Abstract
Genetic variation at the FKBP5 locus has been reported to affect clinical outcomes in patients treated with antidepressant medications in several studies. However, other reports have not confirmed this association. FKBP5 may regulate the sensitivity of the hypothalamic–pituitary–adrenal axis. We tested two FKBP5 single nucleotide polymorphisms (rs1360780 and rs3800373) in a sample of 246 geriatric patients treated for 8 weeks in a double-blind randomized comparison trial of paroxetine and mirtazapine. These two polymorphisms had previously been reported to predict efficacy in depressed patients treated with selective serotonin reuptake inhibitors such as paroxetine, and those treated with mirtazapine, an agent with both serotonergic and noradrenergic actions. However, we found no significant associations between these FKBP5 genetic variants and clinical outcomes. Neither mean Hamilton Depression Rating Scale scores nor time to remission or response were predicted by FKBP5 genetic variation. These results suggest that FKBP5 is unlikely to play a major role in determining antidepressant treatment outcomes in geriatric patients.
Keywords: antidepressive agents, pharmacogenetics, aged, FK506 binding protein 5
INTRODUCTION
There is continuing interest in identifying DNA markers that might predict response to antidepressant medications. Although many genetic associations with treatment response in depression have been reported, few have been replicated across samples. A seemingly robust finding was reported by Binder et al. [2004], who found an association between polymorphisms in the FK506 binding protein 5 (FKBP5) gene that encodes a glucocorticoid receptor-regulating co-chaperone of Hsp90, and response to antidepressant treatment in 294 individuals suffering from depressive disorders. The hypothalamic–pituitary–adrenal (HPA) axis has been implicated in antidepressant response [Binder et al., 2009], and FKBP5 regulates glucocorticoid receptor sensitivity [Binder et al., 2008; Zhang et al., 2008]. However, subsequent attempts to replicate this finding have resulted in both positive and negative associations between FKBP5 polymorphisms and antidepressant response. The original Binder cohort contained adult patients of widely different ages. To our knowledge, there are no published data on the effect of FKBP5 polymorphisms on treatment response in older adults with major depression. We attempted to replicate the Binder et al. [2004] finding in a cohort of 246 elderly patients with major depression who were treated with either paroxetine or mirtazapine in a double-blind, randomized clinical trial setting for 8 weeks. We found no evidence that variation at the FKBP5 locus affects outcomes in elderly patients treated with paroxetine or mirtazapine.
How to Cite this Article.
Sarginson JE, Lazzeroni LC, Ryan HS, Schatzberg AF, Murphy GM Jr. 2010. FKBP5 Polymorphisms and Antidepressant Response in Geriatric Depression. Am J Med Genet Part B 153B:554–560.
MATERIALS AND METHODS
The methods for recruiting the study population and collecting the clinical data have been previously described [Schatzberg et al., 2002; Murphy et al., 2003]. Briefly, the study group included 246 outpatients from 18 clinics in the U.S. with major depressive disorder who were treated with either mirtazapine or paroxetine in an 8-week, double-blind, randomized trial. IRB approval was obtained at each site, and all patients provided written informed consent. All patients (paroxetine N = 124; mirtazapine N = 122) were 65 years of age or older and free of major medical problems for at least 3 months. According to self-report, 20 patients were from minority backgrounds. For mirtazapine, these included one Asian, four African Americans, and two patients from other minority backgrounds. For paroxetine, there were two Asians, six African Americans, and five from other minority backgrounds. All other subjects were of self-reported European Caucasian ancestry. At screening, all met DSM-IV criteria for major depression (single or recurrent), had Mini Mental State Examination (MMSE) [Folstein et al., 1975] scores above the 25th percentile for their age, and had a Hamilton Depression Rating Scale 17-item [Hamilton 1967] (HDRS-17) score of at least 18. Patients were excluded for clinically significant laboratory abnormalities, unstable medical conditions, drug or alcohol abuse, psychosis, recent suicide attempt, and psychiatric conditions other than major depression, or antidepressant treatment within 7 days of commencing the study.
Initial treatment was 15 mg mirtazapine (one active capsule and one placebo capsule) or 20 mg of paroxetine (two 10 mg capsules) given each evening. On day 14, doses were increased to 30 mg of mirtazapine or paroxetine. At days 28 and 42, dose increases to 45 mg of mirtazapine or 40 mg of paroxetine were allowed if the patient had not achieved Clinical Global Impression Scale [Guy 1976] change scores indicating “much improved” or “very much improved.” Patients were evaluated after 1–4, 6, and 8 weeks of treatment. Mood was rated using the HDRS-17 and also the HDRS-21.
DNA extraction from whole blood was previously described [Murphy et al., 2003]. Genotyping was carried out using a Taqman SNP genotyping assay (Applied Biosystems, Foster City, CA; rs1360780: C___8852038_10 assay; rs3800373: C__27489960_10 assay) under standard conditions. We did not genotype the rs4713916 SNP, which showed a weaker effect in the Binder et al. [2004] study, and is in substantial linkage disequilibrium (LD) with rs1360780 and rs3800373 (D’ = ~0.8 in the Binder cohort).
Both the rs1360780 and rs3800373 variants were evaluated for conformity to Hardy–Weinberg equilibrium in our sample. LD between the two SNPs was assessed using Haploview version 4.0, using the full sample. The assessment was repeated using Caucasians only to check for population stratification effects on LD. Regional LD was assessed using the unrelated individuals from the HapMap CEU cohort.
A comparison across the genotype groups for demographic and clinical characteristics was carried out using Chi-squared tests or ANOVA. A complete comparison of demographic and clinical characteristics between drug treatment groups can be found in Schatzberg et al. [2002].
To test for genetic effects on treatment outcomes, we used SAS 9.1.3 routine PROC MIXED (SAS Institute, Cary, NC). For each SNP, two general linear models were used to assess differences in the mean change in HDRS-17 over time between genotypes. The first model was fit with genotype as the main affect and baseline HDRS-17 score as an additional fixed effect, and included a genotype by time interaction term and a quadratic term for time. The second model contained all the components of the first model as well as including amount of medication taken as a covariate and interaction terms for dose by time and dose by genotype. Amount of medication taken was based on pill counting at each clinic visit and is a measure of compliance. To test for remission, Kaplan–Meier survival analyses were performed. Remission was defined as a reduction of the HDRS-17 score to less than 7.
Although the HDRS-17 was the primary outcome measure in our clinical trial, to make a direct comparison to the Binder et al. [2004] results, we also performed analyses using the HDRS-21 score. Chi-squared tests were used to compare genotype frequencies at week 2 between those showing a 25% reduction in HDRS-21 and those not reaching this criterion. Survival analyses using a 50% reduction in HDRS-21 as an indicator of response or an HDRS-21 score of less than 10 as a criterion for remission were also carried out.
For all statistical testing, separate analyses were carried out for those treated with paroxetine and those treated with mirtazapine, initially using all subjects, and then Caucasians only to limit the potential for confounding due to population stratification.
Because we aimed to test whether the results of Binder et al. [2004] could be replicated in our sample, we made no a priori correction for type I error, in order to minimize the possibility of false negatives.
RESULTS
The genotype distributions of both rs1360780 and rs3800373 genotypes were shown to be in Hardy–Weinberg equilibrium. The rs1360780 and rs3800373 variants were found to be in strong, but not complete LD in the full cohort (D’ = 0.94, r2 = 0.83).
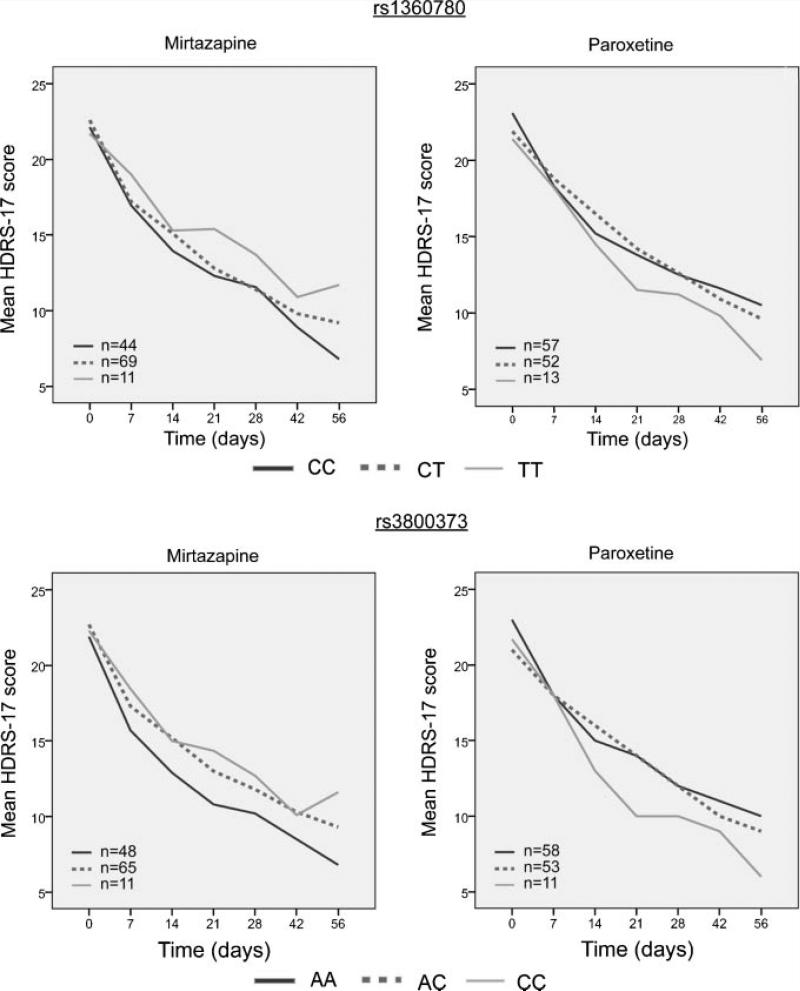
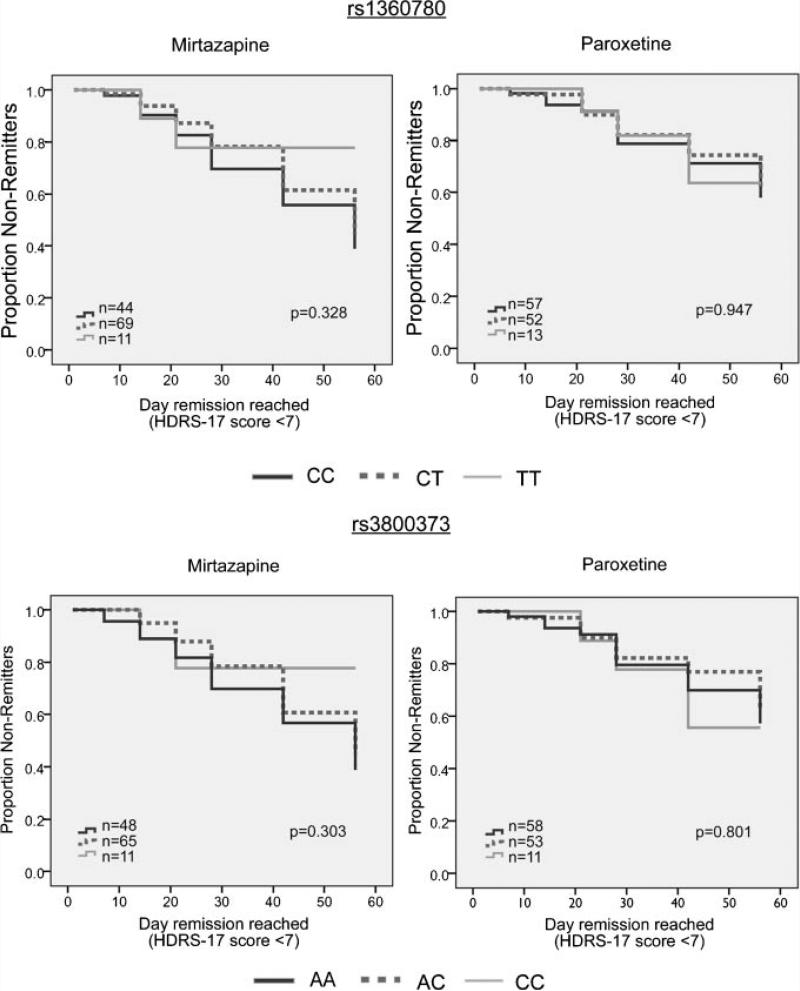
Potential confounding factors were investigated and no significant differences in age, sex, ethnicity, baseline HDRS-17 scores, or final drug dosage when comparing genotype groups within each treatment cohort were observed (Table I), except for slightly higher baseline HDRS-17 scores for carriers of the rs3800373 genotype among paroxetine-treated subjects. HDRS-17 score was the primary outcome measure for our clinical trial. Mixed model analysis of HDRS-17 scores over time, which included baseline scores as a covariate, did not yield any significant genotype effects for either of the two SNPs (Fig. 1). Likewise, survival analysis using time to an HDRS-17 score of 7 or less as a criterion for remission showed no effects of genotype (Fig. 2). Results were no different with the exclusion of non-Caucasian individuals.
TABLE I.
Demographic and Clinical Characteristics of the Sample
| Paroxetine |
Mirtazapine |
|||||||
|---|---|---|---|---|---|---|---|---|
| rs1360780 | CC (n = 57) | CT (n = 52) | TT (n = 13) | P-value | CC (n = 44) | CT (n = 69) | TT (n = 11) | P-value |
| Age (years, SD) | 73 (5.01) | 71 (5.21) | 74 (4.44) | 0.151 | 72 (5.98) | 72 (5.73) | 71 (4.21) | 0.734 |
| Gender (F/M) | 28/29 | 30/22 | 7/6 | 0.699 | 22/22 | 35/34 | 4/7 | 0.667 |
| Ethnicity (Caucasian/other) | 53/4 | 44/8 | 12/1 | 0.294 | 41/3 | 66/3 | 10/1 | 0.391 |
| Final daily dose (mg, SD) | 30.89 (9.56) | 28.80 (9.94) | 28.00 (9.56) | 0.419 | 32.2 (11.64) | 30.44 (11) | 31.74 (11.72) | 0.718 |
| Baseline HDRS-17 (SD) | 23.09 (3.71) | 21.92 (3.33) | 21.38 (3.5) | 0.118 | 21.84 (3.54) | 22.61 (3.56) | 21.73 (2.9) | 0.459 |
| Baseline HDRS-21 (SD) | 24.86 (4.63) | 23.35 (4.04) | 22.85 (3.76) | 0.114 | 23.32 (4.11) | 24.19 (4.25) | 23.00 (3.19) | 0.447 |
| Paroxetine |
Mirtazapine |
|||||||
|---|---|---|---|---|---|---|---|---|
| rs3800373 | AA (n = 58) | CA (n = 53) | CC (n = 11) | P-value | AA (n = 48) | CA (n = 65) | CC (n = 11) | P-value |
| Age (years, SD) | 72 (4.93) | 72 (5.41) | 74 (3.96) | 0.285 | 71.09 (4.18) | 72 (5.67) | 72 (6.06) | 0.855 |
| Gender (F/M) | 29/29 | 29/24 | 7/4 | 0.681 | 5/6 | 31/34 | 25/23 | 0.869 |
| Ethnicity (Caucasian/other) | 54/4 | 45/8 | 10/1 | 0.296 | 10/1 | 62/3 | 45/3 | 0.385 |
| Final daily dose (mg, SD) | 28.00 (7.82) | 28.80 (9.94) | 30.89 (9.56) | 0.419 | 31.74 (11.72) | 30.56 (11.13) | 31.88 (11.45) | 0.817 |
| Baseline HDRS-17 (SD) | 23.22 (3.73) | 21.66 (3.19) | 21.73 (2.90) | 0.049 | 21.55 (2.88) | 22.68 (3.52) | 21.85 (3.58) | 0.365 |
| Baseline HDRS-21 (SD) | 25.09 (4.77) | 22.96 (3.67) | 23.27 (3.90) | 0.029 | 22.82 (3.13) | 24.26 (4.18) | 23.33 (4.21) | 0.361 |
FIG. 1.
Mean HDRS-17 scores at each assessment point, stratified by genotype and by drug treatment. Mixed-model analysis showed no significant genotype by time interactions.
FIG. 2.
Survival curves showing time to reach HDRS-17 score of 7 or less, stratified by genotype and by drug treatment. Kaplan–Meier survival analysis log-rank P values are shown.
When using an HDRS-21 score of less than 10 as a criterion for remission, Kaplan–Meier analysis showed that among mirtazapine-treated patients, there was a marginally significant (P = 0.04) difference among the rs3800373 genotypes, with A allele carriers showing faster remission (Supplemental Fig. 1). Results for rs1360780 were similar. No differences in treatment response due to genotype were noted in the paroxetine treated cohort. Carriers of the rs1360780 C allele showed a trend toward faster response (50% reduction in HDRS-21 score) in the mirtazapine treated cohort (P = 0.089; Supplemental Fig. 2). A similar result (P = 0.096) was observed for rs3800373.
Using data from week 2 of the study, Chi-squared testing showed that frequencies in the genotype groups did not differ between those with 25% reduction in HDRS-21 and those not reaching this criterion for response, except for a marginally significant association for rs1360780 (Table II). Patients with the CC genotype were more frequent in the responder group than in the non-responder group (P < 0.046) for mirtazapine. Power analysis showed that at week 2 our combined sample of 246 subjects had 89% power to detect an effect of the magnitude reported by Binder et al. in their Max Planck sample, and 93% in their Ludwig Maximillian sample at the 0.05 level of significance.
TABLE II.
Comparison of Response Rates Between the Present Results and Those of Binder et al. [2004] at 2-Week Time Point
| Responder |
Percent responder | Non-responder |
Percent non-responder | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CC | CA | AA | CC | CA | AA | P-value | |||
| rs3800373 | |||||||||
| Combined | 9% (11) | 45% (56) | 46% (57) | 60% (124) | 9% (7) | 52% (42) | 40% (32) | 40% (81) | 0.627 |
| Mirtazipine | 6% (4) | 49% (34) | 46% (32) | 63% (70) | 12% (5) | 59% (24) | 29% (12) | 37% (41) | 0.166 |
| Paroxetine | 13% (7) | 41% (22) | 46% (25) | 57% (54) | 5% (2) | 45% (18) | 50% (20) | 43% (40) | 0.430 |
| Max-Planck Institute of Psychiatry | 14% (21) | 33% (48) | 53% (77) | 63% (146) | 0% (0) | 41% (35) | 59% (51) | 37% (86) | 0.00003 |
| Ludwig Maximillian University | 9% (4) | 53% (23) | 37% (16) | 51% (43) | 2% (1) | 36% (15) | 62% (26) | 49% (42) | 0.053 |
| Responder |
Percent responder | Non-responder |
Percent non-responder | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | TT | CT | CC | P value | |||
| rsl360780 | |||||||||
| Combined | 8% (10) | 48% (60) | 44% (54) | 60% (124) | 12% (10) | 51% (41) | 37% (30) | 40% (81) | 0.478 |
| Mirtazipine | 4% (3) | 53% (37) | 43% (30) | 63% (70) | 15% (6) | 61% (25) | 24% (10) | 37% (41) | 0.046 |
| Paroxetine | 13% (7) | 43% (23) | 44% (24) | 57% (54) | 10% (4) | 40% (16) | 50% (20) | 43% (40) | 0.835 |
| Max-Planck Institute of Psychiatry | 15% (22) | 34% (50) | 51% (75) | 63% (147) | 1% (1) | 42% (36) | 57% (49) | 37% (86) | 0.00048 |
| Ludwig Maximillian University | 12% (5) | 56% (24) | 33% (14) | 50% (43) | 5% (2) | 37% (16) | 58% (25) | 50% (43) | 0.02 |
Response is defined as a 25% reduction in HDRS-21 score. Results are given as percent responder or non-responder across rows, with the number of genotype carriers shown in parentheses. Binder et al. [2004] results are indicated by rows labeled as Max-Planck, Institute of Psychiatry, and Ludwig Maximillian University. The format of this table is modeled after that of Binder et al. [2004].
Given that older patients may respond more slowly to antidepressants than younger ones, we also tested for an association between FKBP5 genotype and 25% reduction in HDRS-21 at week 3 of treatment. The results at week 3 were similar to those for week 2 (Supplemental Table I), and did not support an association between FKBP5 genotype and clinical improvement. We also analyzed the effects of genotype on HDRS-21 scores over time using the mixed model analysis. There were no significant genotype effects for either SNP on change in HDRS-21 score over time (Supplemental Fig. 3). Similar results were obtained for both the full cohort and the Caucasians alone.
DISCUSSION
These results do not support the hypothesis that variation in the FKBP5 locus is useful in predicting response to antidepressant treatment. Our results differ from those of Binder et al. [2004], who found an association between FKBP5 variation and response to antidepressant treatment in a German cohort of 294 individuals suffering from depressive disorders. The strongest association in their data was to rs1360780, a SNP located in the second intron of FKBP5. Patients with the rs1360780 TT homozygous genotype showed the fastest response over a 5-week treatment course. In our study, analysis of HDRS-17 scores showed no genetic effects on antidepressant efficacy. The only marginally significant associations were observed with the HDRS-21 scale, and these were actually in the opposite direction from those reported by Binder et al. [2004]. That is, patients with the TT genotype for rs1360780 responded more slowly to mirtazapine, as did carriers of the CC genotype for rs3800373. Further, these associations would not withstand correction for multiple testing. Testing for genetic effects at week 2 of treatment, as they did, produced one marginally significant result, an enrichment of the non-responder group with the rs1360780 TT genotype among mirtazapine treated patients. In contrast, Binder et al. found an enrichment of non-responder group with the rs1360780 CC genotype. Power analysis showed that given our sample size, we should have been readily able to detect a genetic effect of the magnitude reported by Binder et al.
Although our results do not replicate those of Binder et al. [2004], our study differed from theirs in that the mean age of subjects in our study was older. In addition, at least some of the patients in the Binder et al. study appear to have been hospitalized, whereas our sample was entirely outpatients. The Binder et al. [2004] sample was heterogeneous, and included patients with major depression, dysthymic disorder, and bipolar disorder, whereas our sample included only patients with major depression. Their was a naturalistic pharmacogenetic study rather than a clinical trial, and patients were treated with antidepressant drugs of the doctor's choice, and some of the patients were also treated with mood stabilizers or antipsychotics in addition to antidepressants, whereas our patients took no psychotropics other than the two study medications. One possibility is that differences in antidepressant response rates among patients reported by Binder et al. [2004] may have been due to clinical variability rather than genetic effects.
A second German naturalistic study [Kirchheiner et al., 2008] supported the finding of Binder et al. [2004] by showing a trend (P = 0.04; odds ratio, 1.8) towards improved drug response for individuals with at least one rs1360780 T allele. The study also showed rs1360780 and rs3800373 to be in strong LD. In addition, Lekman et al. [2008] found an association between rs1360780 and a diagnosis of major depression in white non-Hispanics in the STAR*D cohort. They also found modest evidence for an association between the FKBP5 SNP rs4713916, which is in strong LD with the markers we studied, and treatment response for the selective serotonin reuptake inhibitor citalopram. A reasonable caveat with all pharmacogenetic studies utilizing the STAR*D sample is that only a subset of the total patient sample contributed DNA. This raises concerns about selection bias in the sample available for genotyping.
Other studies have not found an association between FKBP5 polymorphisms and treatment outcomes. A Spanish study [Papiol et al., 2007] using citalopram found no evidence for FKBP5 genotypes as predictors of treatment outcomes, nor did a Taiwanese study [Tsai et al., 2007] utilizing fluoxetine. A German case–control study by Gawlik et al. [2006] reported a nominal association (P = 0.045) between a haplotype consisting of rs1360780T and rs3800373T and duration of affective psychosis, but no association with age at onset, age of first hospitalization, or frequency and period of in-patient treatment. Pharmacogenetic effects were not tested by Gawlik et al. [2006].
It is possible that differences between our study and that of Binder et al. [2004] could be due to the differing age distributions of the two samples. Structural and functional changes in the brain with normal aging are well documented [Mrak et al., 1997; Sullivan and Pfefferbaum, 2007; Grady, 2008], as well as age-related changes in HPA axis function and other endocrine functions [Chahal and Drake 2007]. Further, at least some cases of geriatric depression may have an ischemic etiology [Taylor et al., 2006]. Whether these age-related changes could negate an effect of FKBP5 genetic variation on antidepressant treatment outcomes is unknown. Some, but not all, studies of antidepressant response in the elderly have indicated a slower response than in younger subjects [Whyte et al., 2004; Mandelli et al., 2007]. However, in our sample, there was no evidence for a marked difference among FKBP5 genotype groups at any time point. In fact, our data cover 8 weeks of treatment, whereas that of Binder et al. [2004] appears to cover 5 weeks of treatment.
In summary, in the first test of FKBP5 polymorphisms as pharmacogenetic predictors in patients treated in a double-blind randomized clinical comparison trial, we found no evidence that these genetic markers contribute to treatment outcomes with paroxetine or mirtazapine. Our study involved only older patients, whereas other studies reporting a positive effect involved younger individuals. Additional work is needed to determine the value, if any, of FKBP5 variation in clinical practice.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charlotte Kremer, Steven Hollander, Heidi Rodrigues, Peter Schot, Yan He, Clara Poon, and Nina Pascoe for assistance. Because the original clinical trial was sponsored by Organon, Inc., we permitted employees of Schering-Plough, Inc. to review the manuscript, but they had no comments or changes.
Grant sponsor: National Institute of Mental Health; Grant sponsor: The Department of Veterans Affairs; Grant sponsor: NARSAD; Grant sponsor: Organon, Inc.
Footnotes
Additional Supporting Information may be found in the online version of this article.
DISCLOSURES
Dr. Sarginson: Past 5 years, none; Current, none. Dr. Lazzeroni: Past 5 years, consulting for Roche Molecular Systems; Current, none. Ms. Ryan: Past 5 years, none; Current, none. Dr. Schatzberg: Past 3 years: Consultant for BrainCells, CeNeRx, Corcept, Eli Lilly, Forest Labs, Merck, Neuronetics, Novartis, Pathway Diagnostics, Pfizer, PharmaNeuroBoost, Quintiles, Sanofi-Aventis, Synosis, Xytis, Wyeth. Equity in BrainCells, CeNeRx, Corcept (co-founder), Forest, Merck, Neurocrine, Pfizer, PharaNeuroBoost, Somaxon, Synosis. Intellectual Property, named inventor on pharmacogenetic use patents on prediction of antidepressant response. Speaking, GlaxoSmithKline, Roche. Past 1 year: Consultant for BrainCells, CeNeRx, Eli Lilly, Neuronetics, Pfizer, PharmaNeuroBoost, Quintiles, Sanofi-Aventis, Xytis. Equity in BrainCells, CeNeRx, Corcept (co-founder), Forest, Merck, Neurocrine, Pfizer, PharaNeuroBoost, Somaxon, Synosis. Intellectual Property, named inventor on pharmacogenetic use patents on prediction of antidepressant response. Speaking, GlaxoSmithKline, Roche. In the past Dr. Schatzberg was a consultant for Organon, Inc. Dr. Murphy: Past 5 years, research support from Organon and Wyeth, consultant for Organon, Genaissance, and Philips Lytle and Allen Matkins law firms. Royalty income from a pharmacogenetic test licensed by Stanford University. Current, none.
REFERENCES
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Kunzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Kohnlein O, Dabitz H, Bruckl T, Muller N, Pfister H, Lieb R, Mueller JC, Lohmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36(12):1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. J Am Med Assoc. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Kunzel HE, Nickel T, Kern N, Pfennig A, Majer M, Uhr M, Ising M, Holsboer F. HPA-axis regulation at in-patient admission is associated with antidepressant therapy outcome in male but not in female depressed patients. Psychoneuroendocrinology. 2009;34(1):99–109. doi: 10.1016/j.psyneuen.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Chahal HS, Drake WM. The endocrine system and ageing. J Pathol. 2007;211(2):173–180. doi: 10.1002/path.2110. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Gawlik M, Moller-Ehrlich K, Mende M, Jovnerovski M, Jung S, Jabs B, Knapp M, Stoeber G. Is FKBP5 a genetic marker of affective psychosis? A case control study and analysis of disease related traits. BMC Psychiatry. 2006;6:52. doi: 10.1186/1471-244X-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady CL. Cognitive neuroscience of aging. Ann NY Acad Sci. 2008;1124:127–144. doi: 10.1196/annals.1440.009. [DOI] [PubMed] [Google Scholar]
- Guy W. ECDEU assessment manual for psychopharmacology, Revised 1976, DHEW Publication No. (ADM) 1976:76–338. [Google Scholar]
- Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol. 1967;6(4):278–296. doi: 10.1111/j.2044-8260.1967.tb00530.x. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Lorch R, Lebedeva E, Seeringer A, Roots I, Sasse J, Brock-moller J. Genetic variants in FKBP5 affecting response to antidepressant drug treatment. Pharmacogenomics. 2008;9(7):841–846. doi: 10.2217/14622416.9.7.841. [DOI] [PubMed] [Google Scholar]
- Lekman M, Laje G, Charney D, Rush AJ, Wilson AF, Sorant AJ, Lipsky R, Wisniewski SR, Manji H, McMahon FJ, Paddock S. The FKBP5-gene in depression and treatment response—An association study in the sequenced treatment alternatives to relieve depression (STAR*D) cohort. Biol Psychiatry. 2008;63(12):1103–1110. doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelli L, Serretti A, Zanardi R, Rossini D, De Ronchi D, Tarricone I, Colombo C. Antidepressant response in the elderly. Psychiatry Res. 2007;152(1):37–44. doi: 10.1016/j.psychres.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Mrak RE, Griffin ST, Graham DI. Aging-associated changes in human brain. J Neuropathol Exp Neurol. 1997;56(12):1269–1275. doi: 10.1097/00005072-199712000-00001. [DOI] [PubMed] [Google Scholar]
- Murphy GM, Jr, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry. 2003;160(10):1830–1835. doi: 10.1176/appi.ajp.160.10.1830. [DOI] [PubMed] [Google Scholar]
- Papiol S, Arias B, Gasto C, Gutierrez B, Catalan R, Fananas L. Genetic variability at HPA axis in major depression and clinical response to antidepressant treatment. J Affect Disord. 2007;104(1–3):83–90. doi: 10.1016/j.jad.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Schatzberg AF, Kremer C, Rodrigues HE, Murphy GM., Jr Double-blind, randomized comparison of mirtazapine and paroxetine in elderly depressed patients. Am J Geriatr Psychiatry. 2002;10(5):541–550. [PubMed] [Google Scholar]
- Sullivan EV, Pfefferbaum A. Neuroradiological characterization of normal adult ageing. Br J Radiol. 2007;80(SpecNo. 2):S99–S108. doi: 10.1259/bjr/22893432. [DOI] [PubMed] [Google Scholar]
- Taylor WD, Steffens DC, Krishnan KR. Psychiatric disease in the twenty-first century: The case for subcortical ischemic depression. Biol Psychiatry. 2006;60(12):1299–1303. doi: 10.1016/j.biopsych.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Hong CJ, Chen TJ, Yu YW. Lack of supporting evidence for a genetic association of the FKBP5 polymorphism and response to antidepressant treatment. Am J Med Genet Part B. 2007;144B(8):1097–1098. doi: 10.1002/ajmg.b.30246. [DOI] [PubMed] [Google Scholar]
- Whyte EM, Dew MA, Gildengers A, Lenze EJ, Bharucha A, Mulsant BH, Reynolds CF. Time course of response to antidepressants in late-life major depression: Therapeutic implications. Drugs Aging. 2004;21(8):531–554. doi: 10.2165/00002512-200421080-00004. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clark AF, Yorio T. FK506-binding protein 51 regulates nuclear transport of the glucocorticoid receptor beta and glucocorticoid responsiveness. Invest Ophthalmol Vis Sci. 2008;49(3):1037–1047. doi: 10.1167/iovs.07-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.