Abstract
An effective exciton Hamiltonian for all amide bands is used to calculate the linear absorption and photon echo spectra of a 17 residue helical peptide (YKKKH17). The cross peak bandshapes are sensitive to the inter-band couplings. Fluctuations of the local amide frequencies of the all amide fundamental and their overtone and combination states are calculated using the multipole electric field induced by environment employing the electrostatic DFT map of N-methyl acetamide. Couplings between neighboring peptide units are obtained using the anharmonic vibrational Hamiltonian of glycine dipeptide (GLDP) at the BPW91/6-31G(d,p) level. Electrostatic couplings between non-neighboring units are calculated by the fourth rank transition multipole coupling (TMC) expansion including 1/R3 (dipole-dipole), 1/R4 (quadrupole-dipole), and 1/R5 (quadrupole-quadrupole and octapole-dipole) interactions.
Keywords: 2-Dimensional Infrared-Spectroscopy, molecular dynamics, Multidimensional Femtosecond Spectroscopies, Amide-I Modes, Amide-II Modes, Amide-III Modes, Amide-A Modes, Molecular-Dynamics, Alpha-Helix, Proteins, 310-Helix, Spectra
1. Introduction
The amide I, II, III and A vibrational modes of proteins have infrared absorption[1,2] around 1200 cm−1, 1500 cm−1, 1700 cm−1, and 3500 cm−1, respectively. These vibrations are localized on the amide bonds. Their sensitivity to hydrogen bonding, dipole-dipole interactions and peptide backbone geometry provides useful indicators of secondary-structural changes [3–7]. The amide I and A are mainly the CO and N-H bond stretch, respectively. The amide II and III bonds are mixtures of the C-N stretch and H-N-C bend. Coherent ultrafast infrared spectroscopy has been applied to probe protein structure [8–10]. Two-dimensional infrared cross-peaks of amide I with other amide modes (A and II) have been reported in model systems of the amide bond [11–13]. Recent simulations [14] showed that the cross-peak bandshapes are sensitive to the correlated hydrogen bond dynamics at the two atom sites where the two amide vibrations reside. This makes them most suitable for probing the local protein environment. The amide A vibration is not resolved in the infrared due to the overlapping broad absorption of water. The cross peaks thus provide a direct window into this vibration.
Most simulation effort had so far focused on the highly-localized amide I vibrations [15–20] which are the easiest to model. Ab initio calculations of all amide band normal modes have been reported [21,22]. The peptide force field depends on frequencies of the local amide vibrations as well as their couplings between neighboring and non-neighboring amide units. Torii and Tasumi had constructed a map of amide I couplings between neighboring amide units based on Hartree-Fock calculations of a glycine dipeptide (GLDP) for various Ramachandran angles (ψ and φ) [15]. They also calculated the amide I couplings between non-neighboring amide units using the transition dipole coupling mechanism (TDC). Neighboring amide I maps have subsequently been developed [23,18] based on normal mode calculations of GLDP at higher computational levels (MP2 and DFT) by employing the Hessian reconstruction. Non-neighboring couplings were calculated by the interaction of vibration-induced partial charges determined by normal mode analysis of NMA [18]. Side-chain contributions to the amide I frequencies of β–hairpins have been investigated as well [19,20]. Hydrogen-bonding with the surrounding water was found to affect the amide I local mode frequencies. All of these studies are restricted to the amide I mode and neglect the anharmonic character of the inter-site couplings as well as contributions of inter-amide couplings to the diagonal and off-diagonal anharmonicity of the amide modes.
We have developed an electrostatic DFT map (EDM) [24,25] that includes the fundamental, the diagonal and off-diagonal anharmonic frequency fluctuations of all amide states in NMA. The map provides a first-principles effective vibrational Hamiltonian which includes the fundamental, overtone, combination frequencies and transition moments of the amide III, II, I and A. It is based on vibrational eigenstate calculations of a 6th order anharmonic DFT potential in the presence of up to 3rd rank multipole (octapole) electric field. We found that higher multipoles of the electric field make significant contributions to the frequency fluctuations and line broadening [24]. The EDM reproduces the experimental amide I and II anharmonicities [13].
Couplings between neighboring peptide units are obtained using a quartic anharmonic vibrational DFT potential of glycine dipeptide (GLDP) at the BPW91/6-31G(d,p) level for various Ramachandran angles. Electrostatic model is used for couplings between non-neighboring units. We have expanded the Transition charge density couplings (TCDC) to 4th rank in multipoles including ∼ R−3 (dipole-dipole), ∼ R−4 (dipole-quadrupole), ∼ R−5 (quadrupole-quadrupole and dipole-octapole) interactions. The higher order multipoles were found critical for smaller R for the relatively delocalized bending amide modes (II and III). These are usually neglected[15,18]. The couplings were expanded as bilinear products of local amide modes(LAMs) of the two sites, by neglecting the electronic anharmonicities.
The peptide Hamiltonian was expanded in a basis of Local Amide States (LAS); the eigenstates of the local Hamiltonian which consist of the localized amide I, II, III and A. These include the fundamental and their overtone and combination states (14 states per each site). The coupling matrix elements are calculated between all amide fundamentals, overtones, and combinations localized in neighboring and non-neighboring peptide units. The LAS anharmonicities contribute to the anharmonicities in these couplings, which are included in the calculations.
2. The Simulation Protocol
The anharmonic vibrational Hamiltonian of the peptide was expanded in the form.
| (1) |
Here qmi represent the 5 local amide modes (LAM) of the amide site m and Mmi and pmi are their masses and momentums. The first term representing the local amide sites is expanded up to 6th order in the local amide modes (LAMs). The anharmonic force field depends on the fluctuating multipole electric field generated by the other parts of the protein and water, it is calculated based on the electrostatic DFT map of the NMA [25] combined with a MD trajectory. The second term represents the couplings between the neighboring amide units, expanded to 4th order in LAMs. Neighboring couplings are obtained by electronic structure calculations of glycine dipeptide (GLDP), parametrized with respect to the Ramachandran angles φ and ψ. The last term includes the Coulombic couplings between non-neighboring units and approximated by the transition multipole couplings up to 4th rank. They are bilinear in LAMs of the two sites electronic anharmonicities are neglected.
The Hamiltonian was first diagonalized by neglecting all inter-unit couplings (only the first term in Eq. (1) is included) to generate the 14 local amide states (LAS) per amide unit (4 amide fundamentals, 4 overtone, and 6 combinations). The effective vibrational Hamiltonian is finally recast using the LAS.
| (2) |
| (3) |
| (4) |
where B̂ma and are the Pauli exciton creation and annihilation operators for to the transition between the ground state and the LAS ma. 14 × 16 creation and annihilation operators are defined in total. Couplings between non-neighboring amide units were expressed to quartic order in B̂ma and .
We have calculated the eigenstates of this effective anharmonic Hamiltonian and simulated the infrared bands of the amide I and A and three-pulse photon echo signals of the amide I and A cross peak region of a 17 residue helical peptide Ac-YAAKAAAAKAAAAKAAH-NH2 (YKKKH17) (Fig. 1) in water. Since YKKKH17 is small and preseves its helical structure in water, it is a good candidate for investigating the 2D photon echo signatures of the amide couplings without further complications. We have used the MD trajectory of a one YKKKH17 and 4330 H2O molecules as reported in Ref. [26]. Four chloride ions were added to make the system neutral. CHARMM27 [27] and TIP3 force fields were used with a 12Å cut-off of the Lennard-Jones interactions and the Ewald sum of the electrostatic interactions. The CHARMM package [27] was used and a 1 ns trajectory was sampled with 1 fs time step.
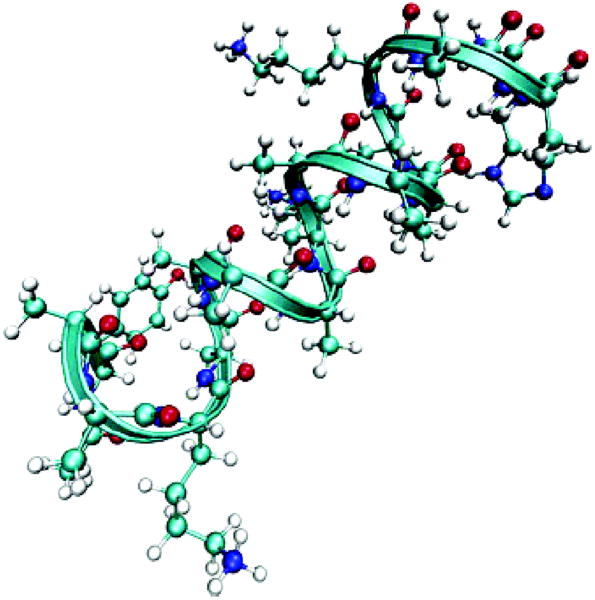
Fig. 1.
Structure of 17 residue helical peptide (YKKKH17) [26].
The eigenstates of each snapshot were calculated by expansion in Hartree products of LAS since YKKKH17 has 16 amide units. Each amide unit has 15 LAS (ground state, + 14 states) and the basis set is | ψ〉 = |a1〉 |a2〉 ⋯ |a16〉 (ai = 0, ⋯, 14). The eigenstates were calculated by numerical diagonalization of the Hamiltonian. The sum over states expressions and the SPECTRON code [26] were used to simulate the spectra. These were averaged over 100 snapshots and a 5.5 cm−1 homogeneous linewidth was added to all transitions.
3. Results and Discussion
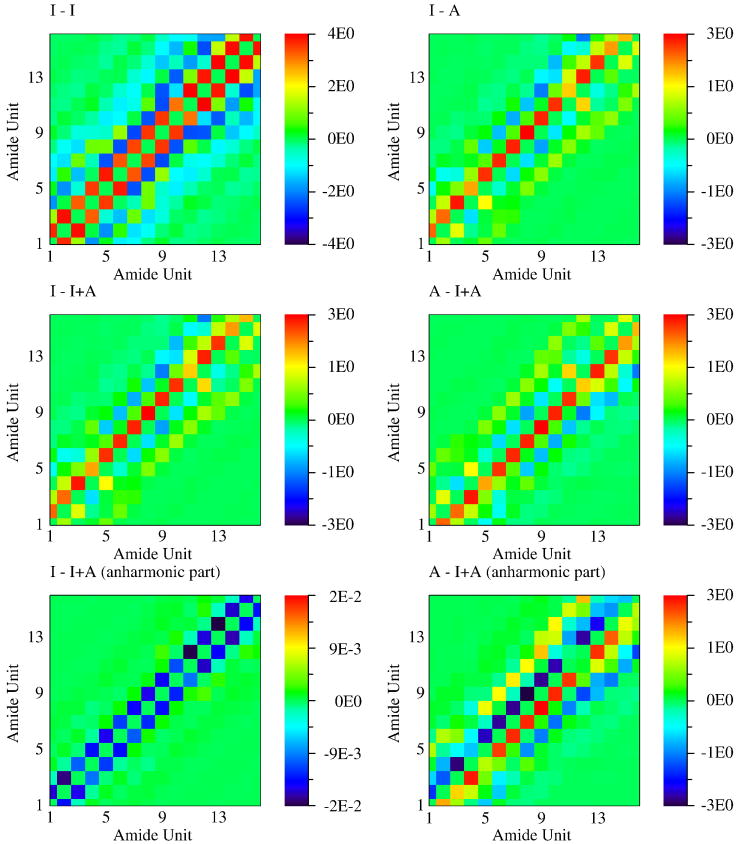
The vibrational couplings between the amide states in different amide units are shown in Fig. 2. The couplings of the amide I fundamental neighboring units are always positive, since their transition dipoles are parallel. Amide I couplings between non-neighboring units are negative since the two dipoles have close to head-to-tail configurations due to the peptide helical structure. The coupling patterns of amide I - A fundamentals are similar to amide I - I couplings, but are weaker due to the smaller transition dipole moment of amide A. The couplings between the amide I or amide A fundamentals and amide I + A combination states show a different pattern.
Fig. 2.
Vibrational couplings between local amide states. sinh(Jmn,ab) of Eq. (3) in cm−1.
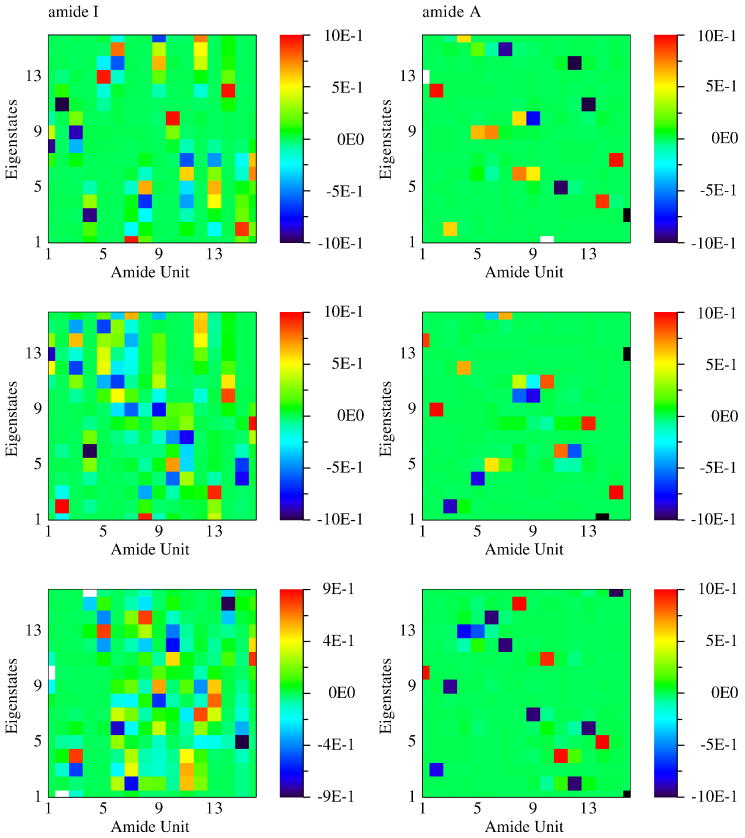
For a single MD snapshot the 16 eigenstates in the frequency region around 1700 cm−1 are mostly linear combinations of 16 local amide I fundamental states. 16 eigenstates in the 3500 cm−1 frequency region come from local amide A fundamental states. The contributions of each local amide vibration to these eigenstates obtained from 3 snapshots are shown in Fig. 3. The Amide I eigenstates are delocalized over 3 to 5 amide bonds. YKKKH17 has bulky residues (tyrosine and histidine) on the both terminals and rather compact residues (mostly alanine with a few lysine) in the middle. The peptide has a α–helical structure on the both terminal, but a 310–helical structure between residue 4 and 11. In the 310–helical part the hydrogen bondings are formed between n and n + 3 residue which cause strong couplings between the units separated by 3. The α–helical part has the hydrogen bondings between n and n + 4 residue resulting in couplings between units separated by 4 amino acid. The amide I eigenstates are thus delocalized along the helical axis rather than the peptide backbone. The amide A eigenstates are more localized on 1 to 2 neighboring amide bonds since their transition dipole moments are smaller and have different directions from amide I. The amide I fundamental eigenstates are more delocalized than the amide A. This may be attributed to the larger transition dipole moments.
Fig. 3.
Contributions of each amide bond to the 16 eigenstates of amide I and A fundamental states obtained from three MD snapshots.
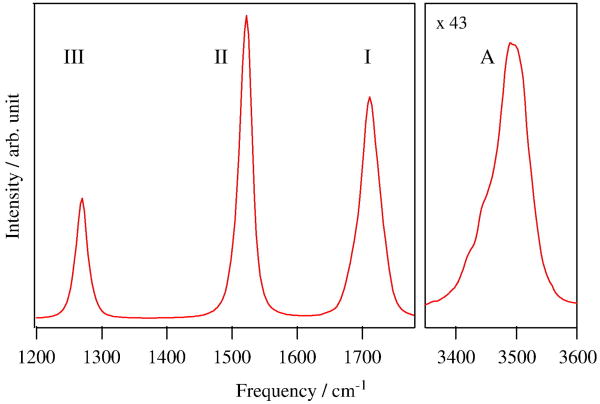
The simulated absorption lineshapes of the amide I, II, III and A regions are shown in Fig. 4. All amide modes have a single peak and a almost symmetric bandshape except for the amide A which has a lower frequency tail. This stems from a helical structure of the peptide which results in a homogeneous environment for all amide units. The amide II intensity is weaker than the amide I, which is consistent with experiment. However the simulated amide II bandwidth is narrower and its peak height is higher than experiment, which is ascribed to that the amide II frequency fluctuation of NMA is underestimated due to insufficient electrostatic sampling ([25]) resulting in the narrower amide II bandwidth. The amide A has the broadest band width due to the largest local amide A frequency fluctuation (Fig. 8 of Ref. [25]). This may be rationalized since the weaker N-H bond is strongly affected by the hydrogen bond formation with water.
Fig. 4.
Simulated linear absorption of the amide I, II, III and A.
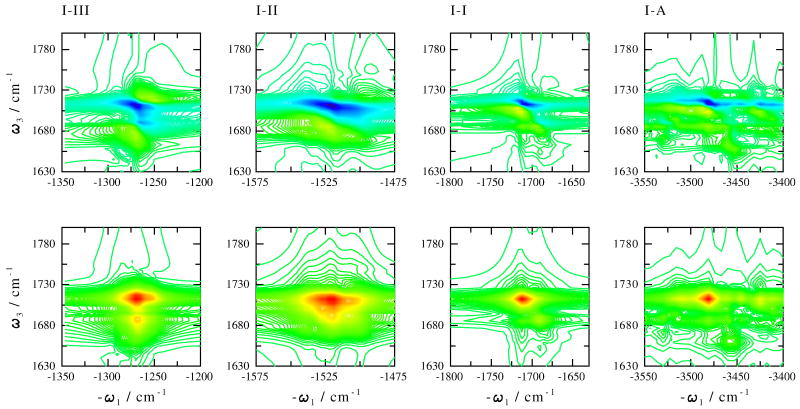
The photon echo cross peaks of the amide I and the amide I, II, III and A are displayed in Fig. 5 (Eq. (5.20), (5.23) and (5.24) in Ref. ([28])). The absolute value spectra of the 4 cross peaks are featureless due to the helical structure. The band width along the ω1 and ω3 direction corresponds to the inhomogeneous frequency distributions of the amide I and the 4 amide modes. The imaginary (absorptive) parts of the signals have more structure. Unlike NMA [14] where positive excited state absorption and negative ground state bleach peaks have similar intensities in a given cross peak, the negative peaks here are stronger. As a result, the negative peaks involve the two pairs of the transitions between the ground state and the amide fundamental states and the orientational averages of the product of these IR transition dipoles moments contributing to the signals are always positive [28]. The combination states give positive peaks and the product of the transition dipole moments can be negative due to the anharmonicities. The negative peaks from different pathways accumulate to form the strong peaks. Positive peaks from different Liouville space paths may cancel out. It should be noted that lifetime broadening could also contribute to the cross peak bandshapes and is expected to broaden more the positive excited state absorption peak. This effect is not included in the present simulation.
Fig. 5.
Photon echo cross peaks of the amide I and all 4 amide modes. Top: imaginary part; bottom: absolute value amplitude. Calculations made using Eq. (5.20), (5.23) and (5.24) in Ref. ([28].)
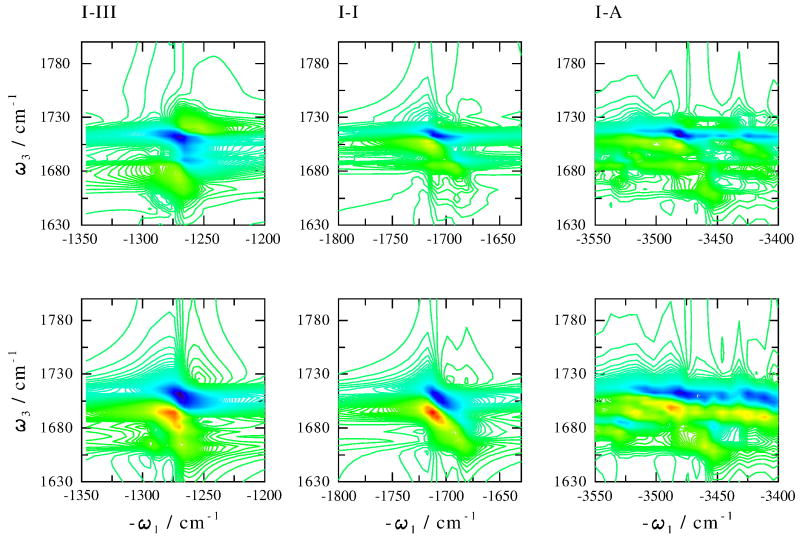
To investigate the bandshape sensitivity to the couplings between the amide modes in different amide units, the amide I - III, I - A cross peaks and the amide I - I diagonal peak were also calculated by turning off these couplings. Comparison with the original calculations is shown in Fig. 6. There is no significant difference in the bandshape and the intensity of the amide I - A cross peak, which is consistent to the highly localized amide A eigenstates (Fig. 3). The positive peaks of amide I - III and I - I regions are stronger when the couplings are switched off. The amide I - I bandshapes significantly depends on the couplings. Both negative and positive peaks are more elongated in the diagonal direction when the couplings are switched off.
Fig. 6.
Photo echo cross peaks with (top) and without (bottom) amide couplings between different amide units, as indicated.
Acknowledgments
The support of the National Institutes of Health grant no. (GM-59230) and the National Science Foundation grant no. (CHE-0446555) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miyazawa T, Shimanouchi T, Mizushima SI. Normal vibrations of n-methylacetamide. J Chem Phys. 1958;29:611–616. [Google Scholar]
- 2.Tsuboi M, Onishi T, Nakagawa I, Shimanouchi T, Mizushima S. Assignments of the vibrational frequencies of glycine. Spectrochimica Acta. 1958;12:253–261. [Google Scholar]
- 3.Torii H, Tasumi M. Model calculations on the amide-i infrared bands of globular proteins. J Chem Phys. 1992;96:3379. [Google Scholar]
- 4.Torii H, Tasumi M. Infrared Spectroscopy of Biomolecules. Wiley-Liss; New York: 1996. [Google Scholar]
- 5.Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved ftir spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 6.Surewicz WK, Mantsch HH. New insight into protein secondary structure from resolution-enhanced infrared-spectra. Biochimica Et Biophysica Acta. 1988;952:115–130. doi: 10.1016/0167-4838(88)90107-0. [DOI] [PubMed] [Google Scholar]
- 7.Jackson M, Haris PI, Chapman D. Fourier-transform infrared spectroscopic studies of lipids, polypeptides and proteins. J Mol Struc. 1989;214:329–355. [Google Scholar]
- 8.Mukamel S. Multidimensional femtosecond correlation spectroscopies of electronic and vibrational excitations. Annu Rev Phys Chem. 2000;51:691–729. doi: 10.1146/annurev.physchem.51.1.691. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser RM. Multidimensional ultrafast spectroscopy. Proc Natl Acad Sci USA. 2007;104:14189–14189. doi: 10.1073/pnas.0706002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochstrasser RM. Two-dimensional spectroscopy at infrared and optical frequencies. Proc Natl Acad Sci USA. 2007;104:14190–14196. doi: 10.1073/pnas.0704079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubtsov IV, Wang J, Hochstrasser RM. Dual frequency 2d-ir of peptide amide-a and amide-i modes. J Chem Phys. 2003;118:7733–7736. [Google Scholar]
- 12.Rubtsov IV, Wang J, Hochstrasser RM. Dual-frequency 2d-ir spectroscopy heterodyned photon echo of the peptide bond. P Nat Acad Sci. 2003;100:5601. doi: 10.1073/pnas.0931292100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeFlores LP, Ganim Z, Ackley SF, Chung HS, Tokmakoff A. The anharmonic vibrational potential and relaxation pathways of the amide i and ii modes of n-methylacetamide. J Phys Chem B. 2006;110:18973–18980. doi: 10.1021/jp0603334. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Mukamel S. Correlated hydrogen bonding fluctuations and vibrational cross peaks in n-methyl acetamide: Simulation based on a complete electrostatic density functional theory map. J Chem Phys. 2006;125:194510. doi: 10.1063/1.2348865. [DOI] [PubMed] [Google Scholar]
- 15.Torii H, Tasumi M. Ab initio molecular orbital study of the amide i vibrational interactions between the peptide groups in di- and tripeptides and considerations on the conformation of the extended helix. J Raman Spec. 1998;29:81–86. [Google Scholar]
- 16.Moran AM, Park SM, Mukamel S. Infrared photon echo signatures of hydrogen bond connectivity in the cyclic decapeptide antamanide. J Chem Phys. 2003;118:9971. [Google Scholar]
- 17.Bour P, Keiderling TA. Empirical modeling of the peptide amide i band ir intensity in water solution. J Chem Phys. 2003;119:11253. [Google Scholar]
- 18.Jansen TL, Dijkstra AG, Watson TM, Hirst JD, Knoester J. Modeling the amide i bands of small peptides. J Chem Phys. 2006;125:044502. doi: 10.1063/1.2218516. [DOI] [PubMed] [Google Scholar]
- 19.Hahn S, Ham S, Cho M. Simulation studies of amide I ir absorption and two-dimensional ir spectra of beta hairpins in liquid water. J Phys Chem B. 2005;109:11789–11801. doi: 10.1021/jp050450j. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Kim SS, Choi JH, Cho M. Theoretical study of internal field effects on peptide amide i modes. J Phys Chem B. 2005;109:5331–5340. doi: 10.1021/jp0461302. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Krimm S. Ab initio-based vibrational analysis of alpha-poly(l-alanine) Biopolymers. 1998;46:283–317. [Google Scholar]
- 22.Kubelka J, Huang R, Keiderling TA. Solvent effects on ir and vcd spectra of helical peptides: Dft-based static spectral simulations with explicit water. J Phys Chem B. 2005;109:8231–8243. doi: 10.1021/jp0506078. [DOI] [PubMed] [Google Scholar]
- 23.Gorbunov RD, Kosov DS, Stock G. Ab initio-based exciton model of amide i vibrations in peptides: Definition, conformational dependence, and transferability. J Chem Phys. 2005;122:224904. doi: 10.1063/1.1898215. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi T, Jansen TL, Zhuang W, Mukamel S. Collective solvent coordinates for the infrared spectrum of hod in d2o based on an ab initio electrostatic map. J Phys Chem A. 2005;109:64–82. doi: 10.1021/jp046685x. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi T, Zhuang W, Mukamel S. Electrostatic dft map for the complete vibrational amide band of nma. J Phys Chem A. 2005;109:9747. doi: 10.1021/jp052324l. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang W, Abramavicius D, Hayashi T, Mukamel S. Simulation protocols for coherent femtosecond vibrational spectra of peptides. J Phys Chem B. 2006;110:3362–3374. doi: 10.1021/jp055813u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm - a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 28.Mukamel S, Abramavicius D. Many-body approaches for simulating coherent nonlinear spectroscopies of electronic and vibrational excitons. Chem Rev. 2004;104:2073. doi: 10.1021/cr020681b. [DOI] [PubMed] [Google Scholar]