primary melanocyte cultures generated from 10 different donors. Expression levels of MITF and DICER were normalized to β-actin and compared (Figure 5D). Highly parallel expression patterns were observed for MITF and DICER mRNAs in primary melanocytes, but not for a control gene (histone deacetylase 1 (HDAC1)). The parallel expression patterns are consistent with the hypothesis that MITF modulates DICER expression.
Targeted DICER knockout results in epidermal and hair follicle melanocyte depletion
Since a tight correlation between DICER and MITF expression was observed in vivo, we chose to study the physiologic significance of DICER in melanocytes in vivo, utilizing a conditional KO strategy. Mice harboring the floxed (fl) Dicer1 allele (Harfe et al., 2005) were bred to mice expressing Cre recombinase driven by the Tyrosinase (Tyr) promoter, which triggers ablation of floxed exons in melanocytes and melanoblasts (Delmas et al., 2003). A Dct-LacZ transgene was also incorporated (Mackenzie et al., 1997) to mark the melanocyte lineage.
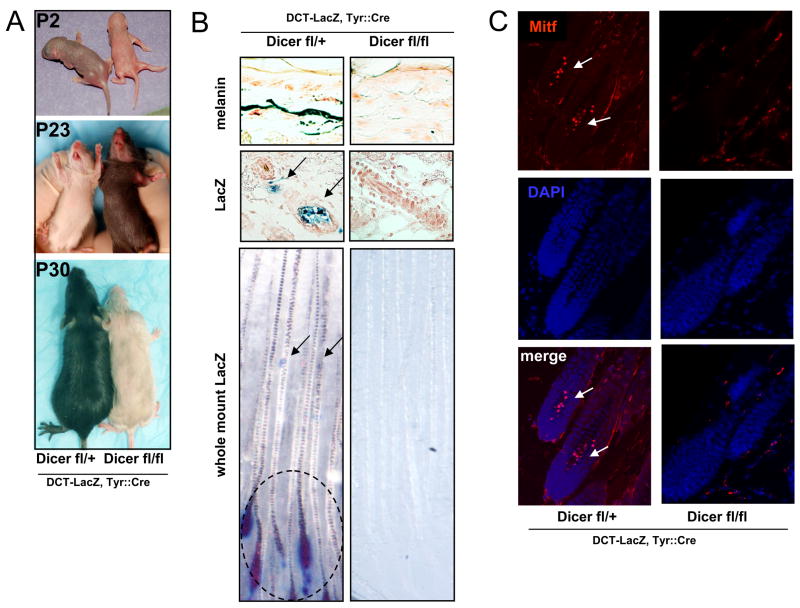
Newborn litters were of the expected size and Mendelian ratios, and appeared to have normal skin and formation of hair follicles irrespective of Dicer expression. As shown in Figure 6A the homozygous Dicer(fl/fl); Tyr::Cre; Dct-lacZ/° mice were easily distinguishable from the Dicer(fl/+); Tyr::Cre; Dct-lacZ/° control littermates, as they were virtually white from birth, except for the tips of the initial hairs on the dorsal trunk, and completely white after the 1st hair follicle cycle. Histopathologic analysis revealed no melanin in the skin of P30 Dicer(fl/fl); Tyr::Cre; Dct-lacZ/° mice as compared to Dicer(fl/+); Tyr::Cre; Dct-lacZ/° control littermates (Figure 6B).
Figure 6. Conditional knockout of DICER in melanocytes leads to melanocyte loss.
(A) Tyr-Cre mediated recombination of the Dicerflox allele in the melanocyte lineage. Homozygous Dicer conditional knockouts (KO) are compared to heterozygote littermates. Dorsal and ventral P2, P23 and P30 view of the indicated littermates.
(B) Fontana-Masson staining of melanin (upper) shows skin regions of the indicated littermates. Blue X-gal staining (middle row) of P42 littermate hair follicles revealed the presence of Dct-lacZ labeled melanocytes (arrows) in the indicated mouse skin sections. Whole mount blue X-gal staining (lower) indicates Dct-lacZ labeled melanocyte stem-cells (arrows) and differentiated melanocytes (circled) in skin sections of control Dicer(fl/+); Tyr::Cre; Dct-lacZ/° mice, but not in homozygously deleted mice.
(C) Immunofluorescence staining of P42 skin sections indicates expression of MITF (red, nuclear) (400x magnification). MITF-stained melanocyte stem-cells and differentiated melanocyte are indicated within hair follicles of control mice, but are absent from Dicer(fl/fl); Tyr::Cre; Dct-lacZ/° mice. DAPI-stained nuclei appear blue.
Profound loss of both melanocyte stem cells and differentiated melanocytes was observed by X-gal staining of skin sections and whole mount staining of homozygous Dicer(fl/fl); Tyr::Cre; Dct-lacZ/° mice, compared to heterozygote Dicer(fl/+); Tyr::Cre; Dct-lacZ/° littermate controls (Figure 6B). Skin sections of 6 week old (P42) mice were also subjected to immunostaining, using Mitf (red) as a melanocytic marker. As shown in Figure 6C no melanocyte/Mitf immunoreactivity was present in the homozygous Dicer(fl/fl); Tyr::Cre; Dct-lacZ/° mice, compared to heterozygous Dicer(fl/+); Tyr::Cre; Dct-lacZ/° control littermates. Multiple melanocyte markers (Dct-LacZ and Mitf) demonstrated that Dicer deficiency produces coat color depigmentation due to melanocyte loss, rather than absence of pigment within viable melanocytes. Thus Dicer deficiency significantly phenocopies the melanocyte survival phenotype of Mitf deficiency (it is uncertain whether the slightly later age of melanocyte disappearance in Dicer(fl/fl); Tyr::Cre mice relative to severe mitf mutants is due to delayed Dicer gene-deletion in this non-germline KO model or age-specific requirement for Dicer activity).
Two additional Cre recombinase-targeted mouse models were also generated, by breeding Dicer(fl/fl) mice to Cre recombinase driven by the dopachrome tautomerase (Dct) promoter (Guyonneau et al., 2004) or a 4-hydroxytamoxifen (OHT)-inducible Cre recombinase-estrogen receptor fusion transgene under the control of the tyrosinase promoter, designated Tyr::CreERt2 (Bosenberg et al., 2006). The Dct-CRE allele resulted in modest fur depigmentation from birth followed by progressive age-dependent depigmentation (Figure S5A-D). To test requirements for Dicer within epidermal (rather than hair follicle) melanocytes, the Dct-CRE allele was also crossed onto the Dicer(fl/fl); K14-SCF transgenic background (which contains epidermal melanocytes due to keratin-14 promoter driven expression of Stem Cell Factor (Kunisada et al., 1998)). Unlike darkly pigmented epidermis in control (Dicer(fl/+); Dct-Cre; K14-SCF; Dct-lacZ/°) littermates, homozygous mutant (Dicer(fl/fl); Dct-Cre; K14-SCF; Dct-lacZ/°) mice exhibited significant epidermal lightening (Fig S5B). The inducible Cre system utilized topical OH-tamoxifen administration on postnatal days 2–5 and resulted in a white phenotype in the treated region (with normal black pigmentation remaining in the adjacent area) and fur depigmentation seen at day P32 (Figure S6A, compare Dicer(fl/fl); Tyr::CreER; Dct-lacZ mice to Dicer(fl/+); TyR::CreER; Dct-lacZ control littermates). As with Dicer(fl/fl); Tyr::Cre mice, histologic analyses of the post-natal/progressive CRE-induced Dicer deletion models demonstrated that the depigmentation was associated with melanocyte cell loss (rather than absence of pigment within viable melanocytes) (Figure S5E–F and Figure S6B). Collectively these data demonstrate an essential role for Dicer in regulating viability of the melanocyte lineage postnatally as well as in earlier developmental stages.
In addition, primary mouse melanocyte cultures were generated from Dicer (fl/fl) mice and exposed to exogenous Cre-adenovirus. In this fashion complete excision of Dicer was induced, resulting in miRNA loss and death of melanocytes (Figure S6C–G). Similarly, siRNA-mediated DICER knockdown (KD) (Figure S7A) in primary human melanocytes also resulted in a decrease in miRNA expression (Figure S7B) as well as melanocyte cell death (Figure S7C–D). These results reveal an essential role of DICER in both human and mouse melanocyte survival.
Regulation of miR-17 and BIM by DICER during melanocyte differentiation
DICER’s survival role in melanocytes may be mediated by multiple miRNAs, but we attempted to identify one such DICER-processed miRNA which may contribute functionally to this viability phenotype. The pro-apoptotic factor BIM (BCL2L11, Isoforms 1 and 6 but not Isoform 9) was previously found to be a direct target of the miRNA-17~92 cluster (Fontana et al., 2008; Koralov et al., 2008; Ventura et al., 2008; Xiao et al., 2008). BIM has been shown to play a critical role in melanocyte survival, since BIM germline deletion profoundly rescues melanocytic cell death (coat-color) in the absence of Bcl-2 (Bouillet et al., 2001). In parallel to the miRNA expression profile, total RNA from stimulated melanocytes was subject to mRNA profiling. We found that miR-17 upregulation in differentiated melanocytes (Figure 2) correlated with BIM mRNA downregulation (Figure S3C).
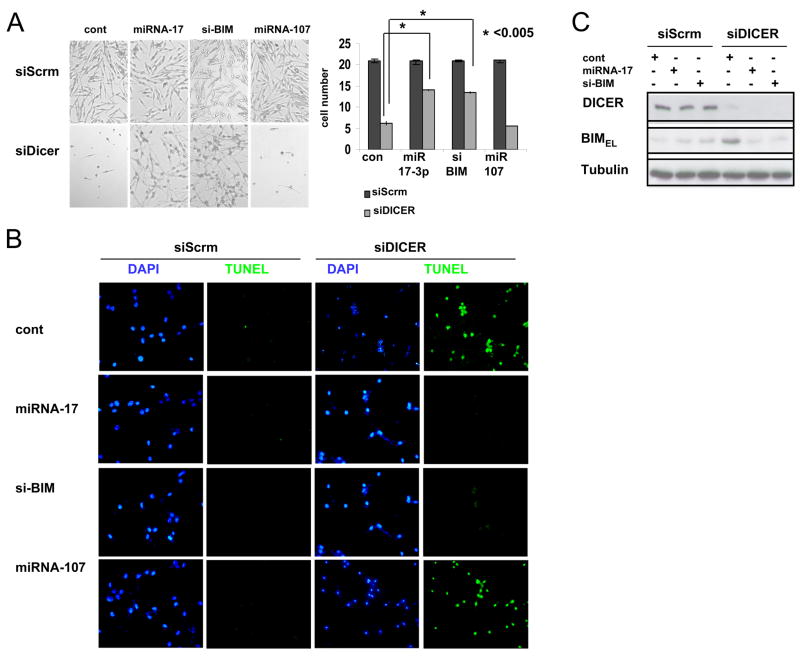
TUNEL assay demonstrated that KD of DICER resulted in significant loss of melanocytes (Figure 7B and Figure S7). Reconstitution of miR-17 or KD of BIM with siBIM in DICER KD cells significantly (albeit incompletely) rescued melanocyte survival/apoptosis (Figure 7A, B and C). miR-17 targeting of BIM may play a significant role in lineage survival, although it is likely that other miRNAs as well as apoptotic regulators also contribute to control of melanocytic survival.
Figure 7. The miRNA-17 cluster rescues melanocyte apoptosis in cells that are DICER-depleted via inhibition of BIM.
(A) Primary human melanocytes were transfected with either siDICER or scrambled siRNA (siScrm). At 48 hrs post-transfection, cells were transfected with either miRNA-17, miRNA-107, siBIM or miRNA/siRNA negative control (cont). Bright field images after 6 days of the indicated oligonucleotide transfections are shown, along with the average cell number of 3 wells from each treatment (A, right panel). Results are triplicates from one experiment (mean ± s.d.) and are representative of three independent experiments.
(B) Apoptosis detection assay (TUNEL) was performed on cells from (A). DAPI-stained nuclei appear blue.
(C) Primary human melanocytes from (A) were subject to immunoblot analysis using. Tubulin levels are shown as loading controls.
Discussion
We have identified a novel mechanism of DICER upregulation via its transcriptional targeting by the melanocytic transcription factor MITF during differentiation. We also demonstrate that DICER plays a crucial role in melanocyte survival involving, in part, post-transcriptional processing of miR-17 which leads to downregulation of the pro-apoptotic protein BIM.
Although it is well known that DICER is ubiquitously expressed among many cell types, regulation of its expression may control miRNA biogenesis in specific cellular contexts. It is plausible that DICER induction may be important for additional differentiation-specific activities, such as melanin production, maturation, or export. However it is also notable that MITF exhibits oncogenic activity in at least two human malignancies: melanoma (where it is amplified in 10–20% of cases) and Clear Cell Sarcoma (where it is activated by the EWS-ATF1 chimeric oncoprotein) (Davis et al., 2006; Garraway et al., 2005). Modulation of melanocyte survival via upregulation of DICER and miR-17, followed by suppression of BIM, may thus contribute to tumor survival, especially since BIM has been shown to play a vital role in mediating melanocytic cell death upon Bcl-2 deletion (Bouillet et al., 2001).
MITF regulates gene expression by binding to DNA as a homodimer or as a heterodimer with Tfe3, Tfeb, or Tfec proteins (MiT family). All these family members, bind to identical DNA sequences (Hemesath et al., 1994; Levy et al., 2006; Steingrimsson et al., 2004). Although MITF expression seems to be restricted to specific cell types, other members of the MiT family are thought to be more ubiquitously expressed. Therefore it will be important to examine potential participation of the other MiT family members in DICER regulation in the context of other tissues and cell types. It was previously shown that DICER cleavage activity is restricted to certain tissues and cell types, albeit by an unknown mechanism (Obernosterer et al., 2006). Here we report that transcriptional upregulation of DICER occurs during melanocyte differentiation due to direct targeting by MITF. These data thus highlight a prospective role of DICER and miRNAs in skin adaptation to sun exposure.
DICER and melanocyte survival
Since numerous human genes are predicted to be negatively regulated by miRNAs (Lewis et al., 2005), it is expected that DICER depletion would affect multiple miRNAs. In our work, restoring miR-17 in DICER-depleted melanocytes only partially rescued melanocytes from apoptosis. Thus additional pathways are very likely to be involved in melanocyte survival. Epidermal melanocytes give rise to malignant melanoma, the most aggressive form of skin cancer (Chin et al., 2006). It was recently shown that BIM expression is inhibited by BRAF signaling in human melanoma (Cartlidge et al., 2008). Somatic activating mutations in BRAF are detected in a major fraction of all melanomas (Davies et al., 2002). Here we identify BIM downregulation within melanocytes as residing downstream of MITF. Neither BIM nor the miR-17 cluster show significant copy number alterations in melanoma according to an analysis of SNP data from 101 melanoma samples (data not shown, False Discovery Rate-adjusted q-value = 1.0; (Lin et al., 2008)). The pathway consisting of cAMP/MITF/DICER/miR-17/BIM thus represents a lineage specific extension of the previously described roles of miR-17 and BIM in cell survival.
miRNA post-transcriptional regulation
Expression of specific miRNAs may be transcriptionally regulated upon cell stimulation (Ozsolak et al., 2008; Sempere et al., 2003). However, like other RNAs, miRNA expression can be also regulated at the post-transcriptional level in either a tissue-specific or a developmentally regulated fashion (Winter et al., 2009). For instance, pre-miR-38, pri-miR-142 and pre let-7 (Ambros et al., 2003; Chang et al., 2009; Heo et al., 2009; Thomson et al., 2006; Viswanathan et al., 2008; Wu et al., 2009; Wulczyn et al., 2007) are known cases of post transcriptional regulation in a tissue and cell type specific manner (Obernosterer et al., 2006). Analysis of gene expression in primary tumors indicates that the downregulation of miRNAs widely observed in cancer (Lu et al., 2005) is associated with a failure of the Drosha processing step (Thomson et al., 2006). The identification of DICER regulation by MITF within melanocytes suggests that similar lineage-specific regulatory mechanisms may also exist in other cell types.
Experimental Procedures
RNA purification and RT-PCR
Total RNA was purified using Trizol (Invitrogen) according to manufacturer’s instructions followed by treatment with RNase-free DNase (Qiagen). For miRs analysis, 10ng total RNA was used in a TaqMan miR assay, miR expression was normalized to RNU48 (Applied Biosystems). For mRNAs analysis, 100ng RNA was subjected to one-step RT-PCR using a QuantiTect RT-PCR kit (Qiagen) and iQ SYBRgreen Supermix (Biorad). Primer sequences are listed in Table S2.
miRNA expression analysis
miR profiling was done using forsk treated and untreated paired samples from 7 donors. Each sample had 8 RNU44 and 8 RNU48 technical replicate measurements for quality control and reference normalization. For each sample, we removed any RNU48 outliers by using the Weisberg t-test at a p-value cutoff of 0.05 (Seely et al., 2003), a method for detecting outliers in small data sets; miR CT values were then normalized by subtracting the mean RNU48 CT value of the resulting trimmed set. Some miR expression values could not be determined by PCR, and we considered only the miRs whose expression could be assayed in at least 3 out of 7 paired samples. Fold-changes in miR expression levels after forsk treatment in the seven samples were pooled together and ranked. Highly ranked miRs were then found; more precisely, p-values for the rank distribution of miR differential expression were computed using the hypergeometric test, as in the Redundant siRNA Activity (RSA) analysis (Konig et al., 2007). For each miR, we also performed a robust paired t-test on a trimmed set obtained by removing at most one outlier sample found by the Weisberg t-test at the 5% significance level (Seely et al., 2003). We obtained a final list of miRs differentially expressed at the 5% significance level of both hypergeometric test and t-test.
Cloning and Luciferase reporter assay
A fragment of the DICER promoter, 2488 nucleotides upstream to the transcription start site (TSS) was amplified from the BAC clone RP11-1143010 (Children’s Hospital Oakland Research Institute). KpnI and HindIII used to clone into PGL-3 basic vector (Promega) upstream of the luciferase reporter gene. Site directed mutagenesis was performed using the quick change method according to supplier’s protocols (Stratagene), on two M-Boxes (1384 and 934 nucleotides upstream of the TSS). The constructs were transfected into UACC62 or 501mel cells in 24-well plates (2 μg DNA/well) using Lipofectamine 2000 (GIBCO BRL) according to the manufacturer’s instructions. Constructs were cotransfected with the pRL-TK plasmids (Promega). 48 hours after transfection cells were lysed and assayed using dual-luciferase reagents (Promega). Promoter activity was normalized to the constitutively expressed Renilla. Primer sequences for cloning and mutagenesis are listed in Table S2.
Mice and whole mount X-gal staining
C57BL/6 mice were crossed with: Dicer(fl/fl) mice (gift from Clifford J. Tabin (Harfe et al., 2005)), Dopachrome tautomerase (Dct)-LacZ transgenic mice (a gift of Ian J Jackson (Mackenzie et al., 1997)), K14-Scf transgenic mice (a gift of T. Kunisada (Kunisada et al., 1998)), Tyr::Cre mice (Delmas et al., 2003) or Tyr::CreER mice (Bosenberg et al., 2006) or Dct-Cre mice (Guyonneau et al., 2004). The generated colony was backcrossed with C57BL/6. Samples were dissected, washed twice with PBS for 5 minutes at room temperature, washed and stained as described (Delmas et al., 2003), over-night staining at 37°C. Samples were then fixed in 2% Paraformaldehyde at 4°C for 30 min.
Immunohistochemistry
Animals were sacrificed by CO2 prior to skin sampling. Samples were formalin fixed paraffin-embedded. Sections were cut at 4 μm, dried at 37°C, deparaffinized in xylene, hydrated in a graded series of ethanol, and subjected to microwave EDTA antigen retrieval. Samples were blocked with 5% BSA/0.5% Tween-20/PBS, and detection of S100, MITF and DICER were achieved using mouse anti-DICER (AbCam), rabbit anti-S100 (Dako), mouse monoclonal C5 anti MITF, followed by incubation with Alexa-488 and Alexa-594 conjugated secondary antibodies (Invitrogen). Nuclear staining was performed with 4′-6-Diamidino-2-phenylindole (DAPI, Vector Laboratories Inc.).
Immunofluorescence image capturing and quantification
Images were captured using an Axio Observer A1 microscope and AxioCam MRc Camera (Carl Zeiss). Camera settings: black and white filter, resolution of full CD 1388×1040 pixels, X40 lens, same exposure time was used for each channel. Pictures were captured using fluorescence laser (450, 500, 600 nm), converted to the indicated color on AxioVision 4.7.1 and saved as 24 bit RGB. For quantification, the Outline tool was used to trace relevant cells and using the 8 bit (0–255 gray levels) signal quantification (represents a densitometric mean) in fluorescence on 3 channels (red, green, and blue –DAPI). Each red/green measurement was normalized to the blue value of the same traced area. Relative red/green expression values of different cells from the same picture were then compared. No compression between cells from different pictures was made, in order to avoid exposure differences.
Melanocyte Apoptosis
Primary melanocytes were seeded on chamber slides (Thermo Fisher Scientific), and transfected as indicated. Six or 8 days post transfection cells were fixed and used for fluorescent Tunnel staining according to manufacture’s protocol (R&D Systems).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge help from Dr. Rod Bronson with mouse pathology, Dr. Carl Novina for useful discussions, Dr. Ian Jackson for Dct-LacZ transgenic mice, Dr. T. Kunisada for K14-SCF transgenic mice, Dr. Clifford J. Tabin for Dicer Floxed mice, Dr. Ruth Halaban for 501mel melanoma cells, Maja Janas, Ana Hernandez, Dr. Jacob Hanna and members of the Fisher lab for help with numerous aspects of this work. JSS was supported in part by the Martin A. and Helen Chooljian Membership. DEF gratefully acknowledges grant support from NIH, The Melanoma Research Alliance, The Doris Duke Charitable Foundation, The Adelson Medical Research Foundation, and the Israel-US Binational Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Asada S, Takahashi T, Isodono K, Adachi A, Imoto H, Ogata T, Ueyama T, Matsubara H, Oh H. Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. American journal of physiology. 2008;295:H2512–2521. doi: 10.1152/ajpheart.00233.2008. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ciotti MT, Serafino A, Calissano P, Cogoni C. Dicer expression and localization in post-mitotic neurons. Brain research. 2007;1175:17–27. doi: 10.1016/j.brainres.2007.07.088. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Molecular cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne JP, Ballotti R. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. The Journal of cell biology. 1998;142:827–835. doi: 10.1083/jcb.142.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, Nogueira C, Horner JW, 2nd, Depinho R, Chin L. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–267. doi: 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM. Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Developmental cell. 2001;1:645–653. doi: 10.1016/s1534-5807(01)00083-1. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cai B, Chang SH, Becker EB, Bonni A, Xia Z. p38 MAP kinase mediates apoptosis through phosphorylation of BimEL at Ser-65. The Journal of biological chemistry. 2006;281:25215–25222. doi: 10.1074/jbc.M512627200. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, McMahon M. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment cell & melanoma research. 2008;21:534–544. doi: 10.1111/j.1755-148X.2008.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annual review of genomics and human genetics. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes & development. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D’Orazio J, Fung CY, Schanbacher CF, Granter SR, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, Larue L. Cre-mediated recombination in the skin melanocyte lineage. Genesis. 2003;36:73–80. doi: 10.1002/gene.10197. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. Beginning to understand microRNA function. Cell research. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Ewings KE, Wiggins CM, Cook SJ. Bim and the pro-survival Bcl-2 proteins: opposites attract, ERK repels. Cell cycle (Georgetown, Tex. 2007;6:2236–2240. doi: 10.4161/cc.6.18.4728. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, et al. Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Guyonneau L, Murisier F, Rossier A, Moulin A, Beermann F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Molecular and cellular biology. 2004;24:3396–3403. doi: 10.1128/MCB.24.8.3396-3403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO reports. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes & development. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Current topics in microbiology and immunology. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Kamada S, Shimono A, Shinto Y, Tsujimura T, Takahashi T, Noda T, Kitamura Y, Kondoh H, Tsujimoto Y. bcl-2 deficiency in mice leads to pleiotropic abnormalities: accelerated lymphoid cell death in thymus and spleen, polycystic kidney, hair hypopigmentation, and distorted small intestine. Cancer research. 1995;55:354–359. [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nature methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Kunisada T, Lu SZ, Yoshida H, Nishikawa S, Nishikawa S, Mizoguchi M, Hayashi S, Tyrrell L, Williams DA, Wang X, et al. Murine cutaneous mastocytosis and epidermal melanocytosis induced by keratinocyte expression of transgenic stem cell factor. The Journal of experimental medicine. 1998;187:1565–1573. doi: 10.1084/jem.187.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. The EMBO journal. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in molecular medicine. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell death and differentiation. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer research. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science (New York, NY. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24:448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Mackenzie MA, Jordan SA, Budd PS, Jackson IJ. Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Developmental biology. 1997;192:99–107. doi: 10.1006/dbio.1997.8738. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Nilsen TW. MicroRNAs, mRNAs, and translation. Cold Spring Harbor symposia on quantitative biology. 2006;71:531–535. doi: 10.1101/sqb.2006.71.043. [DOI] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell cycle (Georgetown, Tex. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad Roles for the miR-17–92 Cluster in Development and Disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer research. 2004;64:509–516. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- Neilson JR, Sharp PA. Small RNA regulators of gene expression. Cell. 2008;134:899–902. doi: 10.1016/j.cell.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science (New York, NY. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC. Bim: a novel member of the Bcl-2 family that promotes apoptosis. The EMBO journal. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA (New York, NY. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Egawa G, Mak SS, Moriyama M, Freter R, Yonetani S, Beermann F, Nishikawa S. Molecular characterization of melanocyte stem cells in their niche. Development (Cambridge, England) 2005;132:5589–5599. doi: 10.1242/dev.02161. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes & development. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, Fisher DE. alpha-Melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. The Journal of biological chemistry. 1998;273:33042–33047. doi: 10.1074/jbc.273.49.33042. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- Schier AF, Giraldez AJ. MicroRNA function and mechanism: insights from zebra fish. Cold Spring Harbor symposia on quantitative biology. 2006;71:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- Seely RJ, Munyakazi L, Haury J, Simmerman H, Rushing WH, Curry TF. Demonstrating the Consistency of Small Data Sets. Application of the Weisberg t-test for Outliers. BioPharm International. 2003:36–42. [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Developmental biology. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annual review of genetics. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nature genetics. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & development. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ. Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell. 1993;75:229–240. doi: 10.1016/0092-8674(93)80065-m. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science (New York, NY. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS ONE. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nature immunology. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura K, Kamada S, Ito S, Nakagawa K, Ichihashi M, Tsujimoto Y. Accelerated disappearance of melanocytes in bcl-2-deficient mice. Cancer research. 1996;56:3546–3550. [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Molecular and cellular biology. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nature reviews. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods in molecular biology (Clifton, NJ. 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. The EMBO journal. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.