Abstract
We introduce Generalized Multilevel Functional Linear Models (GMFLMs), a novel statistical framework for regression models where exposure has a multilevel functional structure. We show that GMFLMs are, in fact, generalized multilevel mixed models (GLMMs). Thus, GMFLMs can be analyzed using the mixed effects inferential machinery and can be generalized within a well researched statistical framework. We propose and compare two methods for inference: 1) a two-stage frequentist approach; and 2) a joint Bayesian analysis. Our methods are motivated by and applied to the Sleep Heart Health Study (SHHS), the largest community cohort study of sleep. However, our methods are general and easy to apply to a wide spectrum of emerging biological and medical data sets. Supplemental materials for this article are available online.
Keywords: Functional principal components, Smoothing, Sleep EEG
1 Introduction
Recording and processing of functional data has become routine due to advancements in technology and computation. Many current studies contain observations of functional data on the same subject at multiple visits. For example, the Sleep Heart Health Study (SHHS) described in Section 7 contains, for each subject, quasi-continuous electroencephalogram (EEG) signals at two visits. In this paper we introduce a class of models and inferential methods for association studies between functional data observed at multiple levels/visits, such as sleep EEG or functional magnetic resonance imaging (fMRI), and continuous or discrete outcomes, such as systolic blood pressure (SBP) or Coronary Heart Disease (CHD). As most of these data sets are very large, feasibility of methods is a primary concern.
Functional regression is a generalization of regression to the case when outcomes or regressors or both are functions instead of scalars. Functional Regression Analysis is currently under intense methodological research [7, 23, 31, 34, 45] and is a particular case of Functional Data Analysis (FDA) [21, 24, 44, 42]. Two comprehensive monographs of FDA with applications to curve and image analysis are [33, 34]. There has been considerable recent effort to apply FDA to longitudinal data, e.g., [14, 37, 47]; see [30] for a thorough review. However, in all current FDA research, the term “longitudinal” represents single-level time series.
FDA was extended to multilevel functional data; see, for example, [2, 13, 19, 29, 28, 40]. However, all these papers have focused on models for functional data and not on functional regression. The multilevel functional principal component analysis (MFPCA) approach in [13] uses functional principal component bases to reduce data dimensionality and accelerate the associated algorithms, which is especially useful in moderate and large data sets. Thus, MFPCA provides an excellent platform for methodological extensions to the multilevel regression case.
We introduce Generalized Multilevel Functional Linear Models (GMFLMs), a novel statistical framework for regression models where exposure has a multilevel functional structure. This framework extends MFPCA in several ways. First, GMFLMs are designed for studies of association between outcome and functional exposures, whereas MFPCA is designed to describe functional exposure only; this extension is needed to answer most common scientific questions related to longitudinal collection of functional/image data. Second, we show that GMFLMs are the functional analog of measurement error regression models; in this context MFPCA is the functional analog of the exposure measurement error models [5]. Third, we show that all regression models with functional predictors contain two mixed effects sub-models: an outcome and an exposure model. Fourth, we propose and compare two methods for inference: 1) a two-stage frequentist approach; and 2) a joint Bayesian analysis. Using the analogy with measurement error models we provide insight into when using a two-stage method is a reasonable alternative to the joint analysis and when it is expected to fail. Our methods are an evolutionary development in a growth area of research. They build on and borrow strength from multiple methodological frameworks: functional regression, measurement error and multilevel modeling. Given the range of applications and methodological flexibility of our methods, we anticipate that they will become one of the standard approaches in functional regression.
The paper is organized as follows. Section 2 introduces the functional multilevel regression framework. Section 3 describes estimation methods based on best linear prediction. Section 4 presents our approach to model selection. Section 5 discusses the specific challenges of a Bayesian analysis of the joint mixed effects model corresponding to functional regression. Section 6 provides simulations. Section 7 describes an application to sleep EEG data from the SHHS. Section 9 summarizes our conclusions.
2 Multilevel functional regression models
In this Section we introduce the GMFLM framework and inferential methods.
2.1 Joint mixed effects models
The observed data for the ith subject in a GMFLM is [Yi, Zi, {Wij(tijm), tijm ∈ [0, 1]}], where Yi is the continuous or discrete outcome, Zi is a vector of covariates, and Wij(tijm) is a random curve in L2[0, 1] observed at time tijm, which is the mth observation, m = 1, …, Mij, for the jth visit, j = 1, …, Ji, of the ith subject, i = 1, …, I. For presentation simplicity we only discuss the case of equally spaced tijm, but our methods can be applied with only minor changes to unequally/random spaced tijm; see [13] for more details.
We assume that Wij(t) is a proxy observation of the true underlying subject-specific functional signal Xi(t), and that Wij(t) = μ(t) + ηj(t) + Xi(t) + Uij(t) + εij(t). Here μ(t) is the overall mean function, ηj(t) is the visit j specific shift from the overall mean function, Xi(t) is the subject i specific deviation from the visit specific mean function, and Uij(t) is the residual subject/visit specific deviation from the subject specific mean. Note that multilevel functional models are a generalization of: 1) the classical measurement error models for replication studies when there is no t variable and ηj(t) = 0; 2) the standard functional models when Ji = 1 for all i, ηj(·) = 0 and Uij(·) = 0; and 3) the two-way ANOVA models when Xi(·) and Uij(·) do not depend on t. This is not important because “a more general model is better”, but because it allows us to borrow and adapt methods from seemingly unrelated areas of Statistics. We contend that this synthesis is both necessary and timely to address the increasing challenges raised by ever larger and more complex data sets.
To ensure identifiability we assume that Xi(t), Uij(t), and εij(t) are uncorrelated, P that Σj ηj(t) = 0 and that εij(t) is a white noise process with variance . Given the large sample size of the SHHS data, we can assume that μ(t) and ηj(t) are estimated with negligible error by W̄··(t) and W̄·j(t) − W̄··, respectively. Here W̄·· (t) is the average over all subjects, i, and visits, j, of Wij(t) and W̄ ·j(t) is the average over all subjects, i, of observation at visit j of Wij(t). We can assume that these estimates have been subtracted from Wij(t), so that Wij(t) = Xi(t) + Uij(t) + εij(t). Note that consistent estimators of W̃ij(t) = Xi(t) + Uij(t) can be obtained by smoothing {t, Wij(t)}. Moreover, consistent estimators for Xi(t) and Uij(t) can be constructed as estimators of and , respectively.
We assume that the distribution of the outcome, Yi, is in the exponential family with linear predictor ϑi and dispersion parameter α, denoted here by EF(ϑi, α). The linear predictor is assumed to have the following form , where Xi(t) is the subject-specific deviation from the visit-specific mean, β(·) ∈ L2[0, 1] is a functional parameter and the main target of inference, is a vector of covariates and γ are fixed effects parameters. If { } and { } are two orthonormal bases in L2[0, 1] then Xi(·), Uij(·) and β(·) have unique representations
| (1) |
This form of the model is impractical because it involves three infinite sums. Instead, we will approximate model (1) with a series of models where the number of predictors is truncated at K = KI,J and L = LI,J and the dimensions K and L increase asymptotically with the total number of subjects, I, and visits per subject, J. A good heuristic motivation for this truncation strategy can be found, for example, in [31]. In section 4 we provide a theoretical and practical discussion of alternatives for estimating K and L. For fixed K and L the multilevel outcome model becomes
| (2) |
Other multilevel outcome models could be considered by including regression terms for the Uij(t) process or, implicitly, for ζijl. However, we restrict our discussion to models of the type (2).
We use MFPCA [13] to obtain the parsimonious bases that capture most of the functional variability of the space spanned by Xi(t) and Uij(t), respectively. MF-PCA is based on the spectral decomposition of the within- and between-visit functional variability covariance operators. We summarize here the main components of this methodology. Denote by and for j ≠ k the total and between covariance operator corresponding to the observed process, Wij(·), respectively. Denote by KX(t, s) = cov{Xi(t), Xi(s)} the covariance operator of the Xi(·) process and by the total covariance operator of the Uij(·) process. By definition, for j ≠ k. Moreover, and , where δts is equal to 1 when t = s and 0 otherwise. Thus, KX(s, t) can be estimated using a method of moments estimator of , say . For t ≠ s a method of moment estimator of , say , can be used to estimate . To estimate one predicts using a bivariate thin-plate spline smoother of for s ≠ t. This method was proposed for single-level FPCA [44] and shown to work well in the MFPCA context [13].
Once consistent estimators of KX(s, t) and are available, the spectral decomposition and functional regression proceed as in the single-level case. More precisely, Mercer’s theorem (see [22], Chapter 4) provides the following convenient spectral decompositions , where are the ordered eigenvalues and are the associated orthonormal eigenfunctions of KX(·,·) in the L2 norm. Similarly, , where are the ordered eigenvalues and are the associated orthonormal eigenfunctions of in the L2 norm. The Karhunen-Loève (KL) decomposition [25, 26] provides the following infinite decompositions and where are the principal component scores with E(ξik) = E(ζijl) = 0, . The zero-correlation assumption between the Xi(·) and Uij(·) processes is ensured by the assumption that cov(ξi, ζijl) = 0. These properties hold for every i, j, k, and l.
Conditional on the eigenfunctions and truncation lags K and L, the model for observed functional data can be written as a linear mixed model. Indeed, by assuming a normal shrinkage distribution for scores and errors, the model can be rewritten as
| (3) |
For simplicity we will refer to and as the level 1 and 2 eigenfunctions and eigenvalues, respectively.
We propose to jointly fit the outcome model (2) and the exposure model (3). Because the joint model is a generalized linear mixed effects model the inferential arsenal for mixed effects models can be used. In particular, we propose to use a Bayesian analysis via posterior Markov Chain Monte Carlo (MCMC) simulations as described in Section 5. An alternative would be to use a two-stage analysis by first predicting the scores from model (3) and then plug-in these estimates into model (2).
2.2 BLUP plug-in versus joint estimation
To better understand the potential problems associated with two-stage estimation we describe the induced likelihood for the observed data. We introduce the following notations ξi = (ξi1, …, ξiK)t and Wi = {Wi1(ti11), …, Wi1(ti1Mi1), …, WiJi(tiJiMiJi)}t. With a slight abuse of notation [Yi|Wi, Zi] = ∫[Yi, ξi|Wi, Zi]dξi, where [·|·] denotes the probability density function of the conditional distribution. The assumptions in models (2) and (3) imply that [Yi, ξi|Wi, Zi] = [Yi|ξi, Zi][ξi|Wi], which, in turn, implies that
| (4) |
Under normality assumptions it is easy to prove that [ξi|Wi] = N{m(Wi), Σi}, where m(Wi) and Σi are the mean and covariance matrix of the conditional distribution of ξi given the observed functional data and model (3). In Section 3 we provide the derivation of m(Wi) and Σi and additional insight into their effect on inference.
For most nonlinear models the induced model for observed data (4) does not have an explicit form. A procedure to avoid this problem is to use a two-stage approach with the following components: 1) produce predictors of ξi, say b ξ̂i, based on the exposure model (3); and 2) estimate the parameters of the outcome model (2) by replacing ξi with ξ̂i. It is reasonable to use the best linear unbiased predictor (BLUP) of ξi, ξ̂i = m(Wi), but other predictors could also be used. For example, for the single-level functional model Müller and Stadtmüller [31] used , which are unbiased predictors of ξik. Such estimators have even higher variance than Σi because they do not borrow strength across subjects. This may lead to estimation bias and misspecified variability. The problem is especially serious in multilevel functional models as we discuss below.
Consider, for example, the outcome model Yi|ξi, Zi ~ Bernoulli(pi), where , and Φ(·) is the cumulative distribution function of a standard normal distribution. Under the normality assumption of the distribution of ξi it follows that the induced model for observed data is Yi|Wi, Zi ~ Bernoulli(qi), where
| (5) |
Thus, using the two-stage procedure, where ξi is simply replaced by mt(Wi), leads to biased estimators with misspecified variability for β and γ. The size of these effects is controlled by βtΣiβ.
There are important potential differences between joint and two-stage analyses in a multilevel functional regression context. Indeed, the term in equation (3) quantifies the visit/subject-specific deviations from the subject specific mean. This variability is typically large and makes estimation of the subject-specific scores, ξi, difficult even when the functions are perfectly observed, that is when . Thus, the effects of variability on bias in a two-stage procedure can be severe, especially when the within-subject variability is large compared to the between-subject variability. In the next section we provide the technical details associated with a two-stage procedure and provide a simple example to build up the intuition.
3 Posterior distribution of subject-specific scores
We now turn our attention to calculating the posterior distribution of subject-specific scores for the MFPCA model (3). While this section is more technical and contains some pretty heavy notation, the results are important because they form the basis of any reasonable inferential procedure in this context, be it two-stage or joint modeling. We first introduce some notation for a subject i. Let Wij = {Wij(tij1), …, Wij(tijMij)}t be the Mij × 1 vector of observations at visit j, be the vector of observations obtained by stacking Wij, be the Mij × 1 dimensional vector corresponding to the kth level 1 eigenfunction at visit j, and be the dimensional vector corresponding to the kth level 1 eigenfunction at all visits. Also, let be the Mij × K dimensional matrix of level 1 eigenvectors obtained by binding the column vectors corresponding to the jth visit and be the dimensional matrix of level 1 eigenfunctions obtained by binding the column vectors . Similarly, we define the vectors and . Finally, let and be the K ×K and L×L dimensional diagonal matrices of level 1 and level 2 eigenvalues, respectively.
If ΣWi denotes the covariance matrix of Wi then its (j, j′)th block matrix is equal to Bi,jj′ where if j ≠ j′ and for 1 ≤ j, j′ ≤ Ji. Moreover, under normality assumptions [ξi|Wi] = N{m(Wi), Σi}, where and . The following results provide simplified expressions for ΣWi, m(Wi) and Σi that greatly reduce computational burden of algorithms.
Theorem 1
Consider the exposure model (3) with a fixed number of observations per visit, i.e. Mij = Mi, at the same subject-specific times for each visit, i.e. tijm = tim for all j = 1, …, Ji. Denote by , by , by 1Ji×Ji the Ji × Ji dimensional matrix of ones, and by ⊗ the Kronecker product of matrices. Then and .
Theorem 2
Assume the balanced design considered in Theorem 1 and denote by . Then and .
Proofs can be found in the accompanying web supplement. Theorem 2 provides a particularly simple description of the conditional distribution ξi|Wi. Moreover, it shows that, conditional on the smoothing matrices Λ(1) and Λ(2), the conditional distribution ξi|Wi is the same as the conditional distribution ξi|W̄i. We now provide a simple example where all calculations can be done explicitly to illustrate the contribution of each individual source of variability to the variability of the posterior distribution ξi|Wi, Σi. As described in section 2.2, this variability affects the size of the estimation bias in a two-stage procedure. Thus, it is important to understand in what applications this might be a problem.
Consider a balanced design model with K = L = 1 and ψ(1)(t) = 1, ψ(2)(t) = 1 for all t. The exposure model becomes a balanced mixed two-way ANOVA model
| (6) |
where, for simplicity, we denoted by ξi = ξi1, ζij = ζij1, and by . In this case the conditional variance Σi is a scalar and, using Theorem 2, we obtain
Several important characteristics of this formula have direct practical consequences. First, Σi ≤ λ1 indicating that Σi is small when the variability at first level, λ1, is small. In this situation one could expect the two-stage procedure to work well. Second, the within-subject/between-visit variability, λ2, is divided by the number of visits, Ji. In many applications λ2 is large compared to λ1 and Ji is small, leading to a large variance Σi. For example, in the SHHS study Ji = 2 and the functional analog of λ2 is roughly 4 times larger than the functional analog of λ1. Third, even when functions are perfectly observed, that is , the variance Σi is not zero. Fourth, in many applications is negligible because the total number of observations for subject i, MiJi, is large. For example, in the SHHS, MiJi ≈ 1600.
4 Model uncertainty
Our framework is faced with two distinct types of model uncertainty related to: 1) the choice of K and L, the dimensions of the two functional spaces in the exposure model (3); and 2) estimating β(t), the functional effect parameter, conditional on K and L, in the outcome model (2).
To address the first problem we focus on estimating K, as estimating L is similar. Note that, as K increases, the models described in (3) form a nested sequence of mixed effects models. Moreover, testing for the dimension of the functional space being equal to K versus K + 1 is equivalent to testing versus , which is testing for the null hypothesis that a particular variance component is equal to zero. This connection provides a paradigm shift for estimating the dimension of the functional space or, more generally, the number of non-zero eigenvalues in PCA. Current methods are based on random matrix theory and require that eigenvalues be bounded away from zero, see, for example, [3, 20]. This is not the correct approach when the null hypothesis is that the eigenvalue is zero.
In this context Staicu, Crainiceanu and Carroll [40] proposed a sequence of Restricted Likelihood Ratio Tests (RLRTs) for zero variance components [10, 12, 41] to estimate K. Müller and Stadtmüller [31] proposed to use either the Akaike’s Information Criterion (AIC) [1] or the Bayesian Information Criterion (BIC) [39]. Moreover, they found these criteria to be more stable and less computationally intensive than methods based on cross-validation [38] or relative difference between the Pearson criterion and deviance [6]. Staicu, Crainiceanu and Carroll [40] show that both AIC and BIC are particular cases of sequential RLRT with non-standard α levels. They also explain that AIC performs well because its associated α level is 0.079, which is different from the standard α = 0.05, but might be reasonable in many applications. In contrast, they recommend against using the BIC in very large data sets, such as in our application, because the corresponding α level becomes extremely small.
In practice we actually prefer an even simpler method for estimating the number of components based on the estimated explained variance. More precisely, let P1 and P2 be two thresholds and define , where . For the cumulative explained variance threshold we used P1 = 0.9 and for the individual explained variance we used P2 = 1/T, where T is the number of grid points. We used a similar method for choosing the number of components at level 2. These choices were slightly conservative, but worked well in our simulations and application. However, the two thresholds should be carefully tuned in any other particular application using simulations.
To address the second problem we note that it can be reduced to a standard model selection problem. Forward, backward, single-variable or all subset selection can be used to identify statistically significant predictors in the outcome model (2). Typical pitfalls reported for these methods are avoided because predictors are mutually orthogonal by construction. In practice, we prefer to do a backward selection combined with sensitivity analysis around the chosen model. More precisely, we obtain an optimal model and the two next best models. For all these models we provide the functional estimates and the log-likelihood differences.
A powerful alternative to estimating β(t) was proposed in a series of papers by Reiss and Ogden [35, 36] for the single-level functional regression case. In short, they project the original (un-smooth) matrix of functional predictors onto a B-spline basis and use the P-spline basis penalty to induce shrinkage directly on the functional parameter. Another alternative is to adapt the forward selection method using pseudo-variables [27, 43], which could work especially well because the estimated eigenvalues are sorted. Both methods could easily be used in our framework. However, they would need to be adapted to a joint analysis context to overcome the bias problem induced by the two-stage analysis described in Section 2.
5 Bayesian inference
Because of the potential problems associated with two-stage procedures, we propose to use joint modeling. Bayesian inference using MCMC simulations of the posterior distribution provides a reasonable, robust, and well tested computational approach for this type of problems. Possible reasons for the current lack of Bayesian methodology in functional regression analysis could be: 1) the connection between functional regression models and joint mixed effects models was not known; and 2) the Bayesian inferential tools were perceived as unnecessarily complex and hard to implement. We clarified the connection to mixed effects models in Section 2.1 and we now show that 2) is not true, thanks to intense methodological and computational research conducted over the last 10–20 years. See, for example, the monographs [4, 8, 16, 18] and the citations therein for a good overview.
To be specific, we focus on a Bernoulli/logit outcome model with functional regressors. Other outcome models would be treated similarly. Consider the joint model with the outcome Yi ~ Bernoulli(pi), linear predictor and functional exposure model (3). The parameters of the model are ω = {(ξi: i = 1, …, I), (ζij: i = 1, …, I; j = 1, …, Ji), β, γ, Λ, }, where ξi was defined in Section 2.2 and ζij = (ζij1, …, ζijL)T. While εi(tijm) are also unknown, we do not incorporate them in the set of parameters because they are automatically updated by .
The priors for ξi and ζij were already defined and it is standard to assume that the fixed effects parameters, β and γ, are apriori independent, with and where and are very large and P is the number of Z covariates. In our applications we used , which we recommend when there is no reason to expect that the components of β and γ could be outside of the interval [− 1000, 1000]. In some applications this priors might be inconsistent with the true value of the parameter. In this situations we recommend re-scaling Wij(tijm) and normalizing, or re-scaling, the Z covariates.
While standard choices of priors for fixed effects parameters exist and are typically non-controversial, the same is not true for priors of variance components. Indeed, the estimates of the variance components are known to be sensitive to the prior specification, see, for example, [11, 15]. In particular, the popular inverse-gamma priors may induce bias when their parameters are not tuned to the scale of the problem. This is dangerous in the shrinkage context where the variance components control the amount of smoothing. However, we find that with reasonable care, the conjugate gamma priors can be used in practice. Alternatives to gamma priors are discussed by, for example, [15, 32], and have the advantage of requiring less care in the choice of the hyperparameters. Nonetheless, exploration of other prior families for functional regression would be well worthwhile, though beyond the scope of this paper.
We propose to use the following independent inverse gamma priors , l = 1, … L, and , where IG(A, B) is the inverse of a gamma prior with mean A/B and variance A/B2. We first write the full conditional distributions for all the parameters and then discuss choices of non-informative inverse gamma parameters. Here we treat and as parameters to be estimated, but a simpler Empirical Bayes (EB) method proved to be a reasonable alternative in practice. More precisely, the EB method estimates and by diagonalizing the functional covariance operators as described in Section 2.1. These estimators are then fixed in the joint model. In the following we present the inferential procedure for the case when and are estimated with obvious simplifications for the EB procedure where they would be fixed.
We use Gibbs sampling [17] to simulate [Ω|D], where D denotes the observed data. A particularly convenient partition of the parameter space and the associated full conditional distributions are described below
where . The first two full-conditionals do not have an explicit form, but can be sampled using MCMC. For Bernoulli outcomes the MCMC methodology is routine. We use the Metropolis-Hastings algorithm with a normal proposal distribution centered at the current value and small variance tuned to provide an acceptance rate around 30–40%. The last four conditionals are explicit and can be easily sampled. However, understanding the various components of these distributions will provide insights into rational choices of inverse gamma prior parameters. The first parameter of the full conditional for is , where I is the number of subjects and it is safe to choose . The second parameter is , where is an estimator of and it is safe to choose . This is especially relevant for those variance components or, equivalently, eigenvalues of the covariance operator, that are small, but estimable. A similar discussion holds for . For we recommend to choose Aε ≤ 0.01 and . Note that method of moments estimators for and are available and reasonable choices of and Bε are easy to propose. These rules of thumb are useful in practice, but they should be used as any other rule of thumb, cautiously. Moreover, for every application we do not recommend to rigidly use these prior parameters but rather tune them according to the general principles described here.
6 Simulation studies
In this section, we compare the performance of the joint analysis procedure with the two-stage procedure through simulation studies. We examine the Bernoulli model with probit link when the functional exposure model is single-level and multilevel.
The outcome data was simulated from a Bernoulli/probit model with linear predictor , for i = 1, …, n, where n = 1000 is the number of subjects. We used the functional predictor Xi(t) = ξiψ1(t), where ξi ~ N (0, λ1) and ψ1(t) ≡ 1, evaluated at M = 15 equidistant time points in [0, 1]. We set β0 = 1, γ = 1 and a constant functional parameter β(t) ≡ β. The zis are taken equally spaced between [−1, 1] with z1 = −1 and zn = 1. Note that the linear predictor can be re-written as Φ−1(pi) = β0 + βξi + ziγ. In the following subsections we conduct simulations with different choices of β and type of functional exposure model. All models are fit using joint Bayesian analysis via MCMC posterior simulations and a two-stage approach using either BLUP or numerical integration [31]. We simulated N = 100 data sets from each model.
6.1 Single-level functional exposure model
Consider the case when for each subject, i, instead of observing Xi(t), one observes the noisy predictors Wi(t), where Wi(t) = Xi(t)+ εi(t), i = 1, …, n and is the measurement error. We set λ1 = 1, consider three values of the signal β = 0.5, 1.0, 1.5 and three different magnitudes of noise σε = 0 (no noise), σε = 1 (moderate) and σε = 3 (very large). Figure 1 shows the boxplots of the parameter estimates β̂ and γ̂. The top and bottom panels provide results for the joint Bayesian analysis and the two-stage analysis with BLUP, respectively. The left and middle panels display the parameter estimates for different magnitudes of noise and the right panel presents the bias of the estimates of β for several true values of β. For the two-stage procedure when the amount of noise, σε, or the absolute value of the true parameter, |β|, increases, the bias increases. These results confirm our theoretical discussion in Section 2.2 and indicate that bias is a problem both for the parameters of the functional variables measured with error and of the perfectly observed covariates. Moreover, bias increases when the true functional effect increases as well as when measurement error increases.
Figure 1.
Joint Bayesian analysis (upper panel) versus two-stage analysis with BLUP (bottom panel): box plots of β̂ and γ̂ for different values of β and σε.
For the case σε = 3, Table 1 displays the root mean squared error (RMSE) and coverage probability of confidence intervals for β and γ. The two-stage approach with scores estimated by numerical integration has a much higher RMSE than the other two methods, which have a practically equal RMSE. However, it would be misleading to simply compare the RMSE for the joint Bayesian analysis and the two-stage procedure based on BLUP estimation. Indeed, the coverage probability for the latter procedure is far from the nominal level and can even drop to zero. This is an example of good RMSE obtained by a combination of two wrong reasons: the point estimate is biased and the variance is underestimated.
Table 1.
Comparison between the two-stage estimates (with numerical integration or BLUP) and Bayesian estimates of β and γ with respect to root mean squared error (RMSE), and coverage probability of the 80% and 50% confidence intervals (80%CI cov. and 50%CI cov.) for σε = 3. The Monte Carlo standard error for the Bayesian analysis was small compared to the RMSE; for β = 0.5 it ranged between (0.002, 0.005) for β and (0.002, 0.004) for γ; for β = 1.5 it ranged between (0.009, 0.043) for β and (0.005, 0.024) for γ.
| Method | β | β̂ | γ̂ | ||||
|---|---|---|---|---|---|---|---|
| RMSE | 80%CI cov. | 50%CI cov. | RMSE | 80%CI cov. | 50%CI cov. | ||
| Numerical integration | 0.5 | 0.20 | 0.00 | 0.00 | 0.10 | 0.79 | 0.46 |
| 1.0 | 0.46 | 0.00 | 0.00 | 0.17 | 0.41 | 0.09 | |
| 1.5 | 0.81 | 0.00 | 0.00 | 0.27 | 0.03 | 0.00 | |
| BLUP | 0.5 | 0.06 | 0.84 | 0.56 | 0.10 | 0.79 | 0.46 |
| 1.0 | 0.16 | 0.26 | 0.11 | 0.17 | 0.41 | 0.09 | |
| 1.5 | 0.40 | 0.01 | 0.00 | 0.27 | 0.03 | 0.00 | |
| Bayesian | 0.5 | 0.07 | 0.85 | 0.58 | 0.11 | 0.77 | 0.54 |
| 1.0 | 0.14 | 0.83 | 0.48 | 0.14 | 0.80 | 0.52 | |
| 1.5 | 0.39 | 0.85 | 0.51 | 0.23 | 0.86 | 0.49 | |
6.2 Multilevel functional exposure model
Consider now the situation when the predictors are measured through a hierarchical functional design, as in SHHS. To mimic the design of the SHHS, we assume J = 2 visits per subject and that the observed noisy predictors Wij(t) are generated from the model Wij(t) = Xi(t) + Uij(t) + εij(t), for each subject i = 1, …, n and visit j = 1, …, J, where and Uij(t) = ζijψ2(t) with ζij ~ N(0, λ2), ψ2(t) ≡ 1. We used various choices of λ1, λ2 and , and compared the two-stage analysis with the scores estimated by BLUP with a joint Bayesian analysis. As in the single-level case, the bias depends on the factor 1 + β2Σi and the only technical difference is the calculation of Σi. Thus, we limit our analyses to the case β = 1 and examine the effects of the other factors that may influence estimation.
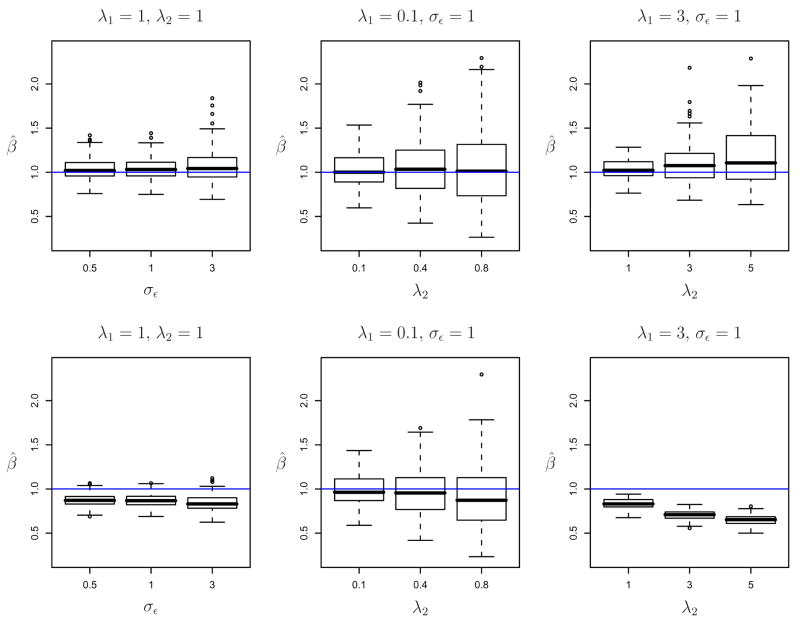
Figure 2 presents the boxplots of the estimates of β using the joint Bayesian analysis (top panels) and the two-stage method with BLUP estimation of scores (bottom panels). The left panels correspond to λ1 = 1, λ2 = 1 and three values of σε, 0.5, 1 and 3. The joint Bayesian inference produces unbiased estimates, while the two-stage procedure produces biased estimates with the bias increasing only slightly with the measurement error variance. This confirms our theoretical results that, typically, in the hierarchical setting the noise magnitude is not the main source of bias. The middle and right panels display results when the measurement error variance is fixed, σε = 1. The middle panels show results for the case when the between-subject variance is small, λ1 = 0.1, and three values of the within-subject variance, λ2 = 0.1, 0.4 and 0.8. The right panels show results for the case when the between-subject variance is large, λ1 = 3, and three values of the within-subject variance, λ2 = 1, 3 and 5. We conclude that bias is small when the between-subject variability, λ1, is small even when the within subject variability, λ2, is much larger than λ1. If λ1 is large then bias is much larger and increases with λ2. In contrast, the joint Bayesian analysis produces unbiased estimators with variability increasing with λ2. The RMSE and coverage probability results were similar to the ones for the single-level case. We have also obtained similar results for γ; results are not reported here, but they are available upon request and can be reproduced using the attached simulation software.
Figure 2.
Joint Bayesian analysis (upper panel) versus two-stage analysis with BLUP (bottom panel): box plots of β̂ for β = 1 and various values of σε and λ’s.
In spite of the obvious advantages of the joint Bayesian analysis, the message is more nuanced than simply recommending this method. In practice, the two-stage method with BLUP estimation of scores is a robust alternative that often produces similar results to the joint analysis with less computational effort. Our recommendation is to apply both methods and compare their results. We also provided insight into why and when inferential differences may be observed, and, especially, how to address such differences.
7 The analysis of sleep data from the SHHS
We now apply our proposed methods to the SHHS data. We considered 3, 201 subjects with complete baseline and visit 2 data with sleep duration that exceeds 4 hours at both visits and we analyzed data for the first 4 hours of sleep. We focus on the association between hypertension (HTN) and sleep EEG δ-power spectrum. Complete descriptions of the SHHS data set and of this functional regression problem can be found in [9, 13]. We provide here a short summary.
A quasi-continuous EEG signal was recorded during sleep for each subject at two visits, roughly 5 years apart. This signal was processed using the Discrete Fourier Transform (DFT). More precisely, if x0, …, xN − 1 are the N measurements from a raw EEG signal then the DFT is , k = 0, …, N − 1, where . If W denotes a range of frequencies, then the power of the signal in that frequency range is defined as . Four frequency bands were of particular interest: 1) δ [0.8–4.0Hz]; 2) θ [4.1–8.0Hz]; 3) α [8.1–13.0Hz]; 4) β [13.1–20.0Hz]. These bands are standard representations of low (δ) to high (β) frequency neuronal activity. The normalized power in the δ band is NPδ = Pδ/(Pδ+Pθ+Pα+Pβ). Because of the nonstationary nature of the EEG signal, the DTF and normalization are applied in adjacent 30 second intervals resulting in the function of time t → NPδ(t), where t indicates the time corresponding to a particular 30 second interval. For illustration, Figure 3 displays the pairs {t, NPδ(t)} for two subjects (gray solid and dashed lines) at baseline and visit 2. Time t = 1 corresponds to the first 30 second interval after sleep onset. Figure 3 also displays the visit-specific average percent δ power across all subjects (solid black line). Our goal is to regress HTN on the subject-specific functional characteristics that do not depend on random or visit-specific fluctuations.
Figure 3.
Gray solid and dashed lines display percent δ-power in 30 seconds intervals for the same 2 subjects at baseline (top panel) and visit 2 (bottom panel). Missing data correspond to wake periods. Solid black line displays visit-specific average δ power over all subjects.
The first step was to subtract from each observed normalized function the corresponding visit-specific population average. Following notations in Section 3, Wij(t) denotes these “centered” functional data for subject i at visit j during the tth 30-second interval. We used model (3) as the exposure model where the subject-level function, , is the actual functional predictor used for HTN.
To obtain the subject- and visit-level eigenfunctions and eigenvalues we used the MFPCA methodology introduced by [13] and summarized in Section 2.1. Table 2 provides the estimated eigenvalues at both levels indicating that 95% of level 1 (subject) variability is explained by the first five eigenfunctions and 80% is explained by the first eigenfunction. Table 2 indicates that there are more directions of variation in the level 2 (visit) space. Indeed, 80% of the variability is explained by the first 7 eigenfunctions and 90% of the variability is explained by the first 14 components (results not shown). The proportion of variability explained by subject-level functional clustering was ρ̂W = 0.213 with a 95% confidence interval: (0.210, 0.236), i.e, 21.3% of variability in the sleep EEG δ-power is attributable to the subject-level variability.
Table 2.
Estimated eigenvalues on both levels for SHHS data. We showed the first 5 components for level 1 (subject level), and 7 components for level 2.
| Level 1 eigenvalues | |||||
|---|---|---|---|---|---|
| Component | 1 | 2 | 3 | 4 | 5 |
| eigenvalue (×10−3) | 12.97 | 1.22 | 0.53 | 0.45 | 0.33 |
| % var | 80.81 | 7.60 | 3.29 | 2.79 | 2.05 |
| cum. % var | 80.81 | 88.40 | 91.70 | 94.48 | 96.53 |
| Level 2 eigenvalues | |||||||
|---|---|---|---|---|---|---|---|
| Component | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| eigenvalue (×10−3) | 12.98 | 7.60 | 7.46 | 6.45 | 5.70 | 4.47 | 3.07 |
| % var | 21.84 | 12.79 | 12.55 | 10.85 | 9.58 | 7.52 | 5.17 |
| cum. % var | 21.84 | 34.63 | 47.17 | 58.02 | 67.61 | 75.13 | 80.30 |
We started with K = 5 and performed a backward selection starting with the full outcome model , where Yi is the HTN indicator variable and no additional covariates were included into the model. Three principal components were eliminated in the following order: PC4 (p-value= 0.49), PC2 (p-value= 0.46), PC3 (p-value= 0.23). The other two principal components (PCs) were retained in the model: PC1 (p-value< 0.001) and PC5 (p-value= 0.0012). For illustration, Figure 4 displays principal components 1, 2, 3, and 5. PC1 is, basically, a vertical shift. Thus, subjects who are positively loaded on it have a higher long-term δ-power than the population average. PC5 is roughly centered around 0 and it has a more interesting behavior: a subject who is positively loaded on PC5 will have a lower percent δ-power (faster brain activity) in the first 45 minutes. This difference is more pronounced in the first 10, 15 minutes of sleep, with the subject “catching-up” to the population average between minute 45 and 60. After 1 hour of sleep the subject will have a higher percent δ-power (slower brain activity) than the average population. After 2 hours, the behavior along this component returns to the population average. Both PC1 and PC5 are very strong predictors of HTN, even though they explain very different proportions of subject-level variability: PC1 (80%) and PC5 (2%). As will be seen below, the parameter of PC5 is negative indicating that subjects who are positively loaded on this component are less likely to have HTN.
Figure 4.
Characteristics of normalized sleep EEC δ-power. Principal components 1, 2, 3 and 5 of the subject-level functional space.
Table 3 provides results for two models, one without confounding adjustment (labeled Model 1) and one with confounding adjustment (labeled Model 2). The confounders in Model 2 are sex, smoking status (with three categories: never smokers, former smokers, and current smokers), age, body mass index (BMI) and respiratory disturbance index (RDI). Each model was fitted using a two-stage analysis with BLUP estimates of scores from the exposure model and a joint Bayesian analysis. We note that there is good agreement between the two methods with the exception of the statistical significance of PC5: the two stage analysis finds it highly significant whereas the Bayesian analysis does not. As expected, the magnitude of association varies with the amount of confounding adjustment. For example, Model 1 estimates that a one standard deviation increase in PC1 scores corresponds to a relative risk e−1.55*0.11 = 0.84 (Table 2 provides the variance of PC1 scores). Model 2, which adjusts for confounders, estimated that a one standard deviation increase in PC1 scores corresponds to a relative risk e−0.85*0.11 = 0.91.
Table 3.
Mean and standard error estimates (within brackets) for parameters of models of association between hypertension and sleep EEG δ-power. Smoking status has three categories: never smokers (reference), former smokers (smk:former) and current smokers (smk.current). For the variable sex, female is the reference group and an asterisks indicates significance at level 0.05.
| Two-stage analysis | Joint analysis | |||
|---|---|---|---|---|
| Model 1 | Model 2 | Model 1 | Model 2 | |
| score 1 | −1.55 (0.28)* | −0.85 (0.30)* | −1.75 (0.33)* | −1.08 (0.40)* |
| score 5 | −7.03 (2.18)* | −4.67 (2.34)* | −7.68 (3.90) | −1.97 (3.80) |
| sex | 0.10 (0.08) | 0.09 (0.08) | ||
| smk:former | −0.18 (0.08)* | −0.19 (0.08)* | ||
| smk:current | −0.10 (0.13) | −0.10 (0.13) | ||
| age | 0.06 (0.00)* | 0.06 (0.00)* | ||
| BMI | 0.06 (0.01)* | 0.06 (0.01)* | ||
| RDI | 0.01 (0.00)* | 0.01 (0.00)* | ||
These results are now easy to explain. The bias of point estimators is likely due to the variability of PC scores. The wider credible intervals obtained from the Bayesian analysis are likely due to the appropriate incorporation of the sources of variability. The negative relationship between smoking and hypertension may seem counterintuitive. However, in this study smokers are younger, have a lower BMI and many other smokers with severe disease were not included in the study [46].
Figure 5 displays results for β(t), the functional association effect between subject-specific deviations, Xi(t), from the visit-specific mean, μ(t)+ ηj(t), and HTN without accounting for confounders. The top panel shows results for the optimal model using a two-stage frequentist analysis. This model includes PCs 1 and 5. The bottom panel shows results for the optimal model using a joint Bayesian analysis. This model includes only PC1, because PC5 was not found to be statistically significant using a joint approach. The differences are visually striking, but they are due to the special shape of PC5 and to the fact that the methods disagree on its importance. Indeed, point estimators of the PC5 component are very close, but Bayesian analysis estimates an 80% larger standard error.
Figure 5.
Results for β(t), the functional association effect between subject-specific deviations, Xi(t), from the visit-specific mean, μ(t) + ηj(t), and HTN in the model without confounders. Two-stage (top panel); joint Bayesian (bottom panel.)
Joint Bayesian analysis is simple, robust and requires minimal tunning. This is possible because MFPCA produces a parsimonious decomposition of the functional variability using orthonormal bases. The use of orthonormal bases leads to reduction in the number of parameters and of posterior correlation among parameters, which lead to excellent mixing properties. For example, the web supplement displays chains for the regression coeffcients indicating independence-like behavior.
8 Discussion
The methodology introduced in this paper was motivated by many current studies where exposure or covariates are functional data collected at multiple time points. The SHHS is just one example of such studies. The GMFLM methodology provides a self contained set of statistical tools that is robust, fast and reasonable for such studies. These properties are due to: 1) the connection between GMFLMs and mixed effects models; 2) the parsimonious decomposition of functional variability in principal directions of variation; 3) the modular way mixed effects models can incorporate desirable generalizations; and 4) the good properties of Bayesian posterior simulations due to the orthogonality of the directions of variation.
The methods described in this paper have a few limitations. First, they require a large initial investment in developing and understanding the multilevel functional structure. Second, they require many choices including number and type of basis functions, distribution of random effects, method of inference, etc. The choices we made are reasonable, but other choices may be more appropriate in other applications. Third, our framework opened many new theoretical problems; addressing all these problems exceeds the scope of the current paper and will be addressed in subsequent papers. Fourth, the computational problems may seem daunting, especially when we propose a joint Bayesian analysis of a data set with thousands of subjects, multiple visits and thousands of random effects. However, we do not think that they are insurmountable; see the software we posted at www.biostat.jhsph.edu/~ccrainic/webpage/software/GFR.zip.
Acknowledgments
Crainiceanu’s and Di’s research was supported by Award Number R01NS060910 from the National Institute Of Neurological Disorders And Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Neurological Disorders And Stroke or the National Institutes of Health.
Contributor Information
Ciprian M. Crainiceanu, Ciprian M. Crainiceanu is Associate Professor, Department of Biostatistics, Johns Hopkins University, Baltimore, Maryland 21205 (E-mail: ccrainic@jhsph.edu)
Ana-Maria Staicu, Ana-Maria Staicu is Assistant Professor, Department of Statistics, North Carolina State University, Raleigh, North Carolina 27695 (E-mail: staicu@stat.ncsu.edu).
Chong-Zhi Di, Chong-Zhi Di is Assistant Professor, Biostatistics Program, Fred Hutchinson Cancer Seattle, WA 98109 (E-mail: cdi@jhsph.edu).
References
- 1.Akaike H. Maximum likelihood identification of Gaussian autoregressive moving average models. Biometrika. 1973;60:255–265. [Google Scholar]
- 2.Baladandayuthapani V, Mallick BK, Hong MY, Lupton JR, Turner ND, Carroll RJ. Bayesian hierarchical spatially correlated functional data analysis with application to colon carcinogenesis. Biometrics. 2008;64:64–73. doi: 10.1111/j.1541-0420.2007.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilodeau M, Brenner D. Theory of Multivariate Statistics. Springer-Verlag; New York: 1999. [Google Scholar]
- 4.Carlin BP, Louis TA. Bayes and Empirical Bayes Methods for Data Analysis. 2. Chapman & Hall/CRC; 2000. [Google Scholar]
- 5.Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models: A Modern Perspective. Chapman & Hall/CRC; New York: 2006. [Google Scholar]
- 6.Chiou JM, Müller HG. Quasi-likelihood regression with unknown link and variance functions. Journal of the American Statistical Association. 1998;93:1376–1387. [Google Scholar]
- 7.Chiou JM, Müller HG, Wang JL. Functional quasi-likelihood regression models with smooth random effects. Journal of the Royal Statistical Society, Series B. 2003;65:405–423. [Google Scholar]
- 8.Congdon P. Applied Bayesian Modelling. Wiley; 2003. [Google Scholar]
- 9.Crainiceanu CM, Caffo B, Di C, Punjabi N. Nonparametric signal extraction and measurement error in the analysis of electroencephalographic activity during sleep. Journal of the American Statistical Association to appear. 2009 doi: 10.1198/jasa.2009.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crainiceanu CM, Ruppert D. Likelihood ratio tests in linear mixed models with one variance component. Journal of the Royal Statistical Society, Series B. 2004;66:165–185. [Google Scholar]
- 11.Crainiceanu CM, Ruppert D, Carroll RJ, Adarsh J, Goodner B. Spatially adaptive Penalized splines with heteroscedastic errors. Journal of Computational and Graphical Statistics. 2007;16(2):265–288. [Google Scholar]
- 12.Crainiceanu CM, Ruppert D, Claeskens G, Wand MP. Exact likelihood ratio tests for penalized splines. Biometrika. 2005;92(1):91–103. [Google Scholar]
- 13.Di C, Crainiceanu CM, Caffo B, Naresh P. Multilevel functional principal component analysis. Annals of Applied Statistics, online access 2008. 2009;3(1):458–488. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, Zhang JT. Two-step estimation of functional linear models with application to longitudinal data. Journal of the Royal Statistical Society, Series B. 2000;62:303–322. [Google Scholar]
- 15.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 2006;1(3):515–533. [Google Scholar]
- 16.Gelman A, Carlin JB, Stern HA, Rubin DB. Bayesian Data Analysis. 2. Chapman & Hall/CRC; 2003. [Google Scholar]
- 17.Geman S, Geman D. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1984;6:721–741. doi: 10.1109/tpami.1984.4767596. [DOI] [PubMed] [Google Scholar]
- 18.Gilks WR, Richardson S, Spiegelhalter DJ. Markov Chain Monte Carlo in Practice. Chapman & Hall/CRC; 1996. [Google Scholar]
- 19.Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- 20.Hall P, Hosseini-Nasab M. On properties of functional principal components analysis. Journal of the Royal Statistical Society, Series B. 2006;68:109–126. [Google Scholar]
- 21.Hall P, Müller HG, Wang JL. Properties of principal component methods for functional and longitudinal data analysis. Annals of Statistics. 2006;34:1493–1517. [Google Scholar]
- 22.Indritz J. Methods in analysis. Macmillan & Colier-Macmillan; 1963. [Google Scholar]
- 23.James GM. Generalized Linear Models with Functional Predictors. Journal of the Royal Statistical Society, Series B. 2002;64:411–432. [Google Scholar]
- 24.James GM, Hastie TG, Sugar CA. Principal component models for sparse functional data. Biometrika. 2000;87:587–602. [Google Scholar]
- 25.Karhunen K. Über lineare Methoden in der Wahrscheinlichkeitsrechnung Suomalainen Tiedeakatemia. 1947 [Google Scholar]
- 26.Loève M. Functions aleatoire de second ordre. Comptes Rendus des Séances de l’Academie des Sciences. 1945:220. [Google Scholar]
- 27.Luo X, Stefanski LA, Boos DD. Tuning variable selection procedures by adding noise. Technometrics. 2006;48:165–175. [Google Scholar]
- 28.Morris JS, Carroll RJ. Wavelet-based functional mixed models. Journal of the Royal Statistical Society, B. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris JS, Vanucci M, Brown PJ, Carroll RJ. Wavelet-based nonparametric modeling of hierarchical functions in colon carcinogenesis. Journal of the American Statistical Association. 2003;98:573–583. [Google Scholar]
- 30.Müller HG. Functional modelling and classification of longitudinal data. Scandivanian Journal of Statistics. 2005;32:223–240. [Google Scholar]
- 31.Müller HG, Stadtmüller U. Generalized Functional Linear Models. The Annals of Statististics. 2005;33(2):774–805. [Google Scholar]
- 32.Natarajan R, Kass RE. Reference Bayesian methods for generalized linear mixed models. Journal of the American Statistical Association. 2000;95:227–237. [Google Scholar]
- 33.Ramsay JO, Silverman BW. Applied Functional Data Analysis. Springer-Verlag; New York: 2005. [Google Scholar]
- 34.Ramsay JO, Silverman BW. Functional Data Analysis. Springer-Verlag; New York: 2006. [Google Scholar]
- 35.Reiss PT, Ogden RT. Functional principal component regression and functional partial least squares. Journal of the American Statistical Association. 2007;102:984–996. [Google Scholar]
- 36.Reiss PT, Ogden RT. Functional generalized linear models with images as predictors. Biometrics, to appear. 2009 doi: 10.1111/j.1541-0420.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- 37.Rice JA. Functional and longitudinal data analysis. Statistica Sinica. 2004;14:631–647. [Google Scholar]
- 38.Rice JA, Silverman BW. Estimating the mean and covariance structure nonparametrically when the data are curves. Journal of the Royal Statistical Society, Series B. 1991;53:233–243. [Google Scholar]
- 39.Schwarz G. Estimating the dimension of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 40.Staicu A-M, Crainiceanu CM, Carroll RJ. Fast methods for spatially correlated multilevel functional data. 2009 doi: 10.1093/biostatistics/kxp058. manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stram DO, Lee JW. Variance components testing in the longitudinal mixed effects model. Biometrics. 1994;50:1171–1177. [PubMed] [Google Scholar]
- 42.Wang N, Carroll RJ, Lin X. Efficient semiparametric marginal estimation for longitudinal/clustered data. Journal of the American Statistical Association. 2005;100:147–157. [Google Scholar]
- 43.Wu Y, Stefanski LA, Boos DD. Controlling variable selection by the addition of pseudovariables. Journal of the American Statistical Association. 2007;102:235–243. [Google Scholar]
- 44.Yao F, Lee TCM. Penalized spline models for functional principal component analysis. Journal of the Royal Statistical Society Series B. 2006;68:3–25. [Google Scholar]
- 45.Yao F, Müller HG, Wang JL. Functional linear regression analysis for longitudinal data. The Annals of Statistics. 2005;33:2873–2903. [Google Scholar]
- 46.Zhang L, Samet J, Caffo B, Punjabi NM. Cigarette smoking and nocturnal sleep architecture. American Journal of Epidemiology. 2006;164(6):529–537. doi: 10.1093/aje/kwj231. [DOI] [PubMed] [Google Scholar]
- 47.Zhao X, Marrron JS, Wells MT. The functional data analysis view of longitudinal data. Statistica Sinica. 2004;14:789–808. [Google Scholar]