Abstract
Variability of phenotypic characteristics in Cryptococcus neoformans var. grubii and var. neoformans as well as Cryptococcus gattii can have diverse effects on the virulence of these fungi and are thus important for pathogenesis. This article summarizes the diverse phenotypic changes that these fungi can manifest. We divide changes into those that affect the entire fungal population and are predominantly induced by environmental signals, and those that involve subpopulations of the fungal population and have to be selected. Last, the article summarizes the experimental evidence that epitopes on the polysaccharide capsule also vary, which may have implications for the pathogenesis as these findings would further diversify the fungal population.
Keywords: pathogenesis, phenotypic switching, phenotypic variability
Cryptococcus neoformans var. neoformans, var. grubii and Cryptococcus gattii are ubiquitous encapsulated yeasts that cause chronic meningoencephalitis, pneumonia and disseminated disease in both immunocompetent as well as immunocompromised individuals. More than 600,000 people worldwide die of cryptococcosis per year [1] and prospective studies have suggested that 10–20% of all deaths in HIV-infected patients in Africa are attributable to cryptococcal infection [2,3]. The majority of clinical isolates are C. grubii although both C. neoformans [4,5] and C. gattii infections also occur [6].
Cryptococci are environmental microbes that accidentally invade the host. Virulence traits of these fungi constitute standard survival mechanisms that are advantageous in the host [7]. Phenotypic variation allows rapid adaptation to a constantly changing environment. Variegated expression of genes can be the result of many different mechanisms and contributes to heterogeneity within populations of genetically identical fungal cells. Such variation affects the host–pathogen interaction and may facilitate evasion of host defenses. Phenotypic changes are common in fungi and are induced by different mechanisms. In this article we will discuss different forms of phenotypic changes in C. neoformans, C. grubii and C. gattii. We divide them into changes that involve the whole fungal population, those that involve only a subset and those showing variability among individual cells. Global changes of phenotypic traits and classic cellular morphology involve the entire fungal population. They are induced by environmental signals such as iron concentration, starvation, temperature, pH change and mating-associated factors. Examples of global changes in phenotypic traits include induction of the polysaccharide capsule and melanization. Examples that involve morphological transitions are hyphal formation and sporulation. Biofilm formation also results from a global phenotypic change to an adherent population, although individual cells within the biofilm may manifest different changes. Phenotypic switching (PS) only occurs in a small percentage of cells. PS of colony morphology is a random, reversible process and is usually not induced by external signals. Last, we discuss the evidence for antigenic variation in the polysaccharide capsule of C. neoformans.
Phenotypic traits & morphological transitions of Cryptococcus species
Cellular changes in fungi can be quite striking and are relevant for pathogenesis in most fungi, including Cryptococcus species and varieties. Examples in other fungi include ‘phase variation’, which allows dimorphic fungi such as Histoplasma capsulatum and Coccidioides immitis to grow as filamentous saprophobic molds at ambient temperatures and as yeasts within the mammalian host [8,9]. Another example is the yeast–hyphal transition in Candida species, whereby the yeast form is associated with dissemination, and the hyphal form with tissue invasion [10–12].
As part of its lifecycle, C. neoformans (both var grubii and var neoformans) and C. gattii manifest both unique phenotypic traits and also morphological transitions common to many fungal species. These processes involve remodeling of the outer surface. C. neoformans, C. grubii and C. gattii have a somewhat unique morphology because, unlike other fungi, they are encapsulated. The induction and remodeling of the cryptococcal polysaccharide capsule is a clinically highly relevant change analogous to capsule induction in encapsulated bacteria [13]. In addition, melanization and biofilm formation are phenotypic traits that can be modified. Change involves the entire fungal population and affects pathogenesis however, it is not unique to these fungal species. Hyphae, pseudohyphae and spore formation occur during mating and monokaryotic fruiting but are not commonly found in vivo. Spore formation is essential to the pathogenesis of cryptococcosis, as spores are the infectious propagule that is inhaled from the environment [14–16]. Expression of morphology-specific genes, many of which are glycosylphosphatidylinositol-anchored, is controlled by diverse signal transduction pathways in C. neoformans and C. grubii, which is reviewed in more detail elsewhere [17–19]. The mitogen activated protein kinase [20–22], high osmolarity glycerol [23,24], calcineurin [25,26], cyclic AMP [27–29], regulation of Ace2p activity and cellular morphogenesis [30] and sterol regulatory element binding proteins [31,32] pathways respond to environmental signals and are key regulators of cellular morphogenesis and proliferation. They are generally conserved among fungi; however, significant differences in these regulatory networks occur even in the closely related C. grubii and C. neoformans [23,33]. In this section we will only briefly review morphological transitions as they have been reviewed in detail elsewhere [34,35], and will instead focus on reviewing phenotypic traits in C. neoformans varieties and species that can be modified during chronic cryptococcosis.
Capsule induction
The cryptococcal polysaccharide capsule is composed primarily of two polysaccharides, glucuronoxylomannan (GXM) and galactox-ylomannan (Figure 1A). The size of capsule is highly variable and dependent on environmental conditions [36]. Within a fungal population, size variability is observed and for some strains the capsule size is more variable and can change during chronic infection [37]. Recent investigations have demonstrated that spores are also coated with GXM on the surface [14]. Most investigations on the polysaccharide capsule are performed with C. neoformans or C. grubii strains. In vivo infection studies demonstrated that the lung environment is a powerful inducer of capsule growth [38]. Capsule enlargement is a controlled reproducible event and does not occur over a certain capsule size in serially passaged yeast cells [39]. Capsule changes during infection are associated with differences in the binding pattern of capsule-specific antibodies [40]. The in vivo C. grubii strain H99 has a larger capsule than cells of C. gattii strain R265, which may explain why these strains elicit different immune responses [41].
Figure 1. Phenotypic variations of Cryptococcus neoformans.
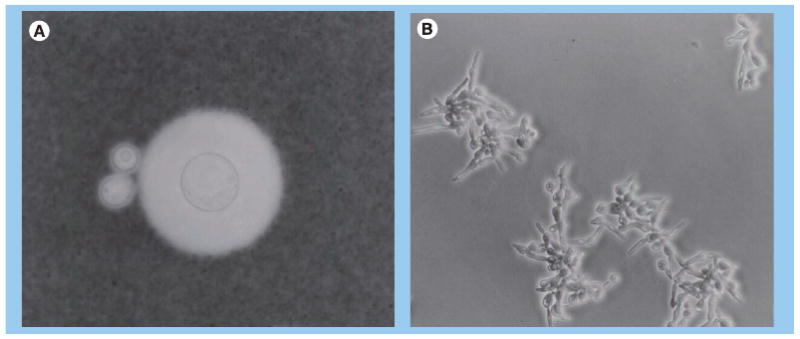
(A) Uninduced capsule and induced capsule of C. neoformans cell. (B) Hyphal formation of C. neoformans.
Serum [42], high CO2 concentration [43], iron deprivation [44,45] and slight alkaline conditions can facilitate capsule enlargement [46]. Many of these inducing conditions are present in the human host. It is thought that capsule growth requires the acidic group of glucuronic acid residues to be ionized, possibly so that they can react with divalent cations for capsule assembly [47]. The ability of cryptococcal strains to respond to serum is affected by species and variety of the strain [48]. Both antibody (Ab) as well as complement-mediated in vitro phagocytosis assays demonstrate that increased capsule volume of both C. neoformans and C. gattii strains negatively affects phagocytosis, which is a major defensive mechanism in the host [48]. In addition, if C. neoformans is carried across the blood–brain barrier in phagocytic cells as suggested, then we would expect that capsule size would affect dissemination [49].
Several genes that are either directly or indirectly involved in capsule biosynthesis have been identified. Most of the null mutants of these genes either lack a capsule altogether or are hypocapsular (for a review see [50]). Accordingly, the majority of these mutants exhibit attenuated virulence when tested in animal models. Of particular interest is the ALL1 gene, which is regulated by PS and thus during chronic infection modifies capsule size in a complex manner. The capsule of the all1Δ mutant in C. neoformans is slightly larger at baseline, but it only induces to approximately two-thirds of the wild-type capsule volume both in vivo and in vitro. Despite the slightly impaired induction, this mutant exhibits enhanced virulence both in the pulmonary as well as in the intracisternal infection model. The mutant grows at the same rate in vivo but affects the host–pathogen interaction, thereby impairing clearance [51]. Capsule formation is a complex and coordinated biosynthetic pathway. Well-studied signaling pathways are involved and include the protein kinase/cAMP and protein kinase C/mitogen activated protein kinase pathways [52]. The most rigorously studied transduction pathway is the cAMP pathway, for which elements have been cloned and disrupted [53]. The protein kinase C pathway does not play a role in capsule induction. Its influence on capsule formation is likely to be indirect, through impaired cell wall organization, as suggested by work in pkc1Δ null mutants [54,55].
The process of capsule assembly has been the subject of debate and the focus of many different studies. The first study used a monoclonal antibody (mAb) and radioactive xylose [56] and concluded that capsule grows by accumulation in the inner part of the capsule, displacing the old capsule to the edge. A different approach using a marker that covalently bonded to the capsule [57,58], suggested that during capsule induction the old polysaccharide fibers remained in a position close to the cell wall. Results from light scatter analysis of capsule-associated polysaccharide implied that capsule growth was achieved by the addition of molecules with a larger effective diameter. It showed that some polysaccharide molecules span the entire diameter of the capsule [59]. It is noteworthy that biophysical investigations have revealed differences in the molecular weight of GXM among strains and in switch variants [60] and also changes of GXM viscosity, which could potentially promote the development of high intracerebral pressure in vivo [61,62].
Hyphal formation
Cryptococcus neoformans, C. grubii and C. gattii typically grow as a haploid yeast, but undergo hyphal differentiation in response to environmental stresses or when confronted with an appropriate mating partner (Figure 1B) [15,63,64]. Diploid cryptococcal strains have been described [65] as well as same-sex mating [66]. Furthermore, different species and strains differ in their ability to mate [67,68]. Hyphal transitions are required for spore formation, which are proposed to be the infectious propagules. They can produce lethal infection in mice at very low doses, are resistant to desiccation and nutrient deprivation, easily aerosolized, and are of an ideal size to lodge in the alveoli of the lung [16]. Spores from the three species differ in size [14]. In addition, C. neoformans produce both intercalary and terminal chlamydospores, which are capable of generating new branches and yeast cells. Their relevance for pathogenesis is unknown [69] and it is not known if they occur in all cryptococcal species.
Factors that enhance hyphal growth include ambient temperatures, nitrogen starvation, dehydrated substrates, darkness and the presence of mating pheromone [70,71]. Dikaryotic hyphae contain two parental nuclei per hyphal compartment, fused clamp connections and are produced during sexual reproduction between a and α cells [63,72]. Monokaryotic hyphae contain one nucleus per hyphal compartment, unfused clamp connections and are produced during fruiting of α and some a cells, mostly in the setting of high ammonium sulfate levels [15,73]. Thus, both pathways lead to hyphal growth and basidiospore production and promote environmental distribution of C. neoformans and ultimately the incidence of cryptococcosis. In addition, cryptic same-sex reproduction can contribute to the production of infectious spores [66,74]. The biological importance of pseudohyphae is not well understood and its definition is unprecise. The observation that cryptoccocal cells in the pseudohyphal form resist phagocytosis by soil ameba [75], the natural predators of cryptococci, suggests that pseudohyphae formation might constitute a survival mechanism [75,76]. Although C. neoformans encounters low glucose conditions in the CNS, India ink preparations of spinal fluid from infected patients usually show mostly yeast forms and no hyphal forms. However, occasional reports of abnormal forms can be found and may be under-reported [77–79]. This suggests that suppression of filamentous growth may be required for growth in the host niche similar to dimorphic fungi [80].
Melanization
Melanization leads to a change in a phenotypic trait that is not necessarily macroscopically evident. It occurs in several fungal species including both C. neoformans and C. gattii. Melanin production requires o-diphenolic or p-diphenolic compounds such as l-3,4-dihydroxyphenylalanine as a substrate. The key enzyme laccase (CNLAC1) has been characterized [81–84] and promotes virulence by inhibiting the oxidative burst in the phagosomal space of macrophages [85]. Deletion mutants of CNLAC1 manifest decreased virulence [86] and melanin-deficient mutant strains are avirulent in murine models of cryptococcal infection [50–51]. Several complex pathways that control the biosynthesis of melanin have been identified [18,87]. Disruption of a gene encoding a cAMP-dependent protein kinase results in both amelanotic and hypocapsular mutants [88], which highlights that other virulence traits are also regulated but makes it difficult to determine if melanization is important but not essential for virulence. All cryptococcal varities and species melanize, although they may differ in the degree of melanization [89,90]. Only rarely have melanin deficient strains been isolated from human specimens [91]. In addition, staining with melanin-specific reagents revealed melanin in the cell walls of cryptococci in human brain tissue and melanin ‘ghosts’ can be recovered from infected mouse tissue [92]. Interestingly, heterogeneity in melanization is described [92]. These results indicate that C. neoformans universally melanizes during human infection and supports the concept that this change in a phenotypic trait may be relevant for pathogenesis.
Biofilm formation
Biofilms are communities of microbes that are attached to surfaces and held together by an extra-cellular matrix, often predominantly consisting of polysaccharides. Biofilm formation is a common phenotypic trait in pathogenic yeasts and makes the yeast cells less susceptible to host defense mechanisms [93]. Biofilm-like structures have also been reported in human cases of cryptococcosis on ventriculoperitoneal shunts [94]. In vitro experiments have demonstrated that biofilm-associated C. neoformans are significantly less susceptible than planktonic cells to azoles, amphotericin B and various microbial oxidants and peptides [95,96]. C. neoformans biofilm development is dependent on the release of capsular polysaccharide to create an exopolysaccharide matrix. Biofilm formation can be inhibited by protective antibodies and not by nonprotective antibodies [97]. Antibodies interfere with capsular polysaccharide release from the fungal cell. Interestingly, lactoferrin – an effector molecule of the innate immune system – inhibits bacterial but not fungal biofilm formation [95]. Furthermore, biofilm-like microcolonies are released by macrophages after antibody-mediated phagocytosis of C. neoformans (var grubii and neoformans) and C. gattii, which would reduce fungal cell dispersion in vivo but would promote cryptococcoma formation [98]. To what extent in vivo biofilm formation contributes to treatment failure is still not clear; however, experimental evidence suggests that they impair the host's ability to eradicate C. neoformans in collaboration with antifungal treatment
Phenotypic switching
Fungal populations manifest distinct epigenetic states at a low frequency in order to maintain adaptability to a changing environment. Colony switching is defined as the spontaneous emergence of colony variants. This phenomenon is mainly described in yeasts because single colonies are hard to distinguish in molds.
Phenotypic switching in Cryptococcus species
Phenotypic switching is described in strains of three species, namely C. grubii, (serotype A SB4, J32) [99], C. neoformans (serotype D 24067A, RC2) [61] and C. gattii (serotype B NP1) [100]. Colony variants arise at a frequency of approximately 1 in 104–105. Smooth colonies (S and SM) of SB4, 24067 and NP1 have a smooth dome, and mucoid (M and MC) colonies have a shiny and mucoid colony surface. Wrinkled (WR), serrated (C) and pseudohyphal colonies exhibit an irregular dome surface with or without serrated margins and are rarely observed in clinical isolates. Most C. neoformans and C. grubii strains manifest a smooth most C. gattii strains exhibit a mucoid colony morphology.
Cryptococcus neoformans strain (serotype D) RC2 switches consistently between the parent SM to the MC variant and another variant strain of ATCC strain 24067 (24067a) switches from an avirulent SM parent to virulent WR and pseudohyphal variants [61,101]. SB4 and J32 are clinical C. grubii strains that also switch but at lower frequencies from S to WR and C (SB4) and M to S (J32) [99]. In C. gattii strain NP1 two colony morphologies, one mucoid (NP1-MC) and one smooth (NP1-SM), were isolated from an immunocompetent patient with meningitis. Switching experiments confirmed that they were the result of PS [100]. PS from NP1-MC to NP1-SM occurred at a frequency of 1 in 2 × 105 colonies whereas reversion occurred 1 in 7 × 105. By contrast, the two rates of switching and reversion in the C. neoformans strain RC2 are comparable and occur at approximately 1 in 104 when 5 × 104 colonies are plated. RC2 is the dominant strain used for research.
Phenotypic switching alters the polysaccharide capsule & other cellular characteristics
All cryptococcal switching strains exhibit changes in the polysaccharide capsule, which is important because changes in this phenotypic trait may affect phagocytosis and rapid destruction by macrophages [102,103]. For RC2 the MC variant has a larger capsule than the SM variant and produces a viscous exopolysaccharide. Hence, in RC2, PS alters biophysical properties of GXM [60,61]. Although not proven, this may result from changes in the spacing of charged β-d-glucupyranosyluronic acid (GlcpA) along the mannose backbone, which are not apparent by nuclear magnetic resonance and explains why the GXMs of the two switch variants manifest the same nuclear magnetic resonance pattern.
In the switching strains SB4 and 24067a, the biochemical composition of the GXM changes by PS. GXM is composed of linear α-d-manno-pyranan chain with β-d-xylopyranosyl (Xylp) and GlcpA side residues. Six (M1–6) structural reporter groups (SRGs) are defined by amount and position of linked Xylp and GlcpA residues. In C. grubii strain SB4 and C. neoformans strain 24067A PS results in changes of SRGs [101]. GXMs derived from SB4-C are composed of M2 and M3 SRGs whereas GXM shed by SB4-SM is composed of only M2. It is noteworthy that the addition of Xylp at the 4–0 position in M3 most likely requires activation of different enzymes, that are traditionally thought to be used only by C. gattii.
The doubling time of C. neoformans strain RC2 is shorter compared with C. gattii strain NP1 but in both switching systems the switch variant (RC2-MC and NP1-SM) grows slower when compared with the parent strain (RC2-SM and NP1-MC). MICs of amphotericin B and fluconazole for switch variants are comparable. Both mucoid switch variants (RC2-MC and NP1-MC) have altered cell walls and exhibit increased sensitivity to lysing enzyme. Cell charge and melanization are not affected by PS in the two switch systems; however, biofilm formation can differ in the switch variants [104].
Phenotypic switching occurs in vivo & directly affects outcome
One of the strengths of C. neoformans as a model organism is that the effects of PS on the pathogenesis of chronic cryptococcosis can be studied in vivo. It was demonstrated that PS of RC2 from a SM parent to a MC variant occurs in vivo during chronic infection in mice [61]. To avoid contamination of the original inoculum with switch variants, the mice were infected with very low inocula and statistical methods were applied that proved that PS occurred in vivo opposed to selection of variants in a heterogenous inoculum. Furthermore, emergence of MC variants was observed in rats that were infected with doses as low as 100 colony-forming units (CFU) [62]. PS occurs at a low rate and in order for switch variants to dominate the pathogen population selection is required. Selection pressure favors the MC switch variant and therefore emergence of this switch variant is only observed in infected mice. Mice infected with MC do not manifest switching to the SM variant, although the in vitro frequency of SM to MC is comparable to that for PS from SM to MC in RC2. Highly relevant for the clinical scenario is the finding that treatment with amphotericin or anticapsular mAb can promote the selection of MC variants in mice [105].
Selection pressures can differ among switching strains and depends on the human niche. In murine infection with C. gattii strain NP1, both phenotypes NP1-SM and NP1-MC were recovered in the lungs, similar to the cerebrospinal fluid of the patient from which the strain was originally grown. By contrast, from the brains of mice infected intravenously or intratrachealy, only the smooth phenotype was recovered regardless of whether the mouse was infected with the NP1-SM or NP1-MC. This supports the notion that in the patient, the PS occurs after dissemination to the CNS [100]. Thus, in contrast to RC2, both phenotypic switch variants in NP1 appear to have selection pressure and in this strain, PS may be necessary for dissemination to the CNS.
Phenotypic switching alters host pathogen interaction & inflammatory response
Phenotypic switching of C. neoformans strains affects virulence by altering the host–pathogen interaction and pathogenesis of the disease. RC2-MC variant is significantly more virulent in all murine and rat animal models. Even in interaction with human monocytes, differences in inflammatory response were noted [106]. In a murine pulmonary infection model, histological analysis demonstrated significant differences in the inflammatory tissue response elicited by RC2-SM and RC2-MC. Specifically, at day 14 postinfection, lungs of RC2-SM mice exhibited moderate inflammatory changes with cellular infiltrates composed primarily of lymphocytes and only a few macrophages [61]. These cellular infiltrates progressed to orderly granuloma formation with little concomitant lung damage by day 28 postinfection. This host immune response is distinct from that elicited by RC2-MC at day 14 and involves changes in Th1 and Th2 cytokine production [107]. Here, infected lungs exhibited extensive cellular infiltrates that extend beyond the peribronchial regions, and were predominantly composed of macrophages and neutrophils with only a few lymphocytes. Near the time of death the inflammatory response increased and resulted in extensive destruction of alveolar membranes. Of high clinical relevance is the finding that the RC2-MC variant was able to promote increased intracranial pressure in a rat model of cryptococcal meningitis [62]. In human infection, increased intracranial pressure is the leading cause of high morbidity and mortality [108]. Investigations of lung-associated macrophages derived from RC2 SM- and MC-infected mice demonstrated that macrophages were alternatively and not classically activated. However, they differed in their level of activation. MC-infected macrophages exhibited high arginine production, IL-6 and MCP production and elicited more Th17-excreting T cells [109]. These findings are consistent with findings in other fungal infection models. Although inflammation is an essential component of the protective response to fungi, its dysregulation may significantly worsen fungal diseases and limit protective, antifungal immune responses. As such, the Th17 pathway may play a damage-promoting inflammatory role previously attributed to uncontrolled Th1 cell responses [110–112].
Virulence is also affected by PS of NP1 [100]. Experiments in mice demonstrated that NP1-SM-infected mice survived significantly longer than NP1-MC infected mice in intratracheal (p = 0.021) as well as in intravenous (p = 0.008) infection models. Consistent with this the CFU in the lung of NP1-SM-infected mice after 14 days was significantly lower (p ≤ 0.03) than NP1-MC-infected mice. The inflammatory response also differed for the NP1-SM and NP1-MC; however, in this strain a damage-promoting, overstimulated inflammatory response was not documented. Histological analysis of the lung sections demonstrated an appropriate and effective inflammatory response in lung tissue infected with NP1-SM. The mononuclear inflammation was composed of lymphocytes and macrophages. By contrast, the lungs of NP1-MC-infected Balb/c mice exhibited minimal inflammatory response and, consistent with the failure to elicit an inflammatory response, a large accumulation of yeast cells in lakes of polysaccharide consistent with cryptococcomas was observed on lung tissue sections. Histological analysis of the brain of NP1-SM- and NP1-MC-infected mice demonstrated multiple cryptococcomas but the cryptococcomas of NP1-SM-infected mice were smaller and elicited more inflammation in brain tissue. In summary, PS mainly affects virulence by altering the host immune response.
Molecular mechanism of PS
The molecular mechanisms mediating PS in C. neoformans are currently not understood. Although karyotype instability was observed in strain 24067A and SB4 [99,113], similar to the switching Candida albicans strain 3153A, it could not consistently be correlated with phenotypic variability and was not reversible. PS was associated with the downregulation of genes. These genes were not located in clusters or in telomeric regions but rather distributed across all chromosomes. Most of the regulated genes are not characterized with respect to function. One gene that is among the most prominently downregulated genes in RC2-MC relative to RC2-SM is ALL1. Interestingly, null mutants of ALL1 exhibit similar phenotypic traits to the MC variant. Specifically, all1Δ exhibits a slightly enlarged capsule at baseline that sheds a more viscous GXM than the wild-type SM parent. Most importantly, in infection models with all1Δ it mimics the hypervirulence of the MC variant including the overstimulated host response [51] and the increased intracranial pressure. Also of note was the finding that epigenetic control of the ‘phenotypic switch state’ appeared to loosen with senescence. Accordingly, it was recently shown that C. neoformans cells (RC2) of advanced generational age exhibit up to 11-fold higher switch rates [114]. In addition, some experimental data suggested that older cells may be more resistant to antifungals and phagocytosis and thus have a selective advantage in vivo and accumulate. This concept is novel and could have broad implications for the pathogenesis of chronic diseases.
Comparison of PS in Cryptococcus with PS in other pathogenic fungi
High-frequency PS has also been studied in diverse Candida species, mainly C. albicans and Candida glabrata [115–118]. The model C. albicans strain WO-1 switches between two colony morphologies, namely white and opaque, at a frequency of 1 in 104–105. Opaque-phase cells are mating competent whereas white-phase cells survive better within the mammalian host, yet can switch to mating-competent cells when required [119]. This phenotypic switch is relevant because clinical C. albicans strains undergo white to opaque switching (WOS) if they are homozygous (a/a or α/α) whereas heterozygous (a/α) strains cannot switch [120,121]. White-phase cells are more virulent in intravenous infection [122] and opaque-phase cells colonize skin more effectively [123]. WOS also affects other virulence traits, including the bud–hyphal transition [124], sensitivity to neutrophils and oxidants [125], antigenicity [126], adhesion [127], secretion of proteinase [128,129], drug susceptibility and phagocytosis by macrophages [130]. All of these altered traits can potentially affect survival in the mammalian host. Wor1 has been identified as a master regulator of WOS, as its deletion blocks opaque cell formation [131]. Interlocking feedback loop networks maintain the epigenetic state of switch variants through cell divisions. This circuit is not present in closely related fungi and could be a recent adaptation in the mammalian host [132,133].
Candida glabrata, another pathogenic yeast, undergoes ‘core switching’ [118] on agar containing CuSO4. Core switching occurs in the majority of clinical strains and results in white, light brown, dark brown, very dark brown and irregular wrinkle colonies. PS may play a fundamental role in virulence of this fungus because dark brown predominates among natural isolates and in mice has a colonization advantage over other colony types [134]. In Candida lusitaniae, PS is associated with emergence of amphotericin resistance (amphotericin B). PS happens at high frequency (1 in 102–104) and may confer a selective advantage in a host that is treated with amphotericin B [135].
Phenotypic switching is a mechanism that facilitates change in important phenotypic traits, as has been demonstrated in C. neoformans, C. grubii and C. gattii. These changes affect host–pathogen interactions and thus contribute to virulence. Although PS shares some basic similarities with PS described in other fungi, the effect of this process in Cryptococcus species and varieties is different from that described in Candida species.
Antigenic variation of the polysaccharide capsule
Traditionally, antigenic variation is achieved by varying expression of surface proteins. Such antigens are often recognized by the host immune response and, therefore, antigenic variation may constitute a mechanism to evade the immune response. Unlike most other fungi, C. neoformans is encapsulated and thus exhibits a different surface. The polysaccharide capsule contains many epitopes, and there is experimental evidence that the capsular surface varies its antigen epitopes.
Antigenic variation of C. neoformans
Antibody staining with capsule-specific antibodies has demonstrated that C. neoformans cells manifest antigenic variation in the polysaccharide capsule during murine infection and transmigration of the blood–brain barrier [37,40,49]. Owing to the complexity of the polysaccharide capsule it is difficult to precisely characterize this antigenic variation. It was proposed that this variation is generated by an infinite combination of polysaccharide triads [60]. Evidence that selection occurs during in vitro passage exists. Specifically, when passaged isolates were analyzed by agglutination assay, flow cytometry and indirect immunofluorescence, it was demonstrated that epitope expression of mAb 18B7 varied (they could be gained or lost) [136]. Analysis of the capsular antigenic properties by mAb binding and Scatchard analysis revealed fluctuations in the binding affinity within the capsule but not in the number of antibody binding sites, suggesting that the spatial organization of high- and low-affinity epitopes within the capsule change according to radial position. It is noteworthy that the structure of the capsule also changes with capsule age, since the capsule of older cells becomes more resistant to γ radiation-induced ablation [58]. In summary, the epitopes of a large polysaccharide such as GXM are manifold and this explains why it is inherently challenging to examine the variability of these epitopes. However, experimental data mainly comparing binding of mAbs to epitopes suggests that antigenic epitopes vary. Except for one gene, which encodes for Cas3p and affects the acetylation of the polysaccharide, and therefore also mAb binding, very little is known about the genetic mechanisms that would control such antigenic variation [137,138].
Comparison with antigenic variation in other pathogenic fungi
In Pneumocystis spp. a gene family called ‘major surface glycoprotein’ (MSG) encodes for surface proteins that undergo antigenic variation [139–145]. In P. carinii 85 distinct MSG genes are organized in clusters [146] and the current model proposes the existence of only one fixed expression. P. carinii populations are dominated by a single MSG gene at the expression site [147,148]. Patients with multiple independent infections are infected by distinct strains [149]. Hence, antigenic variation may be a survival strategy in this host-dependent fungus to avoid eradication by the host. Another example are the EPA genes in C. glabrata. EPA1 is a glycosylphosphatidylinositol-anchored cell-wall protein with 23 paralogues. Most of these EPA genes are located in subtelomeric positions, where they are transcriptionally silenced [150]. Transcription of some subtelomeric EPA genes can be derepressed by limitation of NAD+ precursors [151]. Null mutations in SIR3, SIR4 and RIF1 lead to expression of many EPA genes, resulting in a hyperadherent phenotype [150,152,153].
In summary, antigenic variation in other fungi involves proteins whereas the outer surface of Cryptococcus species is dominated by a polysaccharide capsule that can also be varied however this process is more complex.
Future perspective
Over the next 10 years, in vitro studies are needed that focus on research investigating the molecular mechanisms that control PS and phenotypic variation since the mechanisms are virtually unknown. Such studies could be significantly aided by novel epigenomic techniques such as Chip. So far, the majority of studies are performed with a few laboratory model strains, many of which have been passaged in vitro for years. These adapted strains may not adequately represent the breadth of phenotypic variations that can be found in clinical C. gatti and C. neoformans strains. It is not know if in vivo strain evolution contributes to outcome. Given the availability of spinal fluids with up to 105 C. neoformans cells per ml, the variability of phenotypic traits should be studied in cells derived directly from the host. Understanding antigenic variation of complex polysaccharides remains a major challenge that will require new methods to be explored. Novel in vivo imaging techniques may allow the design of studies that are concentrated on elucidating mechanism of lung to CNS dissemination in rat and mouse animal models. The emergence of a new C. gattii strain that causes infection in immunocompetent individuals in the northwestern states of the USA [154] demand more research on this species. With rising antifungal resistance [155] and the devastating prognosis of chronic cryptococcosis [1,156–159], vaccine development to prevent reactivation of C. neoformans would be highly desirable. This will, however, require better understanding of the relevance, the selection and the molecular regulation of phenotypic changes in vivo as vaccination could promote selection of variants.
Executive summary.
This article discusses how phenotypic traits in Cryptococcus species can vary and how the variability affects the pathogenesis of chronic cryptococcosis.
Global changes of phenotypic traits occur in the whole fungal population under certain conditions. These changes are capsule induction, melanization, biofilm formation and hyphal/spore formation. Capsule induction is unique to Cryptococcus species. The other phenotypic traits are also encountered in other fungi.
Phenotypic switching involves only a small subpopulation but usually these variants are selected and thus can become dominant because the switched phenotypes are stable epigenetic states. This process is also found in several Candida species; however, the phenotypic traits affected are very different. In Candida species, this process commonly changes cellular morphologies but can also change antifungal resistance and copper metabolism. By contrast, in Cryptococcus the main phenotypic trait affected is the polysaccharide capsule.
Antigenic variation in Cryptococcus affects the antigens within the polysaccharide capsule and is very complex, highly variable and affects antibody binding. This process is very different from other fungi where antigenic variation affects the modification of surface proteins.
Acknowledgments
This work was supported by grant AI059681-05 to BC Fries, by a Pilot and Feasibility grant from the Center for AIDS Research at the Albert Einstein College of Medicine and Montefiore Medical Center funded by the NIH (NIH AI-51519) and by the Einstein AIDS International Training and Research Program (D43 TW001403).
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]; ▪ Estimates the current number of patients with cryptococcal meningitis worldwide and concludes that more than 500,000 people die with this disease in sub-Saharan Africa.
- 2.Okongo M, Morgan D, Mayanja B, Ross A, Whitworth J. Causes of death in a rural, population-based human immunodeficiency virus type 1 (HIV-1) natural history cohort in Uganda. Int J Epidemiol. 1998;27:698–702. doi: 10.1093/ije/27.4.698. [DOI] [PubMed] [Google Scholar]
- 3.French N, Gray K, Watera C, et al. Cryptococcal infection in a cohort of HIV-1-infected Ugandan adults. AIDS. 2002;16:1031–1038. doi: 10.1097/00002030-200205030-00009. [DOI] [PubMed] [Google Scholar]
- 4.Dromer F, Mathoulin S, Dupont B, Laporte A. Epidemiology of cryptococcosis in France: a 9-year survey (1985–1993). French Cryptococcosis Study Group. Clin Infect Dis. 1996;23:82–90. doi: 10.1093/clinids/23.1.82. [DOI] [PubMed] [Google Scholar]
- 5.Dromer F, Varma A, Ronin O, Mathoulin S, Dupont B. Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J Clin Microbiol. 1994;32:2364–2371. doi: 10.1128/jcm.32.10.2364-2371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J Infect Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.San-Blas G, Travassos LR, Fries BC, et al. Fungal morphogenesis and virulence. Med Mycol. 2000;38:79–86. [PubMed] [Google Scholar]
- 9.Kugler S, Schurtz Sebghati T, Groppe Eissenberg L, Goldman WE. Phenotypic variation and intracellular parasitism by Histoplasma capsulatum. Proc Natl Acad Sci USA. 2000;97:8794–8798. doi: 10.1073/pnas.97.16.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whiteway M. Transcriptional control of cell type and morphogenesis in Candida albicans. Curr Opin Microbiol. 2000;3:582–588. doi: 10.1016/s1369-5274(00)00144-2. [DOI] [PubMed] [Google Scholar]
- 11.Soll DR. Candida commensalism and virulence: the evolution of phenotypic plasticity. Acta Trop. 2002;81:101–110. doi: 10.1016/s0001-706x(01)00200-5. [DOI] [PubMed] [Google Scholar]
- 12.Romani L, Bistoni F, Puccetti P. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr Opin Microbiol. 2003;6:338–343. doi: 10.1016/s1369-5274(03)00081-x. [DOI] [PubMed] [Google Scholar]
- 13.Swartley L, Marfin A, Edupuganti S, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci USA. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell. 2009;8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates that spores derived from Cryptococcus neoformans and Cryptococcus grubii differ in shape and already exhibit polysaccharide on the surface.
- 15.Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Describes monkaryotic fruiting in C. neoformans and how this could lead to spore formation and contribute to the environmental distribution of this pathogen.
- 16.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates that spores of C. neoformans are highly infectious particles that cause infection at a very low inoculum.
- 17.Lengeler KB, Davidson RC, D'Souza C, et al. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alspaugh JA, Perfect JR, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza CA, Heitman J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev. 2001;25:349–364. doi: 10.1111/j.1574-6976.2001.tb00582.x. [DOI] [PubMed] [Google Scholar]
- 20.Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, ating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 21.Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- 22.Yue C, Cavallo LM, Alspaugh JA, et al. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahn YS, Kojima K, Cox GM, Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruz MC, Fox DS, Heitman J. Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J. 2001;20:1020–1032. doi: 10.1093/emboj/20.5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeng S, Ko YJ, Kim GB, et al. Comparative transcriptome analysis reveals novel roles of the Ras- and cAMP-signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell. 2010;9(3):360–378. doi: 10.1128/EC.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell. 2004;3:1476–1491. doi: 10.1128/EC.3.6.1476-1491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alspaugh JA, Pukkila-Worley R, Harashima T, et al. Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell. 2006;17:3768–3780. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moranova Z, Kawamoto S, Raclavsky V. Hypoxia sensing in cryptococcus neoformans: biofilm-like adaptation for dormancy? Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2009;153:189–193. doi: 10.5507/bp.2009.031. [DOI] [PubMed] [Google Scholar]
- 33.Cruz MC, Sia RA, Olson M, Cox GM, Heitman J. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect Immun. 2000;68:982–985. doi: 10.1128/iai.68.2.982-985.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol. 2009;9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Alspaugh JA, Davidson RC, Heitman J. Morphogenesis of Cryptococcus neoformans. Contrib Microbiol. 2000;5:217–238. doi: 10.1159/000060352. [DOI] [PubMed] [Google Scholar]
- 36.Littman ML, Tsubura E. Effect of degree of encapsulation upon virulence of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1959;101:773–777. doi: 10.3181/00379727-101-25090. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Hermoso D, Dromer F, Janbon G. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect Immun. 2004;72:3359–3365. doi: 10.1128/IAI.72.6.3359-3365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates that the polysaccharide capsule changes in vivo and these changes involve epitopes on the capsule surface that bind antibodies.
- 38.Rivera J, Feldmesser M, Cammer M, Casadevall A. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect Immun. 1998;66:5027–5030. doi: 10.1128/iai.66.10.5027-5030.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaragoza O, Telzak A, Bryan RA, Dadachova E, Casadevall A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol Microbiol. 2006;59:67–83. doi: 10.1111/j.1365-2958.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- 40.Charlier C, Chretien F, Baudrimont M, Mordelet E, Lortholary O, Dromer F. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood–brain barrier. Am J Pathol. 2005;166:421–432. doi: 10.1016/S0002-9440(10)62265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ First article to describe changes of the polysaccharide capsule that are associated with crossing of the blood–brain barrier; a key step in the pathogenesis of cryptococcosis.
- 41.Cheng PY, Sham A, Kronstad JW. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun. 2009;77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaragoza O, Fries BC, Casadevall A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO(2) Infect Immun. 2003;71:6155–6164. doi: 10.1128/IAI.71.11.6155-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacon BE, Cherniak R, Kwon-Chung KJ, Jacobson ES. Structure of the 0-deacteylated glucuronoxylomannan from Cryptococcus neoformans Cap70 as determined by 2D NMR spectroscopy. Carbohydr Res. 1996;283:95–110. doi: 10.1016/0008-6215(95)00397-5. [DOI] [PubMed] [Google Scholar]
- 45.Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, Jacobson ES. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 46.Zaragoza O, Casadevall A. Experimental modulation of capsule size in Cryptococcus neoformans. Biol Proced Online. 2004;6:10–15. doi: 10.1251/bpo68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nimrichter L, Frases S, Cinelli LP, et al. Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot Cell. 2007;6:1400–1410. doi: 10.1128/EC.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaragoza O, Taborda CP, Casadevall A. The efficacy of complement-mediated phagocytosis of Cryptococcus neoformans is dependent on the location of C3 in the polysaccharide capsule and involves both direct and indirect C3-mediated interactions. Eur J Immunol. 2003;33:1957–1967. doi: 10.1002/eji.200323848. [DOI] [PubMed] [Google Scholar]
- 49.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol. 2009;6:8, 133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Comprehensive and recent review of the polysaccharide capsule.
- 51.Jain N, Li L, Hsueh YP, et al. Loss of allergen 1 confers a hypervirulent phenotype that resembles mucoid switch variants of Cryptococcus neoformans. Infect Immun. 2009;7:7, 128–140. doi: 10.1128/IAI.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Characterizes a gene, ALL1, that is differentially regulated in switching strains and its downregulation is associated with hypervirulence and the development of high intracerebral pressure in vivo. This is the first gene whose regulation has been linked to elevated intracerebral pressure.
- 52.Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–380. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pukkila-Worley R, Gerrald QD, Kraus PR, et al. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2008;7:1685–1698. doi: 10.1128/EC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerik KJ, Donlin MJ, Soto CE, et al. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol Microbiol. 2005;58:393–408. doi: 10.1111/j.1365-2958.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- 56.Pierini LM, Doering TL. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol Microbiol. 2001;41:105–115. doi: 10.1046/j.1365-2958.2001.02504.x. [DOI] [PubMed] [Google Scholar]
- 57.Maxson ME, Cook E, Casadevall A, Zaragoza O. The volume and hydration of the Cryptococcus neoformans polysaccharide capsule. Fungal Genet Biol. 2007;44:180–186. doi: 10.1016/j.fgb.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Maxson ME, Dadachova E, Casadevall A, Zaragoza O. Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule. Eukaryot Cell. 2007;6:95–109. doi: 10.1128/EC.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frases S, Pontes B, Nimrichter L, Viana NB, Rodrigues ML, Casadevall A. Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc Natl Acad Sci USA. 2009;106:1228–1233. doi: 10.1073/pnas.0808995106. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Used dynamic light scattering analysis of capsular polysaccharide and optical tweezers to explore the architecture of the capsule. This study demonstrates a linear correlation between polysaccharide effective diameter and microscopic capsular diameter, implying that capsule growth is achieved by the addition of molecules with larger effective diameter, such that some molecules can span the entire diameter of the capsule.
- 60.McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell. 2007;6:1464–1473. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ First study to demonstrate that phenotypic switching alters biophysical parameters of the capsular polysaccharide.
- 61.Fries BC, Taborda CP, Serfass E, Casadevall A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J Clin Invest. 2001;108:1639–1648. doi: 10.1172/JCI13407. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates that phenotypic switching of C. neoformans occurs during chronic infection and alters outcome. Hence, phenotypic switching is not merely an in vitro phenomenon but relevant for the pathogenesis of chronic cryptococcossis.
- 62.Fries BC, Lee SC, Kennan R, Zhao W, Casadevall A, Goldman DL. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect Immun. 2005;73:1779–1787. doi: 10.1128/IAI.73.3.1779-1787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwon-Chung KJ. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 64.Ellis DH, Pfeiffer TJ. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet. 1990;336:923–925. doi: 10.1016/0140-6736(90)92283-n. [DOI] [PubMed] [Google Scholar]
- 65.Litvintseva AP, Lin X, Templeton I, Heitman J, Mitchell TG. Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog. 2007;3:e114. doi: 10.1371/journal.ppat.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates that the majority outbreak C. gattii clone in Vancouver Island (BC, Canada) appears to have descended from two a mating-type parents. Thus, these studies demonstrate how cryptic same-sex reproduction can enable expansion of a human pathogen to a new geographical niche and contribute to the ongoing production of infectious spores.
- 67.Nielsen K, Heitman J. Sex and virulence of human pathogenic fungi. Adv Genet. 2007;57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- 68.Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell. 2007;6:949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin X, Heitman J. Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot Cell. 2005;4:1746–1754. doi: 10.1128/EC.4.10.1746-1754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwong-Chung KB, Bennett JE. Distribution of α and α mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978;108(4):337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- 71.Rutherford JC, Lin X, Nielsen K, Heitman J. Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot Cell. 2008;7:237–246. doi: 10.1128/EC.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Erke KH. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;128:445–455. doi: 10.1128/jb.128.1.445-455.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]; ▪ Reveals how sexual reproduction can occur between partners of the same mating type and that hallmarks of mating occur during fruiting, including diploidization and meiosis. The key meiotic regulator Dmc1 is required for efficient fruiting. Fusion and meiosis can occur between nonisogenic a strains, enabling genetic exchange. Given the preponderance of a strains in nature the discovery that C. neoformans a cells can sexually reproduce via fruiting, without fusing with a partner of opposite mating type, provides a mechanism for a long-term survival advantage for cells.
- 75.Neilson JB, Ivey MH, Bulmer GS. Cryptococcus neoformans pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun. 1978;20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neilson JB, Fromtling RA, Blumer GS. Pseudohyphal forms of Cryptococcus neoformans: decreased survival in vivo. Mycopathologia. 1981;73:57–59. doi: 10.1007/BF00443015. [DOI] [PubMed] [Google Scholar]
- 77.Bava J, Solari R, Isla G, Troncoso A. Atypical forms of Cryptococcus neoformans in CSF of an AIDS patient. J Infect Dev Ctries. 2008;2:403–405. doi: 10.3855/jidc.207. [DOI] [PubMed] [Google Scholar]
- 78.Freed ER, Duma RJ, Shadomy HJ, Utz JP. Meningoencephalitis due to hyphae-forming Cryptococcus neoformans. Am J Clin Pathol. 1971;55:30–33. doi: 10.1093/ajcp/55.1.30. [DOI] [PubMed] [Google Scholar]
- 79.Pappalardo MC, Paschoal RC, Melhem MS. AIDS-associated central nervous system cryptococcosis: a Brazilian case study. AIDS. 2007;21:1971–1972. doi: 10.1097/01.aids.0000287549.79368.33. [DOI] [PubMed] [Google Scholar]
- 80.Rooney PJ, Klein BS. Linking fungal morphogenesis with virulence. Cell Microbiol. 2002;4:127–137. doi: 10.1046/j.1462-5822.2002.00179.x. [DOI] [PubMed] [Google Scholar]
- 81.Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ikeda R, Shinoda T, Morita T, Jacobson ES. Characterization of a phenol oxidase from Cryptococcus neoformans var. neoformans. Microbiol Immunol. 1993;37:759–764. doi: 10.1111/j.1348-0421.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 83.Erickson T, Liu L, Gueyikian A, Zhu X, Gibbons J, Williamson PR. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol Microbiol. 2001;42:1121–1131. doi: 10.1046/j.1365-2958.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 84.Jacobson ES, Emery HS. Temperature regulation of the cryptococcal phenoloxidase. J Med Vet Mycol. 1991;29:121–124. doi: 10.1080/02681219180000201. [DOI] [PubMed] [Google Scholar]
- 85.Liu L, Tewari RP, Williamson PR. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect Immun. 1999;67:6034–6039. doi: 10.1128/iai.67.11.6034-6039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.D'Souza CA, Alspaugh JA, Yue C, et al. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alvarado-Ramirez E, Torres-Rodriguez JM, Sellart M, Vidotto V. Laccase activity in Cryptococcus gattii strains isolated from goats. Rev Iberoam Micol. 2008;25:150–153. doi: 10.1016/s1130-1406(08)70035-4. [DOI] [PubMed] [Google Scholar]
- 90.Nosanchuk JD, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mandal P, Banerjee U, Casadevall A, Nosanchuk JD. Dual infections with pigmented and albino strains of Cryptococcus neoformans in patients with or without human immunodeficiency virus infection in India. J Clin Microbiol. 2005;43:4766–4772. doi: 10.1128/JCM.43.9.4766-4772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nosanchuk JD, Rosas AL, Lee SC, Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet. 2000;355:2049–2050. doi: 10.1016/S0140-6736(00)02356-4. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates that C. neoformans is melanized in vivo in brain tissue.
- 93.Anderson GG, O'Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 94.Ingram CW, Haywood HB, 3rd, Morris VM, Allen RL, Perfect JR. Cryptococcal ventricular-peritoneal shunt infection: clinical and epidemiological evaluation of two closely associated cases. Infect Control Hosp Epidemiol. 1993;14:719–722. doi: 10.1086/646675. [DOI] [PubMed] [Google Scholar]
- 95.Martinez LR, Casadevall A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect Immun. 2006;74:6118–6123. doi: 10.1128/IAI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martinez LR, Casadevall A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 2006;50:1021–1033. doi: 10.1128/AAC.50.3.1021-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez LR, Christaki E, Casadevall A. Specific antibody to Cryptococcus neoformans glucurunoxylomannan antagonizes antifungal drug action against cryptococcal biofilms in vitro. J Infect Dis. 2006;194:261–266. doi: 10.1086/504722. [DOI] [PubMed] [Google Scholar]
- 98.Alvarez M, Saylor C, Casadevall A. Antibody action after phagocytosis promotes Cryptococcus neoformans and Cryptococcus gattii macrophage exocytosis with biofilm-like microcolony formation. Cell Microbiol. 2008;10:1622–1633. doi: 10.1111/j.1462-5822.2008.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goldman DL, Fries BC, Franzot SP, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ First study that demonstrates phenotypic switching in C neoformans.
- 100.Jain N, Li L, McFadden DC, et al. Phenotypic switching in a Cryptococcus neoformans variety gattii strain is associated with changes in virulence and promotes dissemination to the central nervous system. Infect Immun. 2006;74:896–903. doi: 10.1128/IAI.74.2.896-903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun. 1999;67:6076–6083. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Showed that phenotypic switching can change the biochemical structure of glucuronoxylomannan.
- 102.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levitz SM, Nong SH, Seetoo KF, Harrison TS, Speizer RA, Simons ER. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect Immun. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martinez LR, Ibom DC, Casadevall A, Fries BC. Characterization of phenotypic switching in Cryptococcus neoformans biofilms. Mycopathologia. 2008;166:175–180. doi: 10.1007/s11046-008-9133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fries BC, Cook E, Wang X, Casadevall A. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob Agents Chemother. 2005;49:350–357. doi: 10.1128/AAC.49.1.350-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pietrella D, Fries B, Lupo P, Bistoni F, Casadevall A, Vecchiarelli A. Phenotypic switching of Cryptococcus neoformans can influence the outcome of the human immune response. Cell Microbiol. 2003;5:513–522. doi: 10.1046/j.1462-5822.2003.00297.x. [DOI] [PubMed] [Google Scholar]
- 107.Guerrero A, Fries BC. Phenotypic switching in Cryptococcus neoformans contributes to virulence by changing the immunological host response. Infect Immun. 2008;76(9):4322–4331. doi: 10.1128/IAI.00529-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Demonstrates how phenotypic switching leads to hypervirulence by altering the host–pathogen interaction. The hypervirulent switch variant elicits a damage-promoting immune response.
- 108.Graybill JR, Sobel J, Saag M, et al. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups. Clin Infect Dis. 2000;30:47–54. doi: 10.1086/313603. [DOI] [PubMed] [Google Scholar]
- 109.Guerrero A, Jain N, Wang X, Fries BC. Cryptococcus neoformans variants generated by phenotypic switching differ in virulence through effects on macrophage activation. Infect Immun. 2010;78:1049–1057. doi: 10.1128/IAI.01049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Romani L, Puccetti P. Controlling pathogenic inflammation to fungi. Expert Rev Anti Infect Ther. 2007;5:1007–1017. doi: 10.1586/14787210.5.6.1007. [DOI] [PubMed] [Google Scholar]
- 111.Romani L, Zelante T, De Luca A, Fallarino F, Puccetti P. IL-17 and therapeutic kynurenines in pathogenic inflammation to fungi. J Immunol. 2008;180:5157–5162. doi: 10.4049/jimmunol.180.8.5157. [DOI] [PubMed] [Google Scholar]; ▪ Demonstrates that IL-17 excretion is associated with a harmful antifungal host response.
- 112.Zelante T, De Luca A, Bonifazi P, et al. IL-23 and the Th17 pathway promote inflammation and impair antifungal immune resistance. Eur J Immunol. 2007;37:2695–2706. doi: 10.1002/eji.200737409. [DOI] [PubMed] [Google Scholar]
- 113.Franzot SP, Mukherjee J, Cherniak R, Chen LC, Hamdan JS, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. Isolation and characterization of senescent C. neoformans and its implications for phenotypic switching and the pathogenesis of chronic cryptococcosis. Eukaryot Cell. 2009;8(6):858–866. doi: 10.1128/EC.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ First study to demonstrate that phenotypic switching is enhanced in senescent C. neoformans cells. Furthermore, these studies demonstrate that older C. neoformans cells are more resistant to antifungals, thus providing a mechanism by which older cells could potentially accumulate in vivo.
- 115.Slutsky B, Buffo J, Soll DR. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]; ▪ First paper to describe phenotypic switching in Candida albicans.
- 116.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lachke SA, Srikantha T, Tsai LK, Daniels K, Soll DR. Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect Immun. 2000;68:884–895. doi: 10.1128/iai.68.2.884-895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lachke SA, Joly S, Daniels K, Soll DR. Phenotypic switching and filamentation in Candida glabrata. Microbiology. 2002;148:2661–2674. doi: 10.1099/00221287-148-9-2661. [DOI] [PubMed] [Google Scholar]
- 119.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]; ▪ First paper that demonstrates how phenotypic switching enables C. albicans to mate.
- 120.Legrand M, Lephart P, Forche A, et al. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol Microbiol. 2004;52:1451–1462. doi: 10.1111/j.1365-2958.2004.04068.x. [DOI] [PubMed] [Google Scholar]
- 121.Lockhart SR, Pujol C, Daniels KJ, et al. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Anderson J, Cundiff L, Schnars B, Gao MX, Mackenzie I, Soll DR. Hypha formation in the white-opaque transition of Candida albicans. Infect Immun. 1989;57:458–467. doi: 10.1128/iai.57.2.458-467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kolotila MP, Diamond RD. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson J, Mihalik R, Soll DR. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kennedy MJ, Rogers AL, Hanselmen LR, Soll DR, Yancey RJ., Jr Variation in adhesion and cell surface hydrophobicity in Candida albicans white and opaque phenotypes. Mycopathologia. 1988;102:149–156. doi: 10.1007/BF00437397. [DOI] [PubMed] [Google Scholar]
- 128.Morrow B, Srikantha T, Soll DR. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vargas K, Messer SA, Pfaller M, et al. Elevated phenotypic switching and drug resistance of Candida albicans from human immunodeficiency virus-positive individuals prior to first thrush episode. J Clin Microbiol. 2000;38:3595–3607. doi: 10.1128/jcm.38.10.3595-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS ONE. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Srikantha T, Daniels KJ, Wu W, et al. Dark brown is the more virulent of the switch phenotypes of Candida glabrata. Microbiology. 2008;154:3309–3318. doi: 10.1099/mic.0.2008/020578-0. [DOI] [PubMed] [Google Scholar]
- 135.Miller NS, Dick JD, Merz WG. Phenotypic switching in Candida lusitaniae on copper sulfate indicator agar: association with amphotericin B resistance and filamentation. J Clin Microbiol. 2006;44:1536–1539. doi: 10.1128/JCM.44.4.1536-1539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cleare W, Cherniak R, Casadevall A. In vitro and in vivo stability of a Cryptococcus neoformans [corrected] glucuronoxylomannan epitope that elicits protective antibodies. Infect Immun. 1999;67:3096–3107. doi: 10.1128/iai.67.6.3096-3107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moyrand F, Chang YC, Himmelreich U, Kwon-Chung KJ, Janbon G. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot Cell. 2004;3:1513–1524. doi: 10.1128/EC.3.6.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Moyrand F, Klaproth B, Himmelreich U, Dromer F, Janbon G. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol Microbiol. 2002;45:837–849. doi: 10.1046/j.1365-2958.2002.03059.x. [DOI] [PubMed] [Google Scholar]; ▪ Describes the relationship between the capsule structure and the pathophysiology of C. neoformans. A genetic screen identified mutant strains producing a structurally modified capsule that differs in binding of capsule-specific monoclonal antibodies.
- 139.Garbe TR, Stringer JR. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994;62:3092–3101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kitada K, Wada M, Nakamura Y. Multi-gene family of major surface glycoproteins of Pneumocystis carinii: full-size cDNA cloning and expression. DNA Res. 1994;1:57–66. doi: 10.1093/dnares/1.2.57. [DOI] [PubMed] [Google Scholar]
- 141.Kovacs JA, Powell F, Edman JC, et al. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J Biol Chem. 1993;268:6034–6040. [PubMed] [Google Scholar]
- 142.Linke MJ, Smulian AG, Stringer JR, Walzer PD. Characterization of multiple unique cDNAs encoding the major surface glycoprotein of rat-derived Pneumocystis carinii. Parasitol Res. 1994;80:478–486. doi: 10.1007/BF00932694. [DOI] [PubMed] [Google Scholar]
- 143.Wada M, Kitada K, Saito M, Egawa K, Nakamura Y. cDNA sequence diversity and genomic clusters of major surface glycoprotein genes of Pneumocystis carinii. J Infect Dis. 1993;168:979–985. doi: 10.1093/infdis/168.4.979. [DOI] [PubMed] [Google Scholar]
- 144.Haidaris PJ, Wright TW, Gigliotti F, Haidaris CG. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;166:1113–1123. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 145.Wright TW, Gigliotti F, Haidaris CG, Simpson-Haidaris PJ. Cloning and characterization of a conserved region of human and rhesus macaque Pneumocystis carinii gpA. Gene. 1995;167:185–189. doi: 10.1016/0378-1119(95)00704-0. [DOI] [PubMed] [Google Scholar]
- 146.Sunkin SM, Stringer SL, Stringer JR. A tandem repeat of rat-derived Pneumocystis carinii genes encoding the major surface glycoprotein. J Eukaryot Microbiol. 1994;41:292–300. doi: 10.1111/j.1550-7408.1994.tb01509.x. [DOI] [PubMed] [Google Scholar]
- 147.Ambrose HE, Keely SP, Aliouat EM, et al. Expression and complexity of the PRT1 multigene family of Pneumocystis carinii. Microbiology. 2004;150:293–300. doi: 10.1099/mic.0.26539-0. [DOI] [PubMed] [Google Scholar]
- 148.Keely SP, Cushion MT, Stringer JR. Diversity at the locus associated with transcription of a variable surface antigen of Pneumocystis carinii as an index of population structure and dynamics in infected rats. Infect Immun. 2003;71:47–60. doi: 10.1128/IAI.71.1.47-60.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Keely SP, Baughman RP, Smulian AG, Dohn MN, Stringer JR. Source of Pneumocystis carinii in recurrent episodes of pneumonia in AIDS patients. AIDS. 1996;10:881–888. doi: 10.1097/00002030-199607000-00011. [DOI] [PubMed] [Google Scholar]
- 150.Castano I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- 151.Domergue R, Castano I, De Las Penas A, et al. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- 152.De Las Penas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iraqui I, Garcia-Sanchez S, Aubert S, et al. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol Microbiol. 2005;55:1259–1271. doi: 10.1111/j.1365-2958.2004.04475.x. [DOI] [PubMed] [Google Scholar]
- 154.Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci USA. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pfaller MA, Diekema DJ, Gibbs DL, et al. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: 10.5-year analysis of susceptibilities of noncandidal yeast species to fluconazole and voriconazole determined by CLSI standardized disk diffusion testing. J Clin Microbiol. 2009;47:117–123. doi: 10.1128/JCM.01747-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; ▪ Study demonstrates that up to 13% of C. neoformans isolates are already resistant to fluconazole.
- 156.Bicanic T, Meintjes G, Wood R, et al. Fungal burden, early fungicidal activity, and outcome in cryptococcal meningitis in antiretroviral-naive or antiretroviral-experienced patients treated with amphotericin B or fluconazole. Clin Infect Dis. 2007;45:76–80. doi: 10.1086/518607. [DOI] [PubMed] [Google Scholar]
- 157.Bicanic T, Muzoora C, Brouwer AE, et al. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis. 2009;49(5):702–709. doi: 10.1086/604716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bicanic T, Wood R, Bekker LG, Darder M, Meintjes G, Harrison TS. Antiretroviral roll-out, antifungal roll-back: access to treatment for cryptococcal meningitis. Lancet Infect Dis. 2005;5:530–531. doi: 10.1016/S1473-3099(05)70197-3. [DOI] [PubMed] [Google Scholar]
- 159.Bicanic T, Wood R, Meintjes G, et al. High-dose amphotericin B with flucytosine for the treatment of cryptococcal meningitis in HIV-infected patients: a randomized trial. Clin Infect Dis. 2008;47:123–130. doi: 10.1086/588792. [DOI] [PubMed] [Google Scholar]


