Abstract
Purpose
Carbonyl reductase 1 (CBR1) reduces the anticancer anthracyclines doxorubicin and daunorubicin into the cardiotoxic metabolites doxorubicinol and daunorubicinol. We evaluated whether the cardioprotectant monoHER inhibits the activity of polymorphic CBR1.
Methods
We performed enzyme kinetic studies with monoHER, CBR1 (CBR1 V88 and CBR1 I88) and anthracycline substrates. We also characterized CBR1 inhibition by the related flavonoids triHER and quercetin.
Results
MonoHER inhibited the activity of CBR1 V88 and CBR1 I88 in a concentration-dependent manner. The IC50 values of monoHER were lower for CBR1 I88 compared to CBR1 V88 for the substrates daunorubicin and doxorubicin (daunorubicin, IC50-CBR1 I88: 164 μM vs. IC50-CBR1 V88: 219 μM; doxorubicin, IC50-CBR1 I88: 37 μM vs. IC50-CBR1 V88: 59 μM; p < 0.001). Similarly, the flavonoids triHER and quercetin exhibited lower IC50 values for CBR1 I88 compared to CBR1 V88 (p < 0.001). MonoHER acted as a competitive CBR1 inhibitor when using daunorubicin as a substrate (Ki = 45 ± 18 μM). MonoHER acted as an uncompetitive CBR1 inhibitor for the small quinone substrate menadione (Ki = 33 ± 17 μM).
Conclusions
The cardioprotectant monoHER inhibits CBR1 activity. CBR1 V88I genotype status and the type of anthracycline substrate dictate the inhibition of CBR1 activity.
Keywords: Human carbonyl reductase 1 (CBR1), monoHER, anthracycline-related cardiotoxicity, genotype, cardioprotectant
INTRODUCTION
The anticancer anthracyclines doxorubicin and daunorubicin are widely used in the clinic to treat a variety of solid and hematological cancers. The clinical utilization of anthracyclines is hampered by the development of anthracycline-related cardiotoxicity in some patients. Several lines of evidence indicate that the anthracycline C-13 alcohol metabolites doxorubicinol and daunorubicinol are key to the pathogenesis of anthracycline-related cardiotoxicity. Anthracycline alcohol metabolites exert cardiotoxicity by a combination of mechanisms including inhibition of Ca+2/Mg2+-ATPase in the sarcoplasmic reticulum and inactivation of the cytoplasmic aconitase/iron regulatory protein-1 complex (1, 2). In humans, the synthesis of cardiotoxic doxorubicinol and daunorubicinol is catalyzed by carbonyl reductase 1 (CBR1). CBR1 is expressed in several tissues (e.g. liver and heart), and the major role of CBR1 during the development of anthracycline-related cardiotoxicity has been documented in various studies (3-6). For example, mice with a null allele of Cbr1 (Cbr1+/-) showed low plasmatic levels of doxorubicinol and significantly lower incidence of anthracycline-related cardiotoxicity compared to animals with two active Cbr1 alleles (Cbr1+/+). Therefore, the pharmacological inhibition of CBR1 activity has been proposed as a promising strategy to minimize the clinical incidence of anthracycline-related cardiotoxicity (7). The CBR1 gene contains a non-synonymous single nucleotide polymorphism (CBR1 V88I, rs:1143663) that appears to be confined to individuals with African ancestry (p = 0.986, q = 0.014). The CBR1 V88I polymorphism encodes for CBR1 protein isoforms (CBR1 V88 and CBR1 I88) with distinctive catalytic and thermodynamic properties. For example, the CBR1 V88 isoform showed significantly higher Vmax for daunorubicin (50%) than CBR1 I88 (Vmax CBR1 V88: 181 ± 13 vs. Vmax CBR1 I88: 121 ± 12 nmol/min.mg, p < 0.05). In agreement, CBR1 V88 synthesized higher levels (47%) of the cardiotoxic C-13 alcohol metabolite daunorubicinol than CBR1 I88. Titration calorimetry studies together with molecular modeling demonstrated that both CBR1 isoforms bind the NADPH cofactor with different affinities (8).
The semi-synthetic flavonoid monoHER (7-monohydroxyethylrutoside. Fig. 1) showed a favorable cardioprotective profile in several models of anthracycline-related cardiotoxicity, and is now being tested in phase II clinical trials (9-11). Recent data suggest that the overall cardioprotective profile of monoHER results from the combination of various activities including chelation of intracellular iron, and scavenging of free radicals (12-15). In the other hand, a seminal report by Wermuth et al. described inhibition of CBR1 activity by various flavonoids including quercetin and rutin (16). In addition, we have demonstrated that the CBR1 V88 and CBR1 I88 isoforms are differentially inhibited by rutin (8). However, it is still unclear as to whether monoHER inhibits the activity of polymorphic human CBR1. Therefore, in this study we first analyzed the potential CBR1 inhibitory activity of monoHER by conducting enzyme inhibition experiments with recombinant CBR1 isoforms (CBR1 V88 and CBR1 I88), and the anthracycline substrates daunorubicin and doxorubicin, respectively. We also tested whether the structurally related flavonoids triHER and quercetin (Fig. 1) inhibited the activity of CBR1 V88 and CBR1 I88. Finally, we characterized the mechanism of CBR1 inhibition by performing kinetic experiments with monoHER and the prototypical CBR1 substrates daunorubicin and menadione (vitamin K3). Together, our data demonstrate that the anthracycline-reductase activity of polymorphic CBR1 is inhibited by the cardioprotectant flavonoid monoHER.
FIGURE 1.
Chemical structures of monoHER (A), triHER (B), and quercetin (C).
MATERIALS AND METHODS
Kinetic studies
7-monohydroxyethylrutoside (monoHER) was kindly provided by Novartis Consumer Health (Nyon, Switzerland). Cloning, expression and purification of recombinant CBR1 V88 and CBR1 I88 were performed as described (8). Two independent CBR1 V88 and CBR1 I88 protein preparations were used for this study. CBR1 enzymatic activities were measured with a validated kinetic method that records the rate of oxidation of the NADPH cofactor at 340 nm (NADPH molar absorption coefficient, 6,220 M-1 cm-1) (17, 18). Under conditions of initial velocity (V0), the method was linear (r2 > 0.90) and reproducible (CV % range: 94 – 111%) for all CBR1 substrates. Kinetic measurements were recorded in a Synergy HT luminometer equipped with thermal control and proprietary software for enzyme kinetic analysis (BioTek, Winooski, VT). Briefly, reactions were incubated at 37°C and monitored for 3 min. Enzymatic velocities (V0) were automatically calculated by linear regression of the Δabs/Δtime points, at an acquisition rate of 4 readings/minute. Assay mixtures (0.300 ml) contained monoHER (20 - 100 μM), NADPH (200 μM, Sigma-Aldrich, St. Louis, MO), potassium phosphate buffer (pH 7.4, 100 mM), CBR1 enzyme, and one of the following substrates: menadione (20 - 500 μM, Sigma-Aldrich), daunorubicin (20 - 650 μM, Sigma-Aldrich), or doxorubicin (20 - 500 μM, Sigma-Aldrich). Negative control experiments with incubation mixtures containing either no CBR1 enzyme or no NADPH cofactor resulted in non-detectable enzymatic activity. CBR1 protein concentrations were determined by recording the absorbance at 280 nm (molar extinction coefficient: 21,500 M-1 cm1, MW 30200, http://us.expasy.org). The concentration of flavonoids (e.g. monoHER, triHER) that inhibited CBR1 activity by 50% (IC50) was obtained by testing various concentrations of the inhibitors (range: 0 – 250 μM) in the presence of fixed substrate concentrations (menadione: 150 μM, daunorubicin: 300 μM, or doxorubicin: 300 μM).
Data Analysis
Enzyme kinetic parameters (Km, Vmax, and IC50) were calculated by nonlinear regression using a one-site binding model (Michaelis-Menten kinetics) with the software GraphPad Prism (version 4.03 Prism Software Inc.). Kinetic data were also analyzed by using Lineweaver-Burk double reciprocal and Dixon plots. Inhibition constants (Ki) were extrapolated from the replots of the slopes (daunorubicin) and y-intercepts (menadione) obtained from typical Lineweaver-Burk plots (19). Ki values were confirmed by non-linear regression analysis of the substrate-velocity curves using competitive and uncompetitive inhibition models, respectively. In all cases goodness of fit was r2 > 0.90. Data were expressed as the mean ± S.D. Statistical comparisons were performed with the Student's t-test and values of p<0.05 were considered significant.
RESULTS AND DISCUSSION
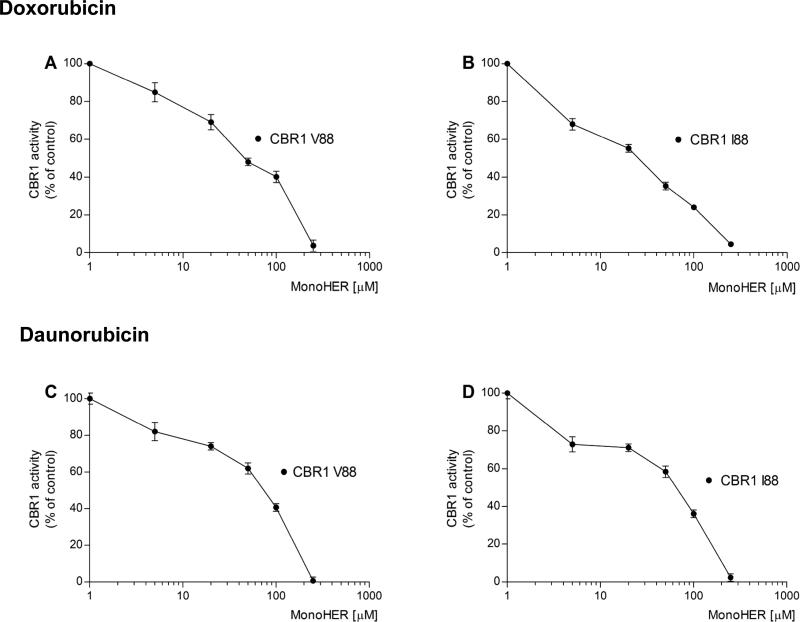
In this study, we first analyzed whether the cardioprotectant flavonoid monoHER inhibits the activity of the CBR1 V88 and CBR1 I88 isoforms. Typical enzyme inhibition experiments with the anthracycline substrates doxorubicin and daunorubicin demonstrated that monoHER inhibits CBR1 V88 and CBR1 I88 activities in a concentration-dependent manner (Fig. 2). Table I shows that for both substrates, the IC50 values of monoHER were significantly lower for CBR1 I88 compared to CBR1 V88 (Table I).
FIGURE 2.
Inhibition of CBR1 V88 (panels A and C) and CBR1 I88 (panels B and D) activities by monoHER with the substrates doxorubicin (top) and daunorubicin (bottom). Data points show the mean ± S.D. of two experiments performed in duplicate with two independent protein preparations for each CBR1 isoform.
TABLE I.
Inhibition of CBR1 V88 and CBR1 I88 by the flavonoids monoHER, triHER and quercetin.
| MonoHERb IC50 (μM) | CBR1 V88a TriHERb IC50 (μM) | Quercetinc IC50 (μM) | MonoHERb IC50 (μM) | CBR1 I88a TriHERb IC50 (μM) | Quercetinc IC50 (μM) | |
|---|---|---|---|---|---|---|
| Doxorubicin | 59 ± 5 | 53 ± 5 | 43 ± 3 | 37 ± 4 | 34 ± 3 | 19 ± 3 |
| Daunorubicin | 219 ± 4 | 383 ± 5 | 25 ± 3 | 164 ± 5 | 214 ± 3 | 14 ± 3 |
Each value represents the mean ± S.D. of two experiments performed in duplicate with two independent protein preparations.
Differences between the IC50 values of each flavonoid inhibitor were significant (p<0.001) when comparing CBR1 V88 vs. CBR1 I88 activities for both substrates.
Differences for monoHER and triHER IC50 values were significant (p<0.001) when comparing the substrates doxorubicin vs. daunorubicin.
Differences for quercetin IC50 values were non-significant (p > 0.05) when comparing the substrates doxorubicin vs. daunorubicin.
We also evaluated the CBR1 inhibitory activities of the flavonoids triHER and quercetin. In line with the previous results, the IC50 values of triHER and quercetin for both substrates (daunorubicin and doxorubicin) were consistently lower for the CBR1 I88 isoform compared to CBR1 V88. Further comparisons revealed that the IC50 of monoHER and triHER were significantly higher for the substrate daunorubicin compared to doxorubicin. These substrate-dependent differences between the IC50 values of both ethyl-hydroxylated flavonoid inhibitors were apparent for CBR1 I88 and CBR1 V88, respectively (Table I). Together, these results show that the pharmacological inhibition of CBR1 activity would be dictated by the type of anthracycline substrate and by CBR1 V88I genotype status.
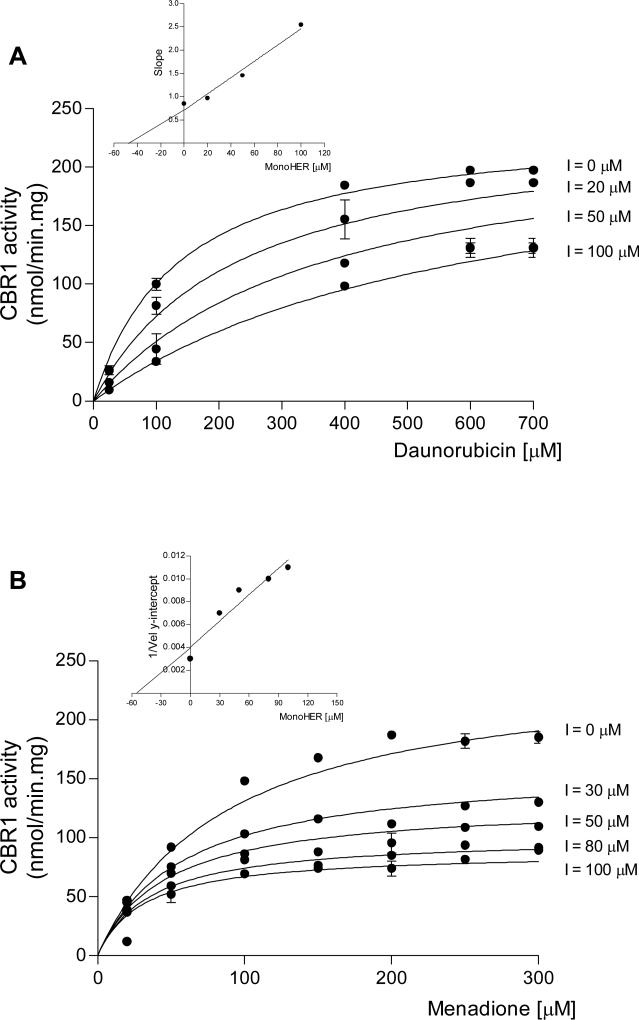
Next, we performed kinetic experiments to characterize the mechanism of CBR1 inhibition by monoHER. We used “wild type” CBR1 (CBR1 V88), and the substrates daunorubicin and menadione. Figure 3 shows inhibition of CBR1 by monoHER. Inhibition was competitive with respect to the substrate daunorubicin. A replot of the slopes of the Lineweaver-Burk plots yielded a Ki value of 45 ± 18 μM (Fig. 3A, inset). In contrast, monoHER acted as an uncompetitive inhibitor of CBR1 activity in the presence of the small quinone substrate menadione (Fig. 3B). Graphical analysis of Dixon plots confirmed the uncompetitive inhibition of CBR1 activity for the substrate menadione by monoHER (not shown). The mechanism of inhibition indicates that monoHER inhibits CBR1 activity by binding to the CBR1-menadione (enzyme-substrate) complex. Replot of the slopes from Lineweaver-Burk plots yielded an inhibition constant of 33 ± 17 μM for monoHER (Fig. 3B, inset). Furthermore, we investigated whether monoHER impacts on the binding of the NADPH cofactor by using fixed substrate concentrations (menadione: 150 μM, and daunorubicin: 400 μM), and varying concentrations of NADPH (range: 25 – 300 μM). Kinetic analysis demonstrated that monoHER inhibits the binding of NADPH in an uncompetitive manner for both substrates. Thus, monoHER inhibits enzymatic catalysis after the formation of the CBR1-substrate complex. The KiNADPH of monoHER for menadione and daunorubicin were 140 ± 37 μM, and 50 ± 15 μM, respectively. These results indicate that in the presence of monoHER, the relative affinity for NADPH is higher for the CBR1-menadione complex compared to the CBR1-daunorubicin complex.
FIGURE 3.
Kinetic analysis of CBR1 inhibition by increasing concentrations of monoHER in the presence of the substrates daunorubicin (panel A), and menadione (panel B). Each point represents the mean ± S.D. of two experiments performed in duplicate with two independent protein preparations. Insets: graphical determination of Ki values. Replot of the slopes from double-reciprocal plots (panel A; r2>0.90), and replot of the 1/velocity y-intercepts from double reciprocal plots (panel B; r2>0.90). Similar Ki values were obtained from non-linear regression analyses (see text).
In conclusion, the cardioprotectant flavonoid monoHER inhibits the activity of polymorphic human CBR1 in a concentration-dependent manner. Our results support the notion that inhibition of CBR1 activity should be considered during the development of novel cardioprotectants against anthracycline-related cardiotoxicity.
ACKNOWLEDGMENTS
National Institutes of Health/National Institute of General Medical Sciences Grant RO1GM73646 to J.G.B supported this work.
List of nonstandard abbreviations
- NADPH
nicotinamide adenine dinucleotide 2'-phosphate
- triHER
5,7,2 trihydroxiethylrutoside (Venoruton®)
REFERENCES
- 1.Minotti G, Recalcati S, Menna P, Salvatorelli E, Corna G, Cairo G, Helmut Sies P, Lester Doxorubicin Cardiotoxicity and the Control of Iron Metabolism: Quinone-Dependent and Independent Mechanisms. Methods Enzymol. 2004;378:340–361. doi: 10.1016/S0076-6879(04)78025-8. [DOI] [PubMed] [Google Scholar]
- 2.Slupe A, Williams B, Larson C, Lee LM, Primbs T, Bruesch AJ, Bjorklund C, Warner DL PJ, Shadle SE, Gambliel HA, Cusack BJ, Olson RD, Charlier HA., Jr. Reduction of 13-deoxydoxorubicin and daunorubicinol anthraquinones by human carbonyl reductase. Cardiovasc. Toxicol. 2005;5:365–376. doi: 10.1385/ct:5:4:365. [DOI] [PubMed] [Google Scholar]
- 3.Forrest GL, Akman S, Doroshow J, Rivera H, Kaplan WD. Genomic sequence and expression of a cloned human carbonyl reductase gene with daunorubicin reductase activity. Mol. Pharmacol. 1991;40:502–507. [PubMed] [Google Scholar]
- 4.Gonzalez B, Akman S, Doroshow J, Rivera H, Kaplan WD, Forrest GL. Protection against Daunorubicin Cytotoxicity by Expression of a Cloned Human Carbonyl Reductase cDNA in K562 Leukemia Cells. Cancer Res. 1995;55:4646–4650. [PubMed] [Google Scholar]
- 5.Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, Boucek RJ. Doxorubicin Cardiotoxicity May Be Caused by Its Metabolite, Doxorubicinol. Proc. Natl. Acad. Sci. U.S.A. 1988;85:3585–3589. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cusack BJ, Mushlin PS, Voulelis LD, Li XD, Boucek RJ, Olson RD. Daunorubicin-Induced Cardiac Injury in the Rabbit: A Role for Daunorubicinol? Toxicol. Appl. Pharmacol. 1993;118:177–185. doi: 10.1006/taap.1993.1023. [DOI] [PubMed] [Google Scholar]
- 7.Mordente A, Meucci E, Giuseppe Ettore M, Bruno G, Giorgio M. Human Heart Cytosolic Reductases and Anthracycline Cardiotoxicity. IUBMB Life. 2001;52:83–88. doi: 10.1080/15216540252774829. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Covarrubias V, Ghosh D, Lakhman SS, Pendyala L, Blanco JG. A Functional Genetic Polymorphism on Human Carbonyl Reductase 1 (CBR1 V88I) Impacts on Catalytic Activity and NADPH Binding Affinity. Drug Metab. Dispos. 2007;35:973–980. doi: 10.1124/dmd.107.014779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willems A, Bruynzeel A, Kedde M, Groeningen C, Bast A, Vijgh W. A phase I study of monohydroxyethylrutoside in healthy volunteers. Cancer Chemother. Pharmacol. 2006;57:678–684. doi: 10.1007/s00280-005-0083-7. [DOI] [PubMed] [Google Scholar]
- 10.Abou El Hassan MAI, Kedde MA, Zwiers UTH, Bast A, van der Vijgh WJF. The cardioprotector monoHER does not interfere with the pharmacokinetics or the metabolism of the cardiotoxic agent doxorubicin in mice. Cancer Chemother. Pharmacol. 2003;51:306–310. doi: 10.1007/s00280-003-0582-3. [DOI] [PubMed] [Google Scholar]
- 11.Bruynzeel AME, Niessen HWM, Bronzwaer JGF, Van Der Hoeven JJM, Berkhof J, Bast A, Van Der Vijgh WJF, Van Groeningen CJ. The effect of monohydroxyethylrutoside on doxorubicin-induced cardiotoxicity in patients treated for metastatic cancer in a phase II study. Br. J. Cancer. 2007;97:1084–1089. doi: 10.1038/sj.bjc.6603994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janisch KM, Williamson G, Needs P, Plumb GW. Properties of Quercetin Conjugates: Modulation of LDL Oxidation and Binding to Human Serum Albumin. Free Radical Research. 2004;38:877–884. doi: 10.1080/10715760410001728415. [DOI] [PubMed] [Google Scholar]
- 13.Kaiserova H, Simunek T, van der Vijgh WJF, Bast A, Kvasnickova E. Flavonoids as protectors against doxorubicin cardiotoxicity: Role of iron chelation, antioxidant activity and inhibition of carbonyl reductase. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2007;1772:1065–1074. doi: 10.1016/j.bbadis.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 14.van Acker FAA, Boven E, Kramer K, Haenen GRMM, Bast A, van der Vijgh WJF. Frederine, a New and Promising Protector Against Doxorubicin-induced Cardiotoxicity. Clin. Cancer Res. 2001;7:1378–1384. [PubMed] [Google Scholar]
- 15.Bast A, Haenen GRMM, Bruynzeel AME, Van Der Vijgh WJF. Protection by flavonoids against anthracycline cardiotoxicity: From chemistry to clinical trials. Cardiovascular Toxicology. 2007;7:154–159. doi: 10.1007/s12012-007-0018-0. [DOI] [PubMed] [Google Scholar]
- 16.Wermuth B. Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J. Biol. Chem. 1981;256:1206–1213. [PubMed] [Google Scholar]
- 17.Bohren KM, Von Wartburg JP, Wermuth B. Kinetics of carbonyl reductase from human brain. Biochem. J. 1987;244:165–171. doi: 10.1042/bj2440165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakhman SS, Ghosh D, Blanco JG. Functional Significance of a Natural Allelic Variant of Human Carbonyl Reductase 3 (CBR3). Drug. Metab. Dispos. 2005;33:254–257. doi: 10.1124/dmd.104.002006. [DOI] [PubMed] [Google Scholar]
- 19.Segel IH. Enzyme Kinetics - Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John Wiley & Sons, Inc.; New York, USA: 1993. [Google Scholar]